Abstract
Introduction
Myopathy, encephalopathy, lactic acidosis, and stroke‐like (MELAS) syndrome, a maternally inherited disorder that is among the most common mitochondrial DNA (mtDNA) diseases, is usually associated with the m.3242A>G mutation of the mitochondrial tRNAleu gene. Very few data are available with respect to prenatal diagnosis of this serious disease. The rate of mutant versus wild‐type mtDNA (heteroplasmy) in fetal DNA is indeed considered to be a poor indicator of postnatal outcome.
Materials and methods
Taking advantage of a novel semi‐quantitative polymerase chain reaction test for m.3243A>G mutant load assessment, we carried out nine prenatal diagnoses in five unrelated women, using two different fetal tissues (chorionic villi v amniocytes) sampled at two or three different stages of pregnancy.
Results
Two of the five women, although not carrying m.3243A>G in blood or extra‐blood tissues, were, however, considered at risk for transmission of the mutation, as they were closely related to MELAS‐affected individuals. The absence of 3243A>G in the blood of first degree relatives was associated with no mutated mtDNA in the cardiovascular system (CVS) or amniocytes, and their three children are healthy, with a follow‐up of 3 months–3 years. Among the six fetuses from the three carrier women, three were shown to be homoplasmic (0% mutant load), the remaining three being heteroplasmic, with a mutant load ranging from 23% to 63%. The fetal mutant load was fairly stable at two or three different stages of pregnancy in CVS and amniocytes. Although pregnancy was terminated in the case of the fetus with a 63% mutant load, all other children are healthy with a follow‐up of 3 months–6 years.
Conclusion
These data suggest that a prenatal diagnosis for MELAS syndrome might be helpful for at‐risk families.
Mitochondrial DNA (mtDNA) mutations cause a wide range of serious genetic diseases with maternal inheritance. Most of these defects result in a progressive disabling neurological syndrome with premature death. Among them, the myopathy, encephalopathy, lactic acidosis, and stroke‐like (MELAS) syndrome (OMIM: 540 000) is one of the most common and serious conditions. MELAS syndrome is mainly caused by the m.3243A>G mutation in the mitochondrial tRNALeu gene1 (Genbank NC001 807), which produces a generalised dysfunction of the mitochondrial respiratory chain. The clinical features are highly variable, not only in terms of age at onset and severity of symptoms but also in relation to the specific organs associated.2 Many patients present with mellitus diabetes, deafness, cardiomyopathy, external ophthalmoplegia and skeletal myopathy, variously associated in different degrees.3 The interfamilial and intrafamilial variability of the clinical phenotype is classically related to the properties of mtDNA segregation: heteroplasmy, “threshold” effect, mitochondrial “bottleneck” and variation in the tissue distribution4.
Owing to the severity of the disease, the high risk of recurrence in siblings and the absence of efficient treatment, couples at risk of transmitting the m.3243A>G mutation often ask for prenatal diagnosis. However, very few data are so far available with respect to the prenatal diagnosis of MELAS syndrome.4,5 Thus, whether prenatal assessment of the fetal mutant load (rate of mutant v wild‐type mtDNA) is a good predictive marker for the postnatal clinical outcome remains a matter of debate. In the few available reports, imprecise assessment of the fetal mutant load, most often studied in a single fetal tissue, hampers consideration of this issue. We report here a sensitive novel technical approach, designed to quantify as accurately as possible the mutant load in a given tissue. The results of prenatal analyses in a series of nine cases are detailed.
Materials and methods
Patients
Nine prenatal analyses were carried out in five unrelated families. Informed consent was obtained from each couple (fig 1).
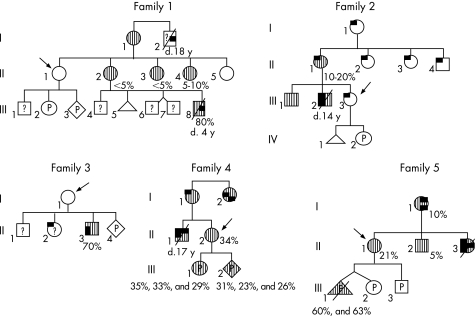
Figure 1 Pedigrees of the families. Postnatal DNA analyses were carried out in white cells (and skeletal muscle for patient II3 from family 3) whereas prenatal studies (P) were carried out on chorionic villous sampling or amniocytes (table 1). The presence and absence of m.3243A>G are denoted as hatched bars and open symbols, respectively. When known, mutant loads are indicated under the appropriate family member. ? ; m.3243A>G not tested. Pregnant patients requesting a prenatal diagnosis are indicated with an arrow. Clinical features are depicted as follows: deafness, diabetes mellitus, myopathy, migraine‐like headache, myoclonus and stroke.
