Abstract
Background: There is a lack of information on prevalence, cause and consequences of slight/mild bilateral sensorineural hearing loss (SNHL) in children. We report the first systematic genetic analysis of the GJB2 gene in a population‐derived sample of children with slight/mild bilateral SNHL.
Methods: Hearing tests were conducted in 6240 Australian elementary school children in Grades 1 and 5. 55 children (0.88%) were found to have a slight/mild sensorineural hearing loss. 48 children with slight/mild sensorineural hearing loss and a matched group of 90 children with normal hearing participated in a genetic study investigating mutations in the GJB2 gene, coding for connexin 26, and the presence of the del(GJB6‐D13S1830) and del(GJB6‐D13S1854) deletions in the GJB6 gene, coding for connexin 30.
Results: Four of 48 children with slight/mild sensorineural hearing loss were homozygous for the GJB2 V37I change. The four children with homozygous V37I mutations were all of Asian background and analysis of SNPs in or near the GJB2 gene suggests that the V37I mutation arose from a single mutational event in the Asian population.
Discussion: Based on the prevalence of carriers of this change we conclude that V37I can be a causative mutation that is often associated with slight/mild sensorineural hearing loss. No other children in the slight/mild hearing loss group had a hearing loss related to a GJB2 mutation. One child with normal hearing was homozygous for the R127H change and we conclude that this change does not cause hearing loss. Two children of Asian background were carriers of the V37I mutation. Our data indicate that slight/mild sensorineural hearing loss due to the GJB2 V37I mutation is common in people of Asian background.
Information on prevalence, cause and consequences of slight or mild bilateral sensorineural hearing loss (SNHL) in children is lacking. Prevalences of 3–5% have been published,1,2,3 and it has been suggested that slight or mild SNHL may contribute to adverse outcomes with respect to language, academic performance and social interactions.4,5,6 We have determined the prevalence in Australian elementary school children (grades 1 and 5) to be 0.88%. Analysis of the effect of the slight or mild SNHL in these children did not provide evidence of marked adverse outcomes when children with mild hearing loss were compared with their normally hearing peers.7
Little is known about the extent to which genetic mutations cause slight or mild SNHL in children. It has been estimated that approximately 60% of cases with moderate, severe and profound prelingual non‐syndromic deafness are genetic, and that 80–85% of these cases are inherited in an autosomal recessive pattern.8,9 Although changes in >40 genes have been associated with dominant and recessive SNHL, mutations in the GJB2 gene (coding for connexin 26) have been shown to be the most common cause of inherited non‐syndromic deafness (Hereditary Hearing Loss Homepage, and Connexins and Deafness Homepage). Inheritance of GJB2 deafness is nearly always recessive. We have estimated that mutations in the GJB2 gene account for the hearing loss in approximately 10–15% of Australians with moderate, severe and profound prelingual non‐syndromic deafness.10
Unlike more severe forms of hearing loss, slight or mild SNHL is often not detected in children or, if detected, rarely investigated in detail. Most GJB2 mutations that have been reported have therefore been mainly associated with more severe hearing losses,11 and their role in milder losses remains unknown.
Universal hearing screening has been introduced in many countries, and as a consequence more children with less severe hearing loss are being identified earlier. In the future, the GJB2 gene will be screened for mutations in many of these children, which will lead to questions of causation and prognosis not just for the probands but also for their carrier siblings. Important unanswered questions are therefore “to what extent do GJB2 mutations contribute to slight/mild SNHL in children?”, “what are the genotype–phenotype correlations?” and “should healthy children identified with slight or mild hearing loss routinely be offered genetic testing?”
To deal with these issues, we report the first systematic genetic analysis of a population‐derived sample of 48 children with slight or mild SNHL and 90 normally hearing controls. The children were identified as part of a large study of the prevalence, aetiology and consequences of slight or mild bilateral SNHL in a representative sample of Australian elementary school children.
Participants and methods
Recruitment and assessment of participants
We studied an unbiased sample of 6581 children (3367 grade 1, 3214 grade 5; 85% response) from 89 elementary schools in Melbourne (population 3.4 million), Australia. Children were recruited via a cross‐sectional cluster sample survey of elementary schools within urban and suburban Melbourne. A stratified random sample of schools was drawn to ensure proportional representation of grade 1 and grade 5 children in each of the three major school sectors (government, Catholic and independent). The study was approved by the Royal Children's Hospital's Ethics in Human Research Committee (EHRC approval number 22056).
