Abstract
Background
Hereditary haemorrhagic telangiectasia (HHT) is an autosomal dominant disease exhibiting multifocal vascular telangiectases and arteriovenous malformations. The majority of cases are caused by mutations in either the endoglin (ENG) or activin receptor‐like kinase 1 (ALK1, ACVRL1) genes; both members of the transforming growth factor (TGF)‐β pathway. Mutations in SMAD4, another TGF‐β pathway member, are seen in patients with the combined syndrome of juvenile polyposis (JP) and HHT (JP‐HHT).
Methods
We sought to determine if HHT patients without any apparent history of JP, who were undergoing routine diagnostic testing, would have mutations in SMAD4. We tested 30 unrelated HHT patients, all of whom had been referred for DNA based testing for HHT and were found to be negative for mutations in ENG and ALK1.
Results
Three of these people harboured mutations in SMAD4, a rate of 10% (3/30). The SMAD4 mutations were similar to those found in other patients with the JP‐HHT syndrome.
Conclusions
The identification of SMAD4 mutations in HHT patients without prior diagnosis of JP has significant and immediate clinical implications, as these people are likely to be at risk of having JP‐HHT with the associated increased risk of gastrointestinal cancer. We propose that routine DNA based testing for HHT should include SMAD4 for samples in which mutations in neither ENG nor ALK1 are identified. HHT patients with SMAD4 mutations should be screened for colonic and gastric polyps associated with JP.
Keywords: hereditary haemorrhagic telangiectasia (HHT); juvenile polyposis (JP); endoglin (ENG); activin receptor‐like kinase 1 (ALK1, ACVRL1); SMAD4
Hereditary haemorrhagic telangiectasia (HHT) is an autosomal dominant disease of vascular dysplasia. The symptoms of HHT include epistaxis, telangiectases, and arteriovenous malformations (AVMs), which are most often found in the lungs, brain, liver, and gastrointestinal tract. There is wide variation in penetrance and severity of these symptoms in patients even within the same family,1 suggesting that environmental or other genetic factors influence the phenotype. Mutations in either one of two genes cause HHT. Mutations in the endoglin (ENG) gene are responsible for HHT12 and mutations in the activin receptor‐like kinase 1 (ALK1) gene for HHT2.3 Both of these genes encode TGF‐β binding proteins and play important roles in regulating endothelial cell growth.4,5
Screening for mutations in either ENG or ALK1 in HHT patient cohorts yields mutation detection rates of between 62% and 93%.6,7,8,9,10,11,12,13,14 The failure to achieve a 100% detection rate is not unique to HHT. This range of mutation detection rates is similar to that found for many other mendelian disorders,15 implying that most, if not all, mutation scans share common detection limitations. In the case of HHT, people may have large deletions or insertions, deep intronic mutations affecting splicing, or regulatory mutations in either ENG or ALK1. A subset of the mutation negative samples may instead have a mutation in the as yet undiscovered HHT3 gene recently linked to chromosome 5.16 Because the majority of HHT patients have been found to have mutations in ENG or ALK1, it is probable that only a small fraction of the remaining cases will be caused by mutations in other genes.
SMAD4 is a key downstream effector of transforming growth factor (TGF)‐β signalling. Mutations in SMAD4 and BMPR1A are known to be causative for juvenile polyposis (JP).17,18 JP is characterised by the presence of five or more hamartomatous gastrointestinal polyps, or any number of polyps in addition to a family history of polyposis.19 There is an increased risk of gastrointestinal cancers associated with these polyps. Recently, SMAD4 was identified as mutated in a subset of HHT patients with JP, a condition termed JP‐HHT syndrome,20 in which juvenile polyps and anaemia are the predominant clinical features. HHT symptoms vary among patients, but all meet the Curaçao criteria for being either definitely or possibly affected.21 There is a high penetrance of AVMs in this JP‐HHT cohort, particularly in young patients. Importantly, each of these patients has symptoms of both JP and HHT.
Both JP and HHT are diseases known to have a range of clinical presentations and to be variably penetrant.1,22 One feature common to both disorders is gastrointestinal (GI) bleeding, potentially confounding the correct diagnosis. Because of this potential for uncertainty in diagnoses, we questioned whether, among a cohort of ENG and ALK1 mutation negative HHT cases, there were occult cases of JP‐HHT, diagnosed as only having the HHT component of the combined phenotype. Using sequence analysis for SMAD4 mutations, we screened 30 people from an unselected group of HHT patients, who had been referred for DNA based testing for HHT and found to be negative for mutations in either ENG or ALK1.