Family 1
Patient I2 presented with severe muscle weakness and died at 18 years of age from respiratory distress. No tissue was available for genetic testing. Patient III8, a grand‐nephew of I2, died at 4 years with muscle weakness, mental retardation and lactic acidosis, whereas his three brothers were healthy at 15 and 11 years (twin pregnancy) of age, respectively. Mutant loads were assessed in leucocytes from III8 (80%), his mother (II2: 5%) and four maternal aunts (II1, II3–5), with only two of them (II3 and II4) being shown to carry m.3243A>G (5% and 10%, respectively). Healthy patient II1 nevertheless requested a prenatal analysis for her two forthcoming pregnancies, owing to an inability to rule out the presence of m.3243A>G in her germ line cells.
Family 2
All known members of this family presented with migraine‐like headaches. MELAS syndrome was diagnosed in patient III2, who died at 14 years of age from a stroke‐like episode. Prenatal diagnosis was requested by patient III3, the proband's symptomatic sister, although m.3243A>G was not detected either in her leucocytes or in skeletal muscle of a miscarriage product (IV1) from her previous pregnancy.
Family 3
A son (II3) born to patient I1 presented at 9 months of age with stroke‐like episodes and myoclonus. m.3243A>G was found at a rate of 70% in a sample of his skeletal muscle. Although II2, an older sister of II3, experienced headaches, a symptom potentially related to MELAS syndrome, her mother declined any DNA testing because of her young age. A prenatal analysis was requested by I1 during her last pregnancy, although no mutation was detected in her leucocytes.
Family 4
The requesting woman (II2) was the sister of proband II1 who died at 17 years of age from a typical MELAS syndrome with several stroke‐like episodes, frequent headache attacks, and deafness. m.3243A>G was found in leucocytes from the proband (II2), his mother (I1), aunt (I2) and sister II2, who was shown to have a 34% mutant load with no clinical features of MELAS at 29 years of age.
Family 5
Healthy patient II1, whose younger sister (II3) died from a typical MELAS syndrome, requested a prenatal diagnosis for the m.3243A>G mutation. Their mother (I1) presented with diabetes mellitus and deafness. The mutation was present at a low level in leucocytes from II1 (21%), her mother (10%) and her asymptomatic brother (II2: 5%). Three prenatal diagnoses were carried out during a twin pregnancy (non‐identical twins, III1 and III2), and during a new pregnancy a few years later.
Procedures
CVS was carried out at 10 weeks gestation. Amniocenteses were carried out between 14 and 20 weeks gestation. Apart from blood sampled in each pregnant woman, hair follicles, oral mucosa cells and urinary tract cells were collected from patients I1 (family 3) and II2 (family 4), as the mutation load is usually higher in these cell types than in leucocytes and is closer to the levels found in critical tissues such as muscle.6
Methods
Fetal m.3243A>G mutant load quantification
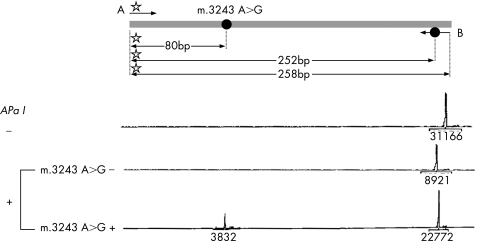
Analyses were carried out on fresh cells. Total DNA was extracted using a classical phenol extraction method. m.3243A>G mutant load was quantified using a polymerase chain reaction (PCR) restriction approach. PCR primers were designed using the software Oligo 6.0. The “forward” primer (A) spanned mtDNA nt3161–nt3178 (5′GCGCCTTCCCCCGTAAAT3′), and the reverse primer (B) spanned nt3421–nt3400. One mismatch (G substituted for A) was incorporated at nucleotide position 3411 within primer B, thus creating an artificial ApaI restriction site as an internal digestion standard. (5′GTTGGGGCCCTTGCGTAGTTG3′). Putative occurrence of a cross hybridisation of oligonucleotide primers to nuclear DNA was ruled out by checking for absence of the PCR amplification using mtDNA‐less RhoO cells.7 Total DNA (1.5 ng) was amplified by PCR (97°C/20 s, 60°C/30 s, 68°C/1 min, for 20 cycles). An unlabelled forward primer was used for the first 19 PCR cycles, whereas the labelled forward primer was added before the last PCR cycle, to prevent an underestimation of the mutant load secondary to heteroduplex formation during the PCR step.8 The 258‐bp amplification product was submitted to ApaI digestion according to the manufacturer's instructions. The fluorescent fragments were studied using the Genescan and Genotyper softwares (Applied Biosystems, Applera, France). The mutant load was quantified as detailed in fig 2. Each assay was carried out in triplicate.