A total of 6240 children underwent otoscopy, followed by screening audiometry under soundproof conditions. A pass result on screening audiometry required the child to respond correctly to two of three presentations in each ear at seven pure‐tone frequencies (0.5, 1, 2, 3, 4, 6 and 8 kHz) presented at 15 dB hearing level (HL). Children who did not meet the pass criteria for screening completed a full audiometric evaluation consisting of pure tone air‐conduction and bone‐conduction threshold tests, tympanometry and acoustic reflex tests.
The air‐conduction pure tone thresholds at 0.5, 1 and 2 kHz were averaged to indicate the low pure tone average and the thresholds at 3, 4 and 6 kHz were averaged to indicate the high pure tone average, classified by severity according to Niskar et al.3 Slight or mild SNHL was defined as low pure tone average or high pure tone average of 16–40 dB HL in the better ear, with the difference between air‐conduction and bone‐conduction thresholds (air–bone gap) <10 dB. Normal hearing was defined as low pure tone average and high pure tone average ⩽15 dB HL in both ears.
Parents of children who met study criteria for slight or mild SNHL (cases) were invited to have their child participating in follow‐up assessments. These children were then matched to two normally hearing children (controls) on age, sex, grade level and school; all cases and controls were asked to provide genetic samples. Exclusion criteria were known intellectual disability or major medical disorder; a known syndromic cause for hearing loss; or hearing loss due solely to otitis media with effusion. As part of the study parents reported their own country of birth. Australian families are not typically asked to identify themselves by race, therefore we defined a child's race on the basis of the country of birth parents reported, according to the National Institutes of Health criteria (National Institutes of Health policy on reporting race and ethnicity data). In line with known immigration patterns over the past century, parents who reported that they were born in Australia were classified as white.
DNA analyses
Buccal cells were collected on Buccal Swab Brushes (Epicentre, Madison, Wisconsin, USA) and DNA isolated using the MasterAmp Buccal Swab DNA Extraction Kit (Epicentre).
The coding region of the GJB2 gene (exon 2) was sequenced, followed by nested polymerase chain reaction (PCR) amplification of approximately 10 ng buccal cell DNA using HotStarTaq DNA Polymerase (Qiagen Pty Ltd, Doncaster, Victoria, Australia). The primers in the first 25‐μl PCR reaction were Cx26‐1F (5′‐TTGGTGTTTGCTCAGGAAGA) and Cx26Dx‐1R (5′‐GGCCTACAGGGGTTTCAAAT). The second PCR was carried out using 1 μl of the first PCR reaction and primers Cx26Dx‐1F (5′‐TGCTTGCTTACCCAGACTCA) and Cx26Dx‐4R (5′‐AGCTGAGCACGGGTTGCCTCAT). PCR conditions were 95°C for 15 min, then 35 cycles of 95°C for 33 s, 55°C for 33 s and 72°C for 100 s, followed by 72°C for 10 min.
The PCR reactions were treated with ExoSAP‐IT (USB Corporation, Cleveland Ohio, USA) and sequenced using the DYEnamic ET Terminator Cycle Sequencing Protecol (GE Healthcare, Castle Hill, New South Wales, Australia). Sequencing primers were Cx26Dx‐1F, Cx26Dx‐4R, Cx26‐30F (5′‐CTGCAGCTGATCTTCGTGTC) and Cx26‐30R (5′‐GAAGATGACCCGGAAGAAGAT) and the cycling conditions were 35 cycles of 95°C for 20 s, 50°C for 15 s and 60°C for 120 s.
All samples were analysed for the presence of the GJB2 IVS1+1 G→A splice site mutation. Approximately 10 ng buccal cell DNA were PCR amplified using primers Cx26_Ex2F (5′‐TCGGCGGCGCCCGGCCCAGGATCCGCCTAG) and Cx26_Ex2F (5′‐TGGCCGGGCAGTCCGGGGCCGGCGGGCTGA), and HotStarTaq DNA Polymerase. PCR conditions were 95°C for 15 min, then 35 cycles of 95°C for 33 s, 55°C for 33 s and 72°C for 100 s, followed by 72°C for 10 min. The resulting 138 bp fragment was digested with restriction endonuclease MboI (Roche, Sydney, New South Wales, Australia) and analysed on a 3% NuSieve agarose gel. The normal sequence resulted in 117 and 21 bp bands. Presence of the Cx26 IVS1+1 G→A splice site mutation results in bands of 87, 30 and 21 bp.