METHODS
Subjects
Of the the subjects in this cohort, 20 were referred to the genetic diagnostic laboratory at the University of Pennsylvania, five were enrolled through the Dutch HHT Center in the Netherlands, and four were referred through HHT Solutions in Toronto, Canada. Individuals were referred to these centres of DNA based diagnostics because of a diagnosis or suspicion of HHT (table 1). One patient with HHT was independently assessed at the hospital clinic in Barcelona, Spain, and referred to Duke University for this study after being diagnosed with colon cancer. All participants in this study were enrolled with their informed consent. Clinical subjects also gave consent for their DNA to be used in further research. Enrolment of participants through Duke University and the Dutch HHT Center was given approval by the institutional review boards of the participating institutions (Duke University Health System Institution Review Board Committee, and METC, Utrecht).
Table 1 Clinical characteristics of the testing cohort.
| Sample | Age (years) | Family history | Visceral lesions | Telangiectasia | Epistaxis | Other | HHT status | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UP001 | 18 | Yes | Yes | N/A | N/A | Possible | ||||||||
| UP002 | 61 | Yes | Yes | N/A | Yes | GI bleeding | Definite | |||||||
| UP003 | 73 | Yes | Yes | Yes | N/A | Definite | ||||||||
| UP004 | 77 | Yes | Yes | Yes | Yes | Definite | ||||||||
| UP005 | 21 | N/A | Yes | N/A | Yes | Possible | ||||||||
| UP006 | 37 | N/A | N/A | Yes, (rectal) | Yes | Possible | ||||||||
| UP007 | 73 | Yes | Yes | N/A | Yes | GI bleeding | Definite | |||||||
| UP008 | 54 | Yes | Yes | Yes | Yes | Definite | ||||||||
| UP009 | 23 | N/A | Yes | Yes | Yes | Definite | ||||||||
| UP010 | 38 | N/A | Yes (lung) | Yes | N/A | Stroke | Possible | |||||||
| UP011 | 78 | N/A | Yes | N/A | Yes | GI bleeding | Possible | |||||||
| UP012 | 74 | Yes | N/A | Yes | Yes | Definite | ||||||||
| UP013 | 52 | Yes | Yes | Yes | Yes | Definite | ||||||||
| UP014 | 69 | N/A | Yes | Yes | Yes | Definite | ||||||||
| UP015* | 37 | No | Yes (lung, GI) | Yes | Yes | GI bleeding, colonic polyps | Definite | |||||||
| UP016 | 36 | Yes | Yes | Yes | Yes | Definite | ||||||||
| UP017 | 69 | Yes | Yes | Yes | Yes | Definite | ||||||||
| UP018 | 32 | Yes | Yes | Yes | N/A | Definite | ||||||||
| UP019 | 87 | Yes | Yes | Yes | Yes | Definite | ||||||||
| UP020 | 56 | N/A | Yes (stomach) | Yes | Yes | Definite | ||||||||
| D1827 | 47 | Yes | Yes (liver) | Yes (inc. GI) | Yes | Confirmed HHT | Definite | |||||||
| DKCL331 | 44 | Yes | Yes (lung) | Yes | Yes | Confirmed HHT | Definite | |||||||
| D0883 | 63 | Yes | No | Yes | Yes | Confirmed HHT | Definite | |||||||
| D1488 | 44 | Yes | Yes (lung) | Yes | Yes | Confirmed HHT | Definite | |||||||
| D3748 | 51 | Yes | No | Yes | Yes | Confirmed HHT | Definite | |||||||
| 5001 | 56 | Yes | Yes (hip) | Yes | Yes | Confirmed HHT, GI bleeding, gastric polyps | Definite | |||||||
| 5031 | 37 | No | Yes (lung, multiple) | No | Yes | Confirmed HHT, migraines | Possible | |||||||
| 5051 | 46 | Yes | No | Yes (chest) | Yes | Confirmed HHT | Definite | |||||||
| 5068* | 39 | No | Yes (lung) | Yes | Yes | Confirmed HHT, liver shunts | Definite | |||||||
| Yes (in children) | ||||||||||||||
| 3892* | 47 | No | Yes (liver) | Yes (lips, tongue) | Yes | Confirmed HHT, anaemiaColon cancer | Definite |
HHT status was‐ as defined by the Curaçao criteria.21 *SMAD4 mutation positive patients. N/A, not available; GI, gastrointestinal.