Figure 2 Determination of m3243 A>G mutant load. m.3243A>G creates an ApaI restriction site. Polymerase chain reaction (PCR) amplification was carried out using primers A ( Rox‐tagged primer) and B (in which an artificial ApaI site was created by mismatch incorporation, as an internal digestion standard). ApaI restriction of the 258‐bp PCR product generated two (252 and 6 bp) and three (172, 80 bp and 6 bp) fragments for wild‐type and mutant species, respectively. After purification and electrophoresis using a ABI3100 genetic analyser (Applied Biosystems), digestion products were analysed by Genescan and Genotyper softwares. Only fluorescent fragments, for example, 258, 252 and 80 bp, were detected. Peak areas are indicated under the baseline. Mutant load was calculated by dividing the 80‐bp peak area by the sum of the 80‐bp and 252‐bp peak areas.
Results
The m.3243A>G mutation load, initially assessed in white cells from all five pregnant women, was additionally measured when possible in other tissues—for example, hair follicles, oral mucosa and urinary tract cells in two of them (table 1). In individual I1 from family 3, m.3243A>G, shown to be absent from white cells, was detected only in urinary tract cells at a barely detectable level. In individual II2 from family 4, the mutant load was higher in non‐blood cells than in white cells, the highest level being in urinary tract cells, in agreement with previous reports.6
Table 1 m.3243A>G prenatal analysis.
| Family | Mutant load (%) | Outcome | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Mother | Fetus | ||||||||
| no | Leucocytes | Hair follicles | Oral mucosa cells | Urinary tract cells | n° | CVS | Amniocytes | ||
| 1 | II1 | 0 | III2 | 0 | 0 | Healthy at 3 years | |||
| III3 | ND | 0 | Healthy ay 3 months | ||||||
| 2 | III3 | 0 | IV2 | 0 | 0 | Healthy at 1 year | |||
| 3 | I1 | 0 | 0 | 0 | 3 (1) | II4 | 0 | 0 | Ongoing pregnancy |
| 4 | II2 | 34 (2) | 40 | 45 (1) | 80 (2) | III1 | 35 (2) | 33 (1), 29 (2) | Healthy at 2 years |
| III2 | 31 (3) | 23 (1), 26 (1) | Ongoing pregnancy | ||||||
| 5 | II1 | 21 (2) | III1 | 60 (2) | 63 (1) | TOP | |||
| III2 | 0 | 0 | Healthy at 6 years | ||||||
| III3 | 0 | 0 | Healthy at 10 months | ||||||
CVS, Chorionic villous sampling; TOP, termination of pregnancy.
Chorionic villi and amniotic fluids were sampled at 10 weeks' gestation and 14 weeks' gestation, respectively
An additional amniocentesis was carried out at 20 weeks' gestation in individuals III1 and III2 from family 4. All assays were carried out in triplicate, apart from an assay in individual II2's hair follicle, which was carried out only once.
Values are expressed as mean (SD).
Mutation research in the CVS and amniocytes was null and void in each woman, without any detectable m.3243A>G mutation in white cells (families 1, 2 and 3). All three children born after these analyses are healthy at 3 years, 3 months (family 1) and 1 year of age (family 2). Patient I1's pregnancy (family 3) is still ongoing.