We also tested for the two GJB6/connexin 30 deletions del(GJB6‐D13S1830) and del(GJB6‐D13S1854), shown on occasion to be associated with GJB2‐related deafness. The PCR‐based tests were carried out as described previously.12
Single‐nucleotide polymorphism analysis
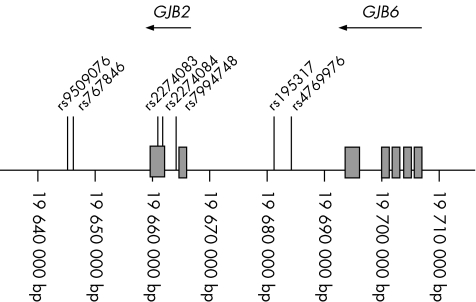
The following single‐nucleotide polymorphisms (SNPs) in or near the coding region of the GJB2 gene (position 18561721–18561040 bp on chromosome 13) were analysed by DNA sequencing: rs9509076 (position 18545640 bp), rs767846 (position 18549205 bp), rs2274083 (position 18561380 bp), rs2274084 (position 18561642 bp), rs7994748 (position 18564130 bp), rs1953517 (position 18581238 bp) and rs4769976 (position 18584298 bp) (fig 1). Allele distribution between the four children homozygotic for the V37I mutation and 15 children of Asian ethnicity with no V37I mutation was evaluated using χ2 and Fisher's exact tests.
Figure 1 Location of single‐nucleotide polymorphism, GJB2 and GJB6 on human chromosome 13q11–q12.
Results
In all, 55 children (0.88%, 95% confidence interval (CI) 0.66 to 1.14) had slight or mild SNHL, of whom 18 were in grade 1 (prevalence 0.53%, 95% CI 0.32 to 0.84) and 37 were in grade 5 (prevalence 1.15%, 95% CI 0.81 to 1.58). The children identified with slight or mild SNHL had air–bone gaps <10 dB, and tympanometry and acoustic reflex test results consistent with normal middle‐ear function. In 48 of the 55 (87%) cases consent was obtained for further investigations including genetic testing. These cases were matched with 96 controls, 90 of whose parents consented to the genetic component of the study. Buccal cell samples were therefore obtained from 48 cases and 90 controls. Table 1 shows the sample characteristic.
Table 1 Characteristics of the sample.
| Slight/mild SNHL | n | Controls | n | |
|---|---|---|---|---|
| Age in years (mean (SD)) | ||||
| Grade 1 | 7.21 (0.45) | 16 | 7.19 (0.46) | 30 |
| Grade 5 | 11.06 (0.46) | 30 | 11.06 (0.49) | 57 |
| Male sex (n (%)) | ||||
| Grade 1 | 9 (50) | 18 | 16 (51.6) | 31 |
| Grade 5 | 17 (56.7) | 30 | 32 (54.2) | 59 |
| Race (n (%)) | 48 | 90 | ||
| Asian | 8 (16.7) | 9 (10.0) | ||
| Black | 0 (0) | 1 (1.1) | ||
| White | 30 (62.5) | 62 (68.9) | ||
| More than one race | 9 (18.8) | 17 (18.9) | ||
| Not reported | 1 (2.1) | 1 (1.1) |
SNHL, sensorineural hearing loss.
GJB2 and GJB6 gene analyses
Known polymorphisms and synonymous nucleotide changes were detected in three cases and five controls (table 2). We found single amino acid changes or deletions in nine children (seven controls and two cases, table 3). The 235delC and the 35delG changes are known GJB2 mutations. The E114G+V27I, V37I, M34T, F191L, V153I and R127H changes have all been described before, but it has not been unequivocally established if they are causative mutations or polymorphisms. The GJB2 IVS1+1 G→A splice site mutation was not found in any of the children.
Table 2 Known polymorphisms and synonymous nucleotide changes identified in GJB2 genes.
| Child ID | Case/control | Change |
|---|---|---|
| H‐8 | Case | V27I |
| H‐50 | Control | G→A at nt 243 |
| H‐97 | Control | C→T at nt 496 |
| H‐101 | Case | C→T at nt 96 |
| H‐104 | Control | Homozygotic G→A at nt 285 |
| H‐112 | Control | Homozygotic G→A at nt 357 |
| H‐127 | Control | Homozygotic V27I |
| H‐132 | Case | T→C at nt 633 |
Table 3 Single amino acid changes or deletions identified in GJB2 genes.
| Child ID | Case/control status | Amino acid change |
|---|---|---|
| H‐43 | Control | F191L |
| H‐44 | Control | E114G+V27I |
| H‐58 | Case | V37I |
| H‐78 | Case | V37I |
| H‐80 | Control | 235delC |
| H‐100 | Control | V153I |
| H‐111 | Control | M34T |
| H‐126 | Control | R127H |
| H‐143 | Control | 35delG |
Two GJB2 changes were found in five children (one control and four cases, table 4). All four cases with slight or mild SNHL were homozygotic for the V37I change and of Asian background. Two were from the grade 1 group and two from the grade 5 group. One child with normal hearing was homozygotic for the R127H change.