Our cohort comprised 30 subjects, of whom 24 (80%) had definite HHT and 6 (20%) had possible HHT, according to the Curaçao diagnostic criteria.21 Ages of the participants ranged from 18 to 87 years (mean 51.3, median 49). The majority of the subjects reported having a positive family history for HHT (19/30, 63%) while four people reported a negative family history. The remaining seven did not respond to the query on their family history. Visceral lesions were present in 25 people, absent in 3, and not reported in 2. Five different participants noted GI bleeding, and individual subjects report having stroke, migraines, gastric polyps, and liver shunts. One individual had both anaemia and colon cancer, and another had GI bleeding and colonic polyps.
The samples included in this study are a subset of 102 ENG/ALK1 mutation negative samples from all of the institutions. The 30 participants comprising this cohort had granted consent for additional research using their DNA. They were not selected for SMAD4 testing based on any reported clinical symptoms.
Diagnostic testing and genetic screening
All 30 subjects were screened by direct DNA sequencing of PCR products consisting of the coding exons and adjacent intronic regions for ENG,ALK1, and SMAD4 from genomic DNA. In all cases, any sequence variations found were reamplified and resequenced to confirm the observed changes. The sequences generated were compared with wild type ENG (GenBank accession no. NT_008470 or BC014271), ALK1 (GenBank NT_029419 or NM_000020), and SMAD4 (GenBank NM_005359). There were 113 control individuals screened for the presence of the SMAD4 missense mutation by using a Sau3A restriction digest assay. The mutation destroys the single Sau3A site in the exon 8 amplimer. In all cases, nucleotides were numbered starting with the A in the initial ATG of the cDNA sequence as c.1, and the starting methionine in the protein sequence as p.1 (http://www.hgvs.org/mutnomen/).
Although all centres used direct DNA sequence analysis to identify point mutations, each diagnostic centre employed a different technique to further examine the DNAs for large scale rearrangements. The 20 samples from the genetic diagnostic laboratory at University of Pennsylvania were tested by quantitative Southern blots. Genomic DNA was digested with RsaI, because most fragments represent individual exons. cDNA probes covering exons 2–14 for ENG and 2–10 for ALK1 were used for the initial analysis. If a change in dosage for any one or more exons was detected, probing with specific individual exonic clones was performed. In addition, genomic probes for exons 1 of both ENG and ALK1 were tested separately. Whole gene deletions were detected by comparison with an internal control (β‐globin gene) run on the same filter.
The four samples from HHT Solutions were tested for changes in exon size and copy number using quantitative multiplex PCR with intronic primers for all 15 ENG exons and 9 ALK1 exons. The fluorescent fragments were analysed using Gene Objects software (version 3.1; Bayer). RNA analysis was used in some cases as an additional test where no mutation was found by sequencing or QM‐PCR.
The five samples from the Dutch HHT Center were probed for exonic deletions or duplications in ENG and ALK1 by MLPA. The DNA from patient 3892 was not examined for genomic rearrangements; however, the coding regions and surrounding intronic regions of BMPR1A were sequenced and compared with a reference sequence (GenBank accession number NM_004329).
RESULTS
Subjects were referred to four different institutions for DNA based diagnostic testing for HHT based on the clinical diagnosis or suspicion of HHT. Of 374 people sent for DNA testing for HHT, 30 had also consented to allow their DNA to be included in research studies. Of these 30, none was known to have JP, although GI bleeding, a feature in common with HHT and JP, was reported for five of the individuals. All 30 of these subjects had been screened for ENG and ALK1 mutations by direct DNA sequencing but no mutations had been found. As part of the attempted molecular diagnosis for HHT, 29 of these people had also been screened for genomic rearrangements of the ENG and ALK1 genes, but no alterations had been uncovered.
We then sequenced the coding exons and adjacent intronic sequences of SMAD4 and identified mutations in three subjects. Individual UP015 was referred to the genetic diagnostic laboratory at the University of Pennsylvania. This 37 year old has telangiectases, epistaxis, and AVMs in the lung and gastrointestinal tract. Although no family history of HHT was noted, this patient fulfils the Curaçao criteria for definite HHT.21 This person was found to have a frameshift mutation in exon 11 of SMAD4, c.1596_97delCCinsT. After this molecular finding, inspection of patient charts revealed that this patient had colonic polyps that had been discoverd during endoscopy.