Different results were observed in women with a detectable level of m.3243A>G mutation (families 4 and 5). Individual II1 (family 5) underwent three prenatal diagnoses. One fetus from her twin pregnancy (non‐identical twins) had a high mutant load (III1: 63%), whereas no mutation was detected in the other one, as shown by the CVS and amniocyte analyses. III1's pregnancy was terminated on parental request, whereas III2 is healthy at 6 years of age. A few years later, a novel prenatal analysis showed the absence of m.3243A>G, both in the CVS and amniocytes. This infant (III3) is healthy at 10 months. In family 4, the first prenatal diagnosis was carried out on the CVS, showing a 35% m.3243A>G level. Two further analyses on amniocytes at 14 and 20 weeks of gestation confirmed the presence of an m.3243A>G mutation with 33% and 29% mutant loads, respectively. During the second prenatal analysis, the rate of mutant DNA was 31% in the CVS at the 12th week of gestation, and 23% and 26% in amniocytes at the 14th and 20th weeks of gestation, respectively. The mother decided not to terminate her pregnancies, even when occurrence of a MELAS syndrome could not be excluded. The first child, a 2‐year‐old girl, is healthy to date, and the second pregnancy is still ongoing.
Discussion
Very few data are available with respect to the prenatal diagnosis of MELAS syndrome.5 Furthermore, in these reports, the predictive value of a given fetal 3243G>A mutant load for the postnatal clinical outcome remains a matter of debate.4,5 Accuracy of the mutant load estimate using a PCR‐ApaI restriction experiment followed by agarose gel electrophoresis remained questionable.3,5 On the basis of a previous report by our group on a fluorescent assay for assessment of heteroplasmy of another mtDNA mutation, namely m.8993T>G,9 we devised a novel PCR test in an attempt to improve quantification of the m.3243A>G mutant load. This test has three advantages.
Artificial introduction of an ApaI restriction site in one of the PCR primers as an internal control ascertains that the PCR product is fully digested.
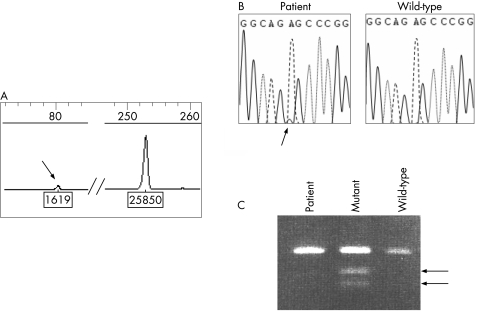
The use of Genescan software makes this test highly sensitive for detection of very low amounts of mutant DNA, as illustrated in fig 3.
Computerised calculation of the respective amounts of the mutant and wild‐type digestion products provides a reliable estimate of the mutant to wild‐type mtDNA ratio (fig 2).
Figure 3 Comparison of sensitivity of three different methods for detection of m.3243A>G in a MELAS‐affected patient. (A) Genescan analysis as described in fig 2. The arrow denotes the mutant mtDNA. Peak area ratio indicates a 6% mutant load. (B) Direct sequencing of the mt tRNALeu gene. Detection of such a low amount of mutant DNA requires absence of any background signal, and sequencing does not enable reliable quantification of the mutant load. (C) Analysis of ApaI–digested PCR product by agarose electrophoresis. Heteroplasmic mutant control DNA (lane 2) was shown to consist of 20% of mutant mtDNA molecules by Genescan analysis, as described in fig 2. The 6% mutant load of the patient could not be detected using this approach.
Furthermore, this technical approach is markedly easier and less expensive to implement than real‐time PCR or Southern blot,10 safer than radioactive methods, all methods extensively used to quantify the level of mutant mtDNA and more quantitative than direct sequencing (fig 3).
By using this test, we carried out nine prenatal analyses in five different families. In the first situation (families 1, 2 and 3), pregnant women (II1, III3, I1, respectively) were shown not to carry the m.3243A>G mutation in their white cells. These three women were, however, considered to be at considerable risk of transmitting the MELAS syndrome to their offspring, on the basis of
the presence of symptoms suggestive of the MELAS syndrome in themselves (III3) or in direct offspring (I1), or
the presence of the mutation in another tissue (I1), or
their positions in respective pedigrees (II1 and III3).