Table 4 Amino acid changes identified in children with two GJB2 changes.
| Child ID | Case/control status | Amino acid changes |
|---|---|---|
| H‐56 | Case | V37I / V37I |
| H‐91 | Case | V37I / V37I |
| H‐93 | Case | V37I / V37I |
| H‐134 | Control | R127H / R127H |
| H‐136 | Case | V37I / V37I |
The del(GJB6‐D13S1854) deletion was not detected in any of the children. The GJB6 deletion del(GJB6‐D13S1830) was seen in two controls (H‐117 and H‐121) and one child (H‐124) with slight or mild SNHL. In none of these three children did we find a GJB2 mutation.
Hearing profiles of children with homozygotic V37I mutations
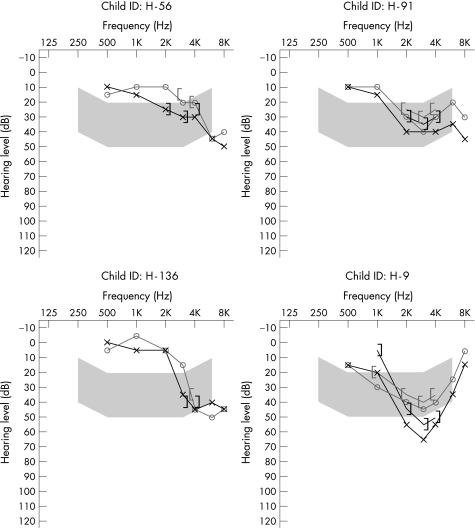
Figure 2 presents the audiograms for the four homozygotic V37I cases. All show a bilateral high‐frequency SNHL involving the 3–6 kHz frequency range, and for three cases lower frequencies have also been affected. One of the children heterozygotic for the V37I mutation had a mild bilateral high‐frequency loss maximal at 6 kHz. The other had a bilateral low‐frequency loss, with normal hearing >2 kHz. Of the 42 cases with no V37I mutation, only two children exhibited a high frequency hearing loss similar to the four homozygotic cases. The remaining 40 audiograms were typically flat or gradually sloping and involving several frequencies.
Figure 2 Audiograms of the four children homozygotic for the V37I mutation.
SNP analysis
SNPs in or near the GJB2 gene were analysed for disequilibrium in the four children homozygotic for the V37I mutation and 15 children of Asian ethnicity with no V371 mutation (table 5). The G allele in rs7994748 and the A allele in rs1953517 are considerably more frequent in chromosomes with the V37I mutation than in control chromosomes.
Table 5 Distribution of single‐nucleotide polymorphism alleles in four children homozygotic for the V37I mutation and 15 controls of Asian background.
| SNP | Allele | V37I chromosomes | Control chromosomes | p Value* |
|---|---|---|---|---|
| rs9509076 | A | 1/8 | 11/30 | 0.19 |
| G | 7/8 | 19/30 | ||
| rs767846 | C | 2/8 | 14/30 | 0.27 |
| T | 6/8 | 16/30 | ||
| rs2274083 | A | 8/8 | 27/30 | 0.48 |
| G | 0/8 | 3/30 | ||
| rs2274084 | A | 0/8 | 2/30 | 0.62 |
| G | 8/8 | 28/30 | ||
| rs7994748 | A | 0/8 | 22/30 | 0.0002 |
| G | 8/8 | 8/30 | ||
| rs1953517 | A | 8/8 | 9/30 | 0.0005 |
| T | 0/8 | 21/30 | ||
| rs4769976 | C | 1/8 | 6/30 | 0.53 |
| G | 7/8 | 24/30 |
SNP, single‐nucleotide polymorphism.
*Determined using Fisher's exact test.
Discussion
Biallelic mutations associated with GJB2 deafness were found in 8.3% (95% CI 2.32 to 19.98) of healthy Australian primary school children with slight or mild bilateral SNHL. However, biallelic V37I mutations are important in Asian children, with 50% (95% CI 15.70 to 84.30) of the Asian children with slight or mild SNHL in our study being homozygotic for this mutation. We found no indication that known recessive GJB2 mutations associated with more severe SNHL cause a slight or mild hearing loss in carrier children.