Individual 5068 (family 530) was referred to HHT Solutions in Toronto. This 39 year old has telangiectases, epistaxis, pulmonary AVMs, and liver shunts, and therefore meets diagnostic criteria as definitely affected with HHT. Although there was no reported past family history of HHT, the subject's children show some presumptive signs of HHT (epistaxis and telangiectases). A missense mutation in exon 8 of SMAD4, c.1081C→T, R361C, was found.
Individual 3892, 47 years old, was diagnosed at 13 years of age with HHT, and had telangiectases of the lips and tongue, epistaxis, and a hepatic AVM. This person has had iron deficiency anaemia for more than 15 years, requiring blood transfusions on multiple occasions, and is on regular iron supplementation. Signet ring cell type colorectal cancer located in the caecum was diagnosed in June 2004. Upon examination, seven hamartomatous polyps were found in the ascending colon and three in the duodenum. There is no history of either HHT or gastrointestinal cancer in the family. This person harboured the same SMAD4 exon 8 missense mutation (c.1081C→T, R361C) seen in patient 5068.
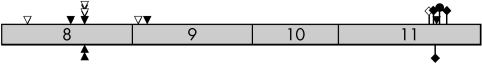
All three mutations identified were found within in the COOH terminus of SMAD4, where all of the mutations in previously reported JP‐HHT cases have been identified (fig 1). One mutation, c.1081C→T (R361C) in exon 8, was found in two unrelated members of our study cohort. This same mutation has been reported in a number of people with JP,23,24,25 and the same base is mutated in a previously reported case of JP‐HHT (c.1081C→G, p.R361G).20 The missense mutation at codon 361 was not identified by us in 113 unaffected population controls (226 alleles) or by Houlston et al in the 50 controls (100 alleles) tested in the case of a JP patient.23 Using two different programs that use evolutionary conservation to investigate the effects of putative missense mutations (SIFT (http://blocks.fhcrc.org/sift/SIFT.html) and PolyPhen (http://genetics.bwh.harvard.edu/cgi‐bin/pph/polyphen.cgi)), the R361C amino acid change is predicted to negatively affect protein function. The third mutation found in this study constitutes a frameshift in exon 11 (c.1596_97delCCinsT). This is a novel change, although frameshift mutations in exon 11 of SMAD4 have also been identified in JP‐HHT patients.20
Figure 1 Schematic diagram of exons 8–11 of SMAD4 showing HHT associated mutations. These four exons encode the MH2 domain of the protein. In confirmed JP‐HHT syndrome patients, no mutations have been identified in the first seven exons of the gene. Shaded boxes, exons; triangles, missense mutations; diamonds, frameshift mutation; circles, nonsense mutations; filled symbols above the shaded exons, previously reported mutations in JP‐HHT patients;20 open symbols, from our unpublished data; marks below the exons, mutations reported here from people exhibiting symptoms of HHT.
The total diagnostic testing cohort from the four centres was 374 HHT patients. ENG or ALK1 mutations were found in 272 people, a detection rate of 72.7%. Thus, 102 subjects (27.3%) were considered ENG/ALK1 mutation negative for this study. The 30 people available to us for research represent approximately 30% of the ENG/ALK1 mutation negative group and 8% of the total HHT cohort. SMAD4 mutations were found in three of the 30, a SMAD4 mutation rate of 10%. Extrapolating to the larger ENG/ALK1 mutation negative group, up to 10 presumptive HHT patients might harbour SMAD4 mutations. This would represent 2–3% (10/374) of the original cohort referred for HHT molecular testing.
DISCUSSION
DNA sequencing performed by numerous groups around the world shows that the majority of HHT patients have mutations in either ENG or ALK1.6,7,8,9,10,11,12,13,14 The mutation detection rates in the scans range from 62% to 93%, a range that matches the detection rates found in scans of other genes in mendelian diseases.15 There are several explanations why a patient might appear mutation negative in a mutation screen. For diseases such as HHT that have highly variable clinical presentations, it is possible that some of the referred patients may not actually have the disease in question. In the present study, presuming that all members of the cohort are affected HHT patients, another explanation could be that these patients may harbour non‐coding mutations, such as alterations in poorly characterised regulatory regions of the gene, including the promoter, introns, and 5′ and 3′ untranslated regions. Additionally, large scale genomic rearrangements involving whole exons, partial gene loss or duplication, and intrachromosomal or interchromosomal rearrangements will bypass detection by DNA sequencing. It is also possible that some patients will have mutations in other genes that lead to the same disease phenotype. A recent report of two unrelated HHT families that exhibit linkage to a region on chromosome 5 demonstrates that at least one other gene is involved in HHT.16 Non‐ENG, non‐ALK1 HHT patients may harbour a mutation in this as yet unidentified gene.