Prenatal analyses consistently failed to detect any mutant load in CVS or amniocytes. All the children born are healthy to date, so the level of m.3243A>G in their blood was not assessed for ethical reasons. These data suggest that the absence of m.3243A>G in maternal white cells might be a good indicator of the absence of the mutation in oocytes. Whereas additional data are obviously required to confirm this, it can, however, be speculated that m.3243A>G‐free relatives of affected individuals might be reassured with respect to the risk for themselves and their offspring of developing the MELAS syndrome, thus preventing implementation of useless and potentially harmful prenatal testing. In the second situation (families 5 and 4), pregnant women carried m.3243A>G in their leucocytes, as well as in all other tested tissues (individual II2), and were symptom‐free at 28 and 24 years of age, respectively. In II1 (21% mutant load in leucocytes family 5), mtDNA segregation throughout the first 12/14 weeks of embryogenesis resulted in fetal mtDNA homoplasmy for two gestations (none of the two fetuses had any detectable mutation). These data corroborate the mitochondrial bottleneck theory,4 speculating that a few mtDNA copies (wild‐type or mutant) become the mitochondrial founders during maturation of primordial germ cells, thus resulting in homoplasmic embryos (0% or about 100% mutant load) at least at the early stages of their development. It can unfortunately be anticipated from data collected in fetus III1 from family 5 and in both fetuses from family 4 that biological laws (if existing) governing mtDNA segregation during gametogenesis and embryo fetal development are more complex than suggested above. Patient II2 embryos from family 4 and embryo III1 from family 5, were indeed heteroplasmic during the first 20 weeks of gestation, with an intermediate mutant load. Apparent stability of the mutant load in the three fetuses of family 5 throughout the first 14 weeks of gestation (table 1), and irrespective of the tested tissue (CVS or amniocytes), supports data from other groups suggesting that (i) mtDNA does not segregate much during embryogenesis4 and (ii) the proportion of mutant mtDNA does not markedly vary among embryo, fetal and extra‐embryonic tissues (placenta).11,12,13 In this respect, it is not clear whether the slight differences in mutant load observed in III1 and III2 (family 4) on three successive samples (35% to 29% and 31% to 23%, respectively) are of any significance with respect to their affect on respiratory chain functioning within a given tissue and at a defined stage of embryo fetal development. It cannot be excluded that the mutation load can vary during pregnancy, and that the trend could worsen in the later stages of pregnancy. Thus, it is worthwhile collecting additional data to ascertain these points, since they could potentially affect the prenatal diagnosis procedure for MELAS, especially regarding the requirement for recurrent assessment of the fetal mutant load or the type of fetal tissue to be analysed. Additionally, the different patterns of m.3243A>G inheritance in fetuses from families 4 and 5 suggest that additional factors such as the maternal mutant load (21% v 34% in leucocytes from pregnant women from family 5 and 4, respectively) might modulate mtDNA segregation during embryogenesis. A similar hypothesis was raised in a recent report,10 suggesting that the outcome of pregnancy is related to the level of mutant mtDNA in m.3243A>G carrier females.
Finally, even if the only child hitherto born after a MELAS prenatal diagnosis positive for m.3243A>G (family 4, patient III1), among the three fetuses shown to carry m.3243A>G, is clinically healthy at 2 years of age (termination of pregnancy and gestation still ongoing for individuals III1 from family 5 and III2 from family 4, respectively), a long‐term follow‐up, of the patients reported here, as well as of additional patients born after a MELAS prenatal diagnosis, is obviously required for a further estimate of the reliability of such a prenatal diagnosis approach before any conclusive recommendation can be made about the use of this technique in clinical practice. This is especially so in patients known to carry an intermediate mutant load in the prenatal period. Taking into account these limitations and provided that (i) appropriate genetic counselling is delivered and (ii) couples consent to recurrent DNA analyses at different stages of pregnancy, we do believe that such a prenatal approach may be helpful to at‐risk families. Different methods of prenatal analyses such as preimplantation genetic diagnosis might constitute an alternative option to conventional prenatal diagnosis and might give some insight into mtDNA segregation during early stages of embryogenesis. We are currently devising such an approach.