This is the largest population study yet to have systematically examined genetic contributions to slight or mild bilateral SNHL. Strengths of the study include the large population‐based sample, high response rates and the careful ascertainment of slight or mild SNHL, all of which minimise bias in the study's findings. However, because the prevalence of slight or mild SNHL was lower than expected, the size of the case group from which to draw conclusions about genetic contributions to slight or mild SNHL was smaller than expected.
All four children with slight or mild SNHL and two biallelic GJB2 changes were homozygotic for the V37I change, which is known to have a high prevalence in people of Asian ethnicity.13,14 This change has previously been reported to be associated with SNHL in the range of 20–90 dB HL,13,15,16 but other data have questioned this conclusion.14 Wattanasirichaigoon et al14 speculated that V37I might be a polymorphism or that people with slight or mild hearing loss were under‐represented in their study owing to selection biases. If V37I is a polymorphism not related to the SNHL but homozygotic in four of eight Asian children with slight or mild SNHL, we can estimate that approximately 70% of all GJB2 alleles in our Asian population should have the V37I change. However, we did not find a single V37I allele in the nine Asian children (18 alleles) in the control group. Our results therefore suggest that V37I is a mutation and that children homozygotic for the V37I mutation usually have slight or mild hearing loss, which is rarely detected. Although the carrier frequency of the V37I mutation in selected European and American populations is <3%,17,20 studies have shown that the carrier frequency in certain Asian populations can be as high as 17%.14 Slight or mild hearing loss is rarely detected, and the connexin 26 gene is therefore not analysed in these children. Only when the V37I mutation is present in children with a more severe hearing loss is it normally noted. This has led to a probably huge underestimation of its contribution to deafness and controversy whether or not it is a harmless polymorphism or causative mutation. The data presented in this paper provide the first indication of the real contribution of the V37I mutation to milder forms of hearing loss.
Do homozygotic V37I mutations usually or always cause hearing loss? Children of Asian ancestry made up 10% of the controls, selected for the study independent of race. Of all the children who underwent screening audiometry and returned a survey, 9.8% were reported to be of Asian ancestry on the hearing survey. To have four homozygotic V37I children with slight or mild SNHL among the approximately 660 Asian children, we calculate the V37I allele prevalence in this group to be about 16%. If homozygotic V37I mutations were commonly associated with normal hearing, then the allele prevalence in the group would be >16%. We did not detect a V37I carrier in the nine Asian controls, supporting our conclusion that homozygotic V37I mutations always or nearly always cause SNHL.
Three of the four affected V37I homozygotes shared a similar audiological profile (bilateral, high frequency at frequencies <3 kHz). Only 2 of the 42 cases without V37I mutation presented with similar audiological profiles.
Six single GJB2 changes were found in five controls. The E114G+V27I, M34T, F191L, V153I and R127H changes have all been described before, but it has not been established if they are mutations or polymorphisms. In fact, the association of GJB2 changes V37I, M34T and R127H to inherited SNHL deafness remains controversial.13,17,18,19,20 Our finding of a normally hearing child homozygotic for the R127H change supports the conclusion that it is a polymorphism. We conclude that as few as 2 (2%) and as many as 7 (8%) of the normally hearing children could be carriers of a GJB2 mutation.
Two of the 90 control children were carriers of the known mutations 35delG and 235delC. This is within the expected range, as the 35delG carrier frequency in the Australian population has previously been estimated to be 1:100 and the overall prevalence of carriers of connexin 26 mutations to be 1:50.10 The V37I amino acid substitution was found as a single change in two cases (one Asian and one “more than one race”). No other GJB2 or GJB6 mutations were detected in these two children. A previous study investigated 52 Australian children with GJB2 mutations and a moderate, severe or profound SNHL. Although two biallelic GJB2 mutations were found in 34 of these children, 18 (35%) had a detectable mutation in only one allele.10 This number is too high to represent carriers of GJB2 mutations with another cause of deafness. Therefore, it was concluded that the GJB2 mutations in many of these 18 children might be associated with the hearing loss. Whether or not the single V37I change contributes to the slight or mild hearing loss in the two cases in this study is therefore not clear. Functional studies have shown that the M34T and V37I mutations, even in the presence of wild‐type connexin 26, reduce gap junction activity.21,22,23 In support of a causal role, we note that the audiogram of one of the children has a sloping high‐frequency loss similar to that of children H‐56, H‐91 and H‐136 (fig 1) homozygotic for the V37I mutation.
We tested the hearing in approximately 6000 children of Caucasian background and none had a slight or mild SNHL due to GJB2 mutations. This suggests that the prevalence of Caucasian carriers of GJB2 mutations resulting in slight or mild SNHL is <1:40 and therefore not markedly more common than GJB2 mutations normally associated with more severe forms of SNHL (1:50) in the Australian population.