A final explanation for these ENG/ALK1 mutation negative HHT cases might be that some of these patients harbour mutations in SMAD4. Based on a molecular diagnosis, these patients would have the JP‐HHT syndrome, but they may have been diagnosed with HHT due to occult or unrecognised manifestations of JP. In this study we queried if SMAD4 mutations were found in the general HHT population. To determine the answer, we sequenced the coding exons and adjacent flanking intronic regions of SMAD4 in a cohort of 30 unrelated people with presumptive HHT. These people had been referred for DNA based diagnostic testing for HHT and all were negative for mutations in ENG and ALK1. One of them had colon cancer, which is sometimes the first symptomatic manifestation of JP, but none of the others was known or reported to have JP. Although GI bleeding was reported in five of these subjects, this was interpreted as a symptom of HHT.
All of the SMAD4 mutations identified in JP‐HHT patients to date are found in the COOH terminus of the protein. The three mutations identified in the subjects here are in the COOH terminus of SMAD4 and are nearly identical in location and type of mutation to those seen in JP‐HHT patients (fig 1). This strongly suggests that the three HHT subjects with SMAD4 mutations in this cohort are affected with JP‐HHT syndrome. Two of these three SMAD4‐HHT subjects are known to have colonic polyps, and one of these two has colorectal cancer. The status of the third individual regarding JP symptoms is unknown.
There is growing evidence of distinct phenotypic differences between patients with HHT1 and HHT2.26,27,28 Similarly, other studies have demonstrated phenotypic differences between JP patients with SMAD4 mutations compared with patients with mutations in BMPR1A,29,30 the second gene known to be involved in JP. JP is also known to be variably penetrant and is associated with an increased risk of gastrointestinal cancer.22,31 Knowing which gene is responsible for the disease in an individual HHT patient will aid in the proper management and care of the patient and, significantly, of related family members.
We recommend that when genetic testing is advised for HHT patients, SMAD4 should be screened if no mutations are found in either ENG or ALK1. We also recommend that screening for colonic and gastric polyps be considered in people in whom neither ENG nor ALK1 mutations have been found, in whom SMAD4 mutations have been uncovered, or in anyone with anaemia that cannot be completely explained by epistaxis or some other cause. This screening will identify those HHT patients with occult polyposis. Early detection of colonic polyps in any patient, but in particular in JP‐HHT patients, could prevent the development of colorectal cancer by finding and removing precancerous polyps.
There appears to be a high rate of de novo cases of JP‐HHT,20 and all three of the SMAD4‐HHT patients in this study reported no family history of HHT. Although a positive family history is one of the criteria for HHT diagnosis,21SMAD4 mutation carriers may often lack this key diagnostic feature, potentially making the clinical diagnosis more difficult. In the cases of presumed HHT without any apparent family history, an argument might be made that SMAD4 mutation analysis should precede analysis of ENG or ALK1, as de novo mutations in these other HHT genes appear to be rare.8,13,14
An HHT patient found to harbour a SMAD4 mutation would be a prime situation where genetic testing can be used to guide clinical management. Because of the increased risk of gastrointestinal cancer associated with JP, it is critically important for a patient with a SMAD4 mutation to be screened for JP. We suggest that HHT patients harbouring a SMAD4 mutation should be considered at high risk of JP‐HHT, requiring more intensive colorectal cancer screening strategies than those recommended for the average risk population.32 Correspondingly, JP patients with SMAD4 mutations similar to those seen in JP‐HHT patients should be examined for the visceral manifestations of HHT which can present suddenly and catastrophically.
SMAD4, ENG, and ALK1 are all members of the TGF‐β signalling pathway, and mutations in the genes encoding them can cause a broad constellation of phenotypes with both distinct and overlapping clinical features. It has been known for a number of years that mutations in SMAD4 cause JP,17 and we have recently shown that certain types of mutations in SMAD4 cause JP‐HHT.20 Here we report that unselected HHT patients have SMAD4 mutations that are strikingly similar to those seen in JP‐HHT patients. In JP cases with the same types of SMAD4 mutations, it remains to be seen if these JP patients exhibit symptoms of HHT due to the paucity of clinical descriptions in the literature. Mutations in ENG have recently been reported in patients with JP,33 and mutations in ALK1 cause some cases of primary pulmonary hypertension.34,35,36 Molecular dissection of the interconnections between these genes and the effects of aberrant signalling through the TGF‐β and other alternative signalling pathways, will help to further elucidate the pathophysiology of these inherited diseases.