Acknowledgements
We acknowledge the Departement de l'Hospitalisation et de l'Organisation des Soins (DHOS) of the French Health Ministry, and l'Assistance Publique‐Hôpitaux de Paris”. JS is supported by a grant from the l'Association Française contre les Myopathies (AFM). This work was supported by a grant from the European Community (MITOCIRCLE project).
Abbreviations
CVS - chorionic villous sampling
MELAS - myopathy, encephalopathy, lactic acidosis, and stroke‐like
mtDNA - mitochondrial DNA
PCR - polymerase chain reaction
Footnotes
Competing interests: None declared.
References
- 1.Goto Y, Nonaka I, Horai S. A mutation in the tRNA (Leu) (UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 1990348651–653. [DOI] [PubMed] [Google Scholar]
- 2.Ciafaloni E, Ricci E, Shanske S, Moraes C, Silvestri G, Hirano M, Simonetti S, Angelini C, Donati M A, Garcia C. MELAS: clinical features, biochemistry, and molecular genetics. Ann Neurol 199231391–398. [DOI] [PubMed] [Google Scholar]
- 3.Manouvrier S, Rotig A, Hannebique G, Gheerbrandt J D, Royer‐Legrain G, Munnich A, Parent M, Grunfeld J P, Largilliere C, Lombes A, Bonnefont J P. Point mutation of the mitochondrial tRNA(Leu) gene (A 3243 G) in maternally inherited hypertrophic cardiomyopathy, diabetes mellitus, renal failure, and sensorineural deafness. J Med Genet 199532654–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poulton J, Marchington D. Progress in genetic counseling and prenatal diagnosis of maternally inherited mtDNA diseases. Neuromuscul Disord 200010484–487. [DOI] [PubMed] [Google Scholar]
- 5.Chou Y J, Ou C Y, Hsu T Y, Liou C W, Lee C F, Tso D J, Wei Y H. Prenatal diagnosis of a fetus harboring an intermediate load of the A3243G mtDNA mutation in a maternal carrier diagnosed with MELAS syndrome. Prenat Diagn 200424367–370. [DOI] [PubMed] [Google Scholar]
- 6.Frederiksen A L, Andersen P H, Kyvik K O, Jeppesen T D, Vissing J, Schwartz M. Tissue specific distribution of the 3243A>G mtDNA mutation. J Med Genet 200643671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parfait B, Rustin P, Munnich A, Rotig A. Co‐amplification of nuclear pseudogenes and assessment of heteroplasmy of mitochondrial DNA mutations. Biochem Biophys Res Commun 199824757–59. [DOI] [PubMed] [Google Scholar]
- 8.Meirelles F V, Smith L C. Mitochondrial genotype segregation during preimplantation development in mouse heteroplasmic embryos. Genetics 1997148877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gigarel N, Ray P F, Burlet P, Frydman N, Royer G, Lebon S, Bonnefont J P, Frydman R, Munnich A, Steffann J. Single cell quantification of the 8993T>G NARP mitochondrial DNA mutation by fluorescent PCR. Mol Genet Metab 200584289–292. [DOI] [PubMed] [Google Scholar]
- 10.Chinnery P F, Howell N, Lightowlers R N, Turnbull D M. MELAS and MERRF. The relationship between maternal mutation load and the frequency of clinically affected offspring. Brain 19981211889–1894. [DOI] [PubMed] [Google Scholar]
- 11.Matthews P M, Hopkin J, Brown R M, Stephenson J B, Hilton‐Jones D, Brown G K. Comparison of the relative levels of the 3243 (A‐‐>G) mtDNA mutation in heteroplasmic adult and fetal tissues. J Med Genet 19943141–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardaioli E, Fabrizi G M, Grieco G S, Dotti M T, Federico A. Heteroplasmy of the A3243G transition of mitochondrial tRNALeu (UUR) in a MELAS case and in a 25‐week‐old miscarried fetus. J Neurol 2000247885–887. [DOI] [PubMed] [Google Scholar]
- 13.White S L, Collins V R, Wolfe R, Cleary M A, Shanske S, DiMauro S, Dahl H H, Thorburn D R. Genetic counseling and prenatal diagnosis for the mitochondrial DNA mutations at nucleotide 8993. Am J Hum Genet 199965474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]