More than 80 different GJB2 mutations have been identified (Connexins and Deafness Homepage). Nevertheless, 35delG, 167delT, 235delC and V37I account for >50% of mutant alleles in Caucasians, Ashkenazi Jews and Asians, respectively. Our analysis of disequilibrium of SNPs in or near the GJB2 gene suggests that the V37I mutation could have arisen from a single mutational event in the Asian population. The founder chromosome would have had the haplotype G–T–A–G–G–A–G (rs9509076–rs767846–rs2274083–rs2274084–rs7994748–rs1953517–rs4769976). Similar findings have been reported for other common GJB2 mutations.24,25,26,27,28 It is not known if the GJB2 mutations have become so prevalent owing to assortative mating, relaxed selection, linguistic homogamy, selective advantage or a combination of these factors.29 In support of selective advantage as the reason for the high prevalence of V37I in Asians, it has been suggested that carriers of connexin 26 mutations have a selective advantage due to epidermal thickening30 or increased cell survival.31 Further, whereas 35delG, 167delT and 235delC normally are associated with more severe forms of deafness, V37I is in most cases associated with a subclinical form of hearing loss and therefore not likely to influence choice of partner.
We conclude that the prevalence of slight or mild SNHL in Australian children is lower than previously reported, that mutations in the GJB2 gene is the underlying cause in about 8% of cases, and that the V37I change is a common cause of slight or mild SNHL in children of Asian ethnicity. Slight or mild SNHL is more common in children of Asian background (1.3%) than in Caucasian children (0.83%). Homozygotic GJB2 V37I mutations explain the prevalence gap. Further studies are needed to determine the exact prevalence and type of slight or mild SNHL caused by the V37I mutation.
Acknowledgements
We thank the contribution of the field workers responsible for data collection and also all the children, families and schools who took part in the study.
Electronic database information
The URLs for data presented herein are as follows:
Connexins and Deafness Homepage (Ballana E, Ventayol M, Rabionet R, Gasparini P, Estivill X), http://davinci.org.es/deafness/
Hereditary Hearing Loss Homepage (Van Camp G, Smith, RJH), http://webhost.ua.ac.be/hhh/
NIH Policy on Reporting Race and Ethnicity Data, http://grants.nih.gov/grants/funding/phs398/instructions2/p2_nih_policy_report_race_ethnicity.htm
Abbreviations
PCR - polymerase chain reaction
SNHL - sensorineural hearing loss
SNP - single‐nucleotide polymorphism
Footnotes
Funding: This study was supported by a Grant R01 DC 005662‐03 from the US National Institute of Deafness and Communication Disorders (NIDCD). H‐HMD is a NHMRC Principal Research Fellow. MW is supported by National Health and Medical Research Council (NHMRC) Career Development Award 284556.
Competing interests: None.
References
- 1.Lee D J, Gomez‐Marin O, Lee H M. Prevalence of childhood hearing loss. The Hispanic Health and Nutrition Examination Survey and the National Health and Nutrition Examination Survey II. Am J Epidemiol 1996144442–449. [DOI] [PubMed] [Google Scholar]
- 2.Bess F H, Dodd‐Murphy J, Parker R A. Children with minimal sensorineural hearing loss: prevalence, educational performance, and functional status. Ear Hear 199819339–354. [DOI] [PubMed] [Google Scholar]
- 3.Niskar A S, Kieszak S M, Holmes A, Esteban E, Rubin C, Brody D J. Prevalence of hearing loss among children 6 to 19 years of age: the Third National Health and Nutrition Examination Survey. JAMA 19982791071–1075. [DOI] [PubMed] [Google Scholar]
- 4.Fria T J, Cantekin E I, Eichler J A. Hearing acuity of children with otitis media with effusion. Arch Otolaryngol 198511110–16. [DOI] [PubMed] [Google Scholar]