ACKNOWLEDGEMENTS
We would first like to thank the patients for their participation in this study. We also thank K Vandezande at HHT Solutions for helpful discussions. A Castells has received funding from the Ministerio de Educación y Ciencia (SAF 04‐07190) and from the Instituto de Salud Carlos III (RC03/02 and RC03/10). D A Marchuk has received funding from the National Institutes of Health (R01 HL49171).
Note added in proof: Further examination of patient records revealed that patient 5068 has colon polyps. This individual also has a liver AVM, migrolines and clubbing of the figures.
Abbreviations
ALK1 - activin receptor‐like kinase 1
AVM - arteriovenous malformation
ENG - endoglin
GI - gastrointestinal
JP - juvenile polyposis
TGF - transforming growth factor
Footnotes
Competing interests: there are no competing interests
References
- 1.Begbie M E, Wallace G M, Shovlin C L. Hereditary haemorrhagic telangiectasia (Osler‐Weber‐Rendu syndrome): a view from the 21st century. Postgrad Med J 20037918–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McAllister K A, Grogg K M, Johnson D W, Gallione C J, Baldwin M A, Jackson C E, Helmbold E A, Markel D S, McKinnon W C, Murrell J, McCornlich M K, Perieak‐Vanee M A, Heutink P, Oostra B A, Haitjema T, Westermann C J J, Porteous M E, Guttmacher A E, Letarte M, Marchuk D A. Endoglin, a TGF‐beta binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat Genet 19948345–351. [DOI] [PubMed] [Google Scholar]
- 3.Johnson D W, Berg J N, Baldwin M A, Gallione C J, Marondel I, Yoon S J, Stenzel T T, Speer M, Pericak‐Vance M A, Diamond A, Guttmacher A E, Jackson C E, Attisano L, Kucherlapati R, Porteous M E, Marchuk D A. Mutations in the activin receptor‐like kinase 1 gene in hereditary haemorrhagic telangiectasia type 2. Nat Genet 199613189–195. [DOI] [PubMed] [Google Scholar]
- 4.Lebrin F, Deckers M, Bertolino P, Ten Dijke P. TGF‐beta receptor function in the endothelium. Cardiovasc Res 200565599–608. [DOI] [PubMed] [Google Scholar]
- 5.ten Dijke P, Hill C S. New insights into TGF‐beta‐Smad signalling. Trends Biochem Sci 200429265–273. [DOI] [PubMed] [Google Scholar]
- 6.Letteboer T G, Zewald R A, Kamping E J, de Haas G, Mager J J, Snijder R J, Lindhout D, Hennekam F A, Westermann C J, Ploos van Amstel J K. Hereditary hemorrhagic telangiectasia: ENG and ALK‐1 mutations in Dutch patients. Hum Genet 20051168–16. [DOI] [PubMed] [Google Scholar]
- 7.Schulte C, Geisthoff U, Lux A, Kupka S, Zenner H P, Blin N, Pfister M. High frequency of ENG and ALK1/ACVRL1 mutations in German HHT patients. Hum Mutat 200525595. [DOI] [PubMed] [Google Scholar]
- 8.Abdalla S A, Cymerman U, Rushlow D, Chen N, Stoeber G P, Lemire E G, Letarte M. Novel mutations and polymorphisms in genes causing hereditary hemorrhagic telangiectasia. Hum Mutat 200525320–321. [DOI] [PubMed] [Google Scholar]
- 9.Kuehl H K, Caselitz M, Hasenkamp S, Wagner S, El‐Harith el H A, Manns M P, Stuhrmann M. Hepatic manifestation is associated with ALK1 in hereditary hemorrhagic telangiectasia: identification of five novel ALK1 and one novel ENG mutations. Hum Mutat 200525320. [DOI] [PubMed] [Google Scholar]
- 10.Draghi F, Precerutti M, Danesino G M, Olivieri C, Valacca C, Danesino C, Pagella F, Semino L, Lanzarini L, Buscarini E, Danesino C. Vascular abnormalities in the fingers of patients affected with hereditary hemorrhagic telangiectasia (HHT) as assessed by color doppler sonography. Am J Med Genet A 2005135106–109. [DOI] [PubMed] [Google Scholar]
- 11.Lesca G, Plauchu H, Coulet F, Lefebvre S, Plessis G, Odent S, Riviere S, Leheup B, Goizet C, Carette M F, Cordier J F, Pinson S, Soubrier F, Calender A, Giraud S. Molecular screening of ALK1/ACVRL1 and ENG genes in hereditary hemorrhagic telangiectasia in France. Hum Mutat 200423289–299. [DOI] [PubMed] [Google Scholar]
- 12.Brusgaard K, Kjeldsen A D, Poulsen L, Moss H, Vase P, Rasmussen K, Kruse T A, Horder M. Mutations in endoglin and in activin receptor‐like kinase 1 among Danish patients with hereditary haemorrhagic telangiectasia. Clin Genet 200466556–561. [DOI] [PubMed] [Google Scholar]
- 13.Lastella P, Sabba C, Lenato G M, Resta N, Lattanzi W, Gallitelli M, Cirulli A, Guanti G. Endoglin gene mutations and polymorphisms in Italian patients with hereditary haemorrhagic telangiectasia. Clin Genet 200363536–540. [DOI] [PubMed] [Google Scholar]
- 14.Cymerman U, Vera S, Karabegovic A, Abdalla S, Letarte M. Characterization of 17 novel endoglin mutations associated with hereditary hemorrhagic telangiectasia. Hum Mutat 200321482–492. [DOI] [PubMed] [Google Scholar]
- 15.van Ommen G J, Bakker E, den Dunnen J T. The human genome project and the future of diagnostics, treatment, and prevention. Lancet 1999354(Suppl 1)SI5–NaN10. [DOI] [PubMed] [Google Scholar]
- 16.Cole S G, Begbie M E, Wallace G M, Shovlin C L. A new locus for hereditary haemorrhagic telangiectasia (HHT3) maps to chromosome 5. J Med Genet 200542577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howe J R, Roth S, Ringold J C, Summers R W, Jarvinen H J, Sistonen P, Tomlinson I P, Houlston R S, Bevan S, Mitros F A, Stone E M, Aaltonen L A. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science 19982801086–1088. [DOI] [PubMed] [Google Scholar]
- 18.Howe J R, Bair J L, Sayed M G, Anderson M E, Mitros F A, Petersen G M, Velculescu V E, Traverso G, Vogelstein B. Germline mutations of the gene encoding bone morphogenetic protein receptor 1A in juvenile polyposis. Nat Genet 200128184–187. [DOI] [PubMed] [Google Scholar]
- 19.Jass J R, Williams C B, Bussey H J, Morson B C. Juvenile polyposis‐‐a precancerous condition. Histopathology 198813619–630. [DOI] [PubMed] [Google Scholar]
- 20.Gallione C J, Repetto G M, Legius E, Rustgi A K, Schelley S L, Tejpar S, Mitchell G, Drouin E, Westermann C J, Marchuk D A. A combined syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia associated with mutations in MADH4 (SMAD4). Lancet 2004363852–859. [DOI] [PubMed] [Google Scholar]
- 21.Shovlin C L, Guttmacher A E, Buscarini E, Faughnan M E, Hyland R H, Westermann C J, Kjeldsen A D, Plauchu H. Diagnostic criteria for hereditary hemorrhagic telangiectasia (Rendu‐Osler‐Weber syndrome). Am J Med Genet 20009166–67. [DOI] [PubMed] [Google Scholar]
- 22.Stemper T J, Kent T H, Summers R W. Juvenile polyposis and gastrointestinal carcinoma. A study of a kindred. Ann Intern Med 197583639–646. [DOI] [PubMed] [Google Scholar]
- 23.Houlston R, Bevan S, Williams A, Young J, Dunlop M, Rozen P, Eng C, Markie D, Woodford‐Richens K, Rodriguez‐Bigas M A, Leggett B, Neale K, Phillips R, Sheridan E, Hodgson S, Iwama T, Eccles D, Bodmer W, Tomlinson I. Mutations in DPC4 (SMAD4) cause juvenile polyposis syndrome, but only account for a minority of cases. Hum Mol Genet 199871907–1912. [DOI] [PubMed] [Google Scholar]