- 5.Wolgemuth K S, Luttrell W E, Kamhi A G, Wark D J. The effectiveness of the Navy's Hearing Conservation Program. Mil Med 1995160219–222. [PubMed] [Google Scholar]
- 6.Roberts J, Hunter L, Gravel J, Rosenfeld R, Berman S, Haggard M, Hall J, Lannon C, Moore D, Vernon‐Feagans L, Wallace I. Otitis media, hearing loss, and language learning: controversies and current research. J Dev Behav Pediatr 200425110–122. [DOI] [PubMed] [Google Scholar]
- 7.Wake M, Tobin S, Cone‐Wesson B, Dahl H H M, Gillam L, McCormick L, Poulakis Z, Rickards F W, Saunders K, Ukoumunne O C, Williams J. Slight/mild sensorineural hearing loss children. Pediatrics. In press [DOI] [PubMed]
- 8.Petit C. Genes responsible for human hereditary deafness: symphony of a thousand. Nat Genet 199614385–391. [DOI] [PubMed] [Google Scholar]
- 9.Van Camp G, Willems P J, Smith R J. Nonsyndromic hearing impairment: unparalleled heterogeneity. Am J Hum Genet 199760758–764. [PMC free article] [PubMed] [Google Scholar]
- 10.Dahl H H, Saunders K, Kelly T M, Osborn A H, Wilcox S, Cone‐Wesson B, Wunderlich J L, Du Sart D, Kamarinos M, Gardner R J, Dennehy S, Williamson R, Vallance N, Mutton P. Prevalence and nature of connexin 26 mutations in children with non‐syndromic deafness. Med J Aust 2001175191–194. [DOI] [PubMed] [Google Scholar]
- 11.Snoeckx R L, Huygen P L, Feldmann D, Marlin S, Denoyelle F, Waligora J, Mueller‐Malesinska M, Pollak A, Ploski R, Murgia A, Orzan E, Castorina P, Ambrosetti U, Nowakowska‐Szyrwinska E, Bal J, Wiszniewski W, Janecke A R, Nekahm‐Heis D, Seeman P, Bendova O, Kenna M A, Frangulov A, Rehm H L, Tekin M, Incesulu A, Dahl H H, du Sart D, Jenkins L, Lucas D, Bitner‐Glindzicz M, Avraham K B, Brownstein Z, del Castillo I, Moreno F, Blin N, Pfister M, Sziklai I, Toth T, Kelley P M, Cohn E S, Van Maldergem L, Hilbert P, Roux A F, Mondain M, Hoefsloot L H, Cremers C W, Lopponen T, Lopponen H, Parving A, Gronskov K, Schrijver I, Roberson J, Gualandi F, Martini A, Lina‐Granade G, Pallares‐Ruiz N, Correia C, Fialho G, Cryns K, Hilgert N, Van de Heyning P, Nishimura C J, Smith R J, Van Camp G. GJB2 mutations and degree of hearing loss: a multicenter study. Am J Hum Genet 200577945–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.del Castillo F J, Rodriguez‐Ballesteros M, Alvarez A, Hutchin T, Leonardi E, de Oliveira C A, Azaiez H, Brownstein Z, Avenarius M R, Marlin S, Pandya A, Shahin H, Siemering K R, Weil D, Wuyts W, Aguirre L A, Martin Y, Moreno‐Pelayo M A, Villamar M, Avraham K B, Dahl H H, Kanaan M, Nance W E, Petit C, Smith R J, Van Camp G, Sartorato E L, Murgia A, Moreno F, del Castillo I. A novel deletion involving the connexin‐30 gene, del(GJB6‐d13s1854), found in trans with mutations in the GJB2 gene (connexin‐26) in subjects with DFNB1 non‐syndromic hearing impairment. J Med Genet 200542588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bason L, Dudley T, Lewis K, Shah U, Potsic W, Ferraris A, Fortina P, Rappaport E, Krantz I D. Homozygosity for the V37I Connexin 26 mutation in three unrelated children with sensorineural hearing loss. Clin Genet 200261459–464. [DOI] [PubMed] [Google Scholar]
- 14.Wattanasirichaigoon D, Limwongse C, Jariengprasert C, Yenchitsomanus P T, Tocharoenthanaphol C, Thongnoppakhun W, Thawil C, Charoenpipop D, Pho‐iam T, Thongpradit S, Duggal P. High prevalence of V37I genetic variant in the connexin‐26 (GJB2) gene among non‐syndromic hearing‐impaired and control Thai individuals. Clin Genet 200466452–460. [DOI] [PubMed] [Google Scholar]
- 15.Kenna M A, Wu B L, Cotanche D A, Korf B R, Rehm H L. Connexin 26 studies in patients with sensorineural hearing loss. Arch Otolaryngol Head Neck Surg 20011271037–1042. [DOI] [PubMed] [Google Scholar]
- 16.Cryns K, Orzan E, Murgia A, Huygen P L, Moreno F, del Castillo I, Chamberlin G P, Azaiez H, Prasad S, Cucci R A, Leonardi E, Snoeckx R L, Govaerts P J, Van de Heyning P H, Van de Heyning C M, Smith R J, Van Camp G. A genotype‐phenotype correlation for GJB2 (connexin 26) deafness. J Med Genet 200441147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelley P M, Harris D J, Comer B C, Askew J W, Fowler T, Smith S D, Kimberling W J. Novel mutations in the connexin 26 gene (GJB2) that cause autosomal recessive (DFNB1) hearing loss. Am J Hum Genet 199862792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Houseman M J, Ellis L A, Pagnamenta A, Di W L, Rickard S, Osborn A H, Dahl H H, Taylor G R, Bitner‐Glindzicz M, Reardon W, Mueller R F, Kelsell D P. Genetic analysis of the connexin‐26 M34T variant: identification of genotype M34T/M34T segregating with mild‐moderate non‐syndromic sensorineural hearing loss. J Med Genet 20013820–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feldmann D, Denoyelle F, Loundon N, Weil D, Garabedian E N, Couderc R, Joannard A, Schmerber S, Delobel B, Leman J, Journel H, Catros H, Ferrec C, Drouin‐Garraud V, Obstoy M F, Moati L, Petit C, Marlin S. Clinical evidence of the nonpathogenic nature of the M34T variant in the connexin 26 gene. Eur J Hum Genet 200412279–284. [DOI] [PubMed] [Google Scholar]
- 20.Roux A F, Pallares‐Ruiz N, Vielle A, Faugere V, Templin C, Leprevost D, Artieres F, Lina G, Molinari N, Blanchet P, Mondain M, Claustres M. Molecular epidemiology of DFNB1 deafness in France. BMC Med Genet 200455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White T W, Deans M R, Kelsell D P, Paul D L. Connexin mutations in deafness (letter). Nature 1998394630–631. [DOI] [PubMed] [Google Scholar]
- 22.Martin P E, Coleman S L, Casalotti S O, Forge A, Evans W H. Properties of connexin26 gap junctional proteins derived from mutations associated with non‐syndromal heriditary deafness. Hum Mol Genet 199982369–2376. [DOI] [PubMed] [Google Scholar]
- 23.Bruzzone R, Veronesi V, Gomes D, Bicego M, Duval N, Marlin S, Petit C, D'Andrea P, White T W. Loss‐of‐function and residual channel activity of connexin26 mutations associated with non‐syndromic deafness. FEBS Lett 200353379–88. [DOI] [PubMed] [Google Scholar]
- 24.Lerer I, Sagi M, Malamud E, Levi H, Raas‐Rothschild A, Abeliovich D. Contribution of connexin 26 mutations to nonsyndromic deafness in Ashkenazi patients and the variable phenotypic effect of the mutation 167delT. Am J Med Genet 20009553–56. [DOI] [PubMed] [Google Scholar]
- 25.Van Laer L, Coucke P, Mueller R F, Caethoven G, Flothmann K, Prasad S D, Chamberlin G P, Houseman M, Taylor G R, Van de Heyning C M, Fransen E, Rowland J, Cucci R A, Smith R J, Van Camp G. A common founder for the 35delG GJB2 gene mutation in connexin 26 hearing impairment. J Med Genet 200138515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan D, Park H J, Ouyang X M, Pandya A, Doi K, Erdenetungalag R, Du L L, Matsushiro N, Nance W E, Griffith A J, Liu X Z. Evidence of a founder effect for the 235delC mutation of GJB2 (connexin 26) in east Asians. Hum Genet 200311444–50. [DOI] [PubMed] [Google Scholar]
- 27.Rothrock C R, Murgia A, Sartorato E L, Leonardi E, Wei S, Lebeis S L, Yu L E, Elfenbein J L, Fisher R A, Friderici K H. Connexin 26 35delG does not represent a mutational hotspot. Hum Genet 200311318–23. [DOI] [PubMed] [Google Scholar]
- 28.Ben‐Yosef T, Friedman T B. The genetic bases for syndromic and nonsyndromic deafness among Jews. Trends Mol Med 20039496–502. [DOI] [PubMed] [Google Scholar]
- 29.Nance W E, Kearsey M J. Relevance of connexin deafness (DFNB1) to human evolution. Am J Hum Genet 2004741081–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer C G, Amedofu G K, Brandner J M, Pohland D, Timmann C, Horstmann R D. Selection for deafness? Nat Med 200281332–1333. [DOI] [PubMed] [Google Scholar]
- 31.Common J E, Di W L, Davies D, Kelsell D P. Further evidence for heterozygote advantage of GJB2 deafness mutations: a link with cell survival. J Med Genet 200441573–575. [DOI] [PMC free article] [PubMed] [Google Scholar]