- 24.Woodford‐Richens K, Bevan S, Churchman M, Dowling B, Jones D, Norbury C G, Hodgson S V, Desai D, Neale K, Phillips R K, Young J, Leggett B, Dunlop M, Rozen P, Eng C, Markie D, Rodriguez‐Bigas M A, Sheridan E, Iwama T, Eccles D, Smith G T, Kim J C, Kim K M, Sampson J R, Evans G, Tejpar S, Bodmer W F, Tomlinson I P, Houlston R S. Analysis of genetic and phenotypic heterogeneity in juvenile polyposis. Gut 200046656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howe J R, Sayed M G, Ahmed A F, Ringold J, Larsen‐Haidle J, Merg A, Mitros F A, Vaccaro C A, Petersen G M, Giardiello F M, Tinley S T, Aaltonen L A, Lynch H T. The prevalence of MADH4 and BMPR1A mutations in juvenile polyposis and absence of BMPR2, BMPR1B, and ACVR1 mutations. J Med Genet 200441484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berg J, Porteous M, Reinhardt D, Gallione C, Holloway S, Umasunthar T, Lux A, McKinnon W, Marchuk D, Guttmacher A. Hereditary haemorrhagic telangiectasia: a questionnaire based study to delineate the different phenotypes caused by endoglin and ALK1 mutations. J Med Genet 200340585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letteboer T G, Mager H J, Snijder R J, Koeleman B P, Lindhout D, Ploos van Amstel H K, Westermann K J. Genotype ‐ phenotype relationship in Hereditary Hemorrhagic Telangiectasia. J Med Genet 200643371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kjeldsen A D, Moller T R, Brusgaard K, Vase P, Andersen P E. Clinical symptoms according to genotype amongst patients with hereditary haemorrhagic telangiectasia. J Intern Med 2005258349–355. [DOI] [PubMed] [Google Scholar]
- 29.Friedl W, Uhlhaas S, Schulmann K, Stolte M, Loff S, Back W, Mangold E, Stern M, Knaebel H P, Sutter C, Weber R G, Pistorius S, Burger B, Propping P. Juvenile polyposis: massive gastric polyposis is more common in MADH4 mutation carriers than in BMPR1A mutation carriers. Hum Genet 2002111108–111. [DOI] [PubMed] [Google Scholar]
- 30.Sayed M G, Ahmed A F, Ringold J R, Anderson M E, Bair J L, Mitros F A, Lynch H T, Tinley S T, Petersen G M, Giardiello F M, Vogelstein B, Howe J R. Germline SMAD4 or BMPR1A mutations and phenotype of juvenile polyposis. Ann Surg Oncol 20029901–906. [DOI] [PubMed] [Google Scholar]
- 31.Merg A, Howe J R. Genetic conditions associated with intestinal juvenile polyps. Am J Med Genet C Semin Med Genet 200412944–55. [DOI] [PubMed] [Google Scholar]
- 32.Gazelle G S, McMahon P M, Scholz F J. Screening for colorectal cancer. Radiology 2000215327–335. [DOI] [PubMed] [Google Scholar]
- 33.Sweet K, Willis J, Zhou X P, Gallione C, Sawada T, Alhopuro P, Khoo S K, Patocs A, Martin C, Bridgeman S, Heinz J, Pilarski R, Lehtonen R, Prior T W, Frebourg T, Teh B T, Marchuk D A, Aaltonen L A, Eng C. Molecular classification of patients with unexplained hamartomatous and hyperplastic polyposis. JAMA 20052942465–2473. [DOI] [PubMed] [Google Scholar]
- 34.Harrison R E, Flanagan J A, Sankelo M, Abdalla S A, Rowell J, Machado R D, Elliott C G, Robbins I M, Olschewski H, McLaughlin V, Gruenig E, Kermeen F, Halme M, Raisanen‐Sokolowski A, Laitinen T, Morrell N W, Trembath R C. Molecular and functional analysis identifies ALK‐1 as the predominant cause of pulmonary hypertension related to hereditary haemorrhagic telangiectasia. J Med Genet 200340865–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abdalla S A, Gallione C J, Barst R J, Horn E M, Knowles J A, Marchuk D A, Letarte M, Morse J H. Primary pulmonary hypertension in families with hereditary haemorrhagic telangiectasia. Eur Respir J 200423373–377. [DOI] [PubMed] [Google Scholar]
- 36.Harrison R E, Berger R, Haworth S G, Tulloh R, Mache C J, Morrell N W, Aldred M A, Trembath R C. Transforming growth factor‐beta receptor mutations and pulmonary arterial hypertension in childhood. Circulation 2005111435–441. [DOI] [PubMed] [Google Scholar]