Abstract
Background
Mutations in exonic splicing enhancer sequences are known to cause splicing errors. Although exonic splicing enhancers have been identified as a stretch of purine‐rich sequences, it has been difficult to precisely pinpoint the determinant nucleotides in these sequences. This article reports that a 4‐bp deletion in exon 38 of the dystrophin gene induced complete exon 38 skipping in vivo. Moreover, the third nucleotide of the deletion was shown to be determinant for the exonic splicing enhancer activity in in vivo splicing analysis of hybrid minigenes encoding mutant exons.
Method
Genomic DNA analysis of a 2‐year‐old boy with a raised level of serum creatine kinase yielded a 4‐bp deletion 11 bp upstream of the 3′ end of exon 38 of the dystrophin gene (c. 5434–5437del TTCA), disrupting a predicted SC35‐binding site.
Result
Interestingly, his dystrophin mRNA was shown to completely lack exon 38 (exon 38− transcript). As the exon 38− transcript coded for a truncated dystrophin protein, this exon skipping was determined to be a modifying factor of his phenotype. In an in vivo splicing assay, a hybrid minigene encoding exon 38 with the 4‐bp deletion was shown to induce complete exon 38 skipping, confirming the deleted region as a splicing enhancer sequence. Site‐directed mutagenesis of the deleted sequence showed that the complete exon 38 skipping was caused by mutation of the third nucleotide position of the deletion (C5436), whereas mutations at the other three nucleotide positions induced partial exon skipping.
Conclusion
Our results underline the potential of understanding the regulation of exonic splicing enhancer sequences and exon skipping therapy for treatment of Duchenne's muscular dystrophy.
For eukaryotic splicing of pre‐mRNA transcripts, a process that removes introns from pre‐mRNA, splice sites are defined by splicing consensus sequences located near intron–exon boundaries. Mutation of splicing consensus sequences can result in exon skipping or the activation of cryptic splice sites, thereby producing aberrant mRNAs. Recently, exonic splicing enhancers were shown to be required for proper splicing.1 Mutations in these sequences were shown to result in aberrantly spliced transcripts, and are currently receiving a great deal of attention as modifiers of genetic mutations.2
Dystrophinopathy, which is caused by mutations in the dystrophin gene, is the most common inherited myopathy, affecting approximately one in 3500 men. This genetic condition shows varying degrees of severity, ranging from the severe Duchenne's muscular dystrophy (DMD) to the milder Becker's muscular dystrophy (BMD). DMD is a rapidly progressive disease that is first recognised during childhood; children with DMD commonly lose their ability to walk before they turn 12 years old. BMD has a slower rate of progression; those with BMD remain ambulatory beyond the age of 16 years and may lead near‐normal lives. In as many as 60% of patients with dystrophinopathy, a partial deletion or duplication of the dystrophin gene can be detected. According to the reading‐frame rule, deletions or duplications that create a shift in the reading frame of dystrophin mRNA lead to the more severe DMD phenotype, whereas the milder BMD phenotype occurs if the open reading frame is preserved after the deletion or duplication.3 Exceptions to the rule, however, have been reported, and the production of aberrant transcripts is considered to be a key modifier of the clinical phenotype of dystrophinopathy.4,5,6,7,8,9,10
The complex structure of the dystrophin gene, which is characterised by a large number of exons (79), extremely large introns (up to 200 kb) and tissue‐specific alternative splicing, suggests an indispensable role for exonic splicing enhancer sequences in normal dystrophin pre‐mRNA splicing. Analyses of splicing errors in cases of dystrophinopathy with exonic mutations in the dystrophin gene first identified splicing enhancer sequences in exon 19.11,12 So far, splicing enhancer sequences, however, have been found only in a limited number of dystrophin gene exons.13,14
In this study, we report that a novel 4‐bp deletion in exon 38 of the dystrophin gene leads to complete exon 38 skipping. Results of an in vivo splicing assay disclosed that this 4‐bp deletion disrupted an exonic splicing enhancer, and that a single nucleotide in the deletion is determinant for proper exon 38 splicing, whereas the other three nucleotides are less essential.
Case and methods
Case
A 2‐year‐old boy (KUHPCG414) was referred to the Kobe University Hospital, Japan, for a genetic diagnosis of his raised serum creatine kinase level. There was no family history of neuromuscular disease. As an infant, he started to crawl at age 6 months and began to walk at age 1.5 years. By chance, his serum creatine kinase level was found to be 3805 IU/l (normal <169 IU/l), although he did not show any muscle weakness. No abnormalities were detected on either radiographs or electrocardiograms. Laboratory investigation of blood yielded a raised level of serum creatine kinase (8642 IU/l). Subsequent analyses showed a wide variation in the serum creatine kinase level, which ranged from 1074 to 10 136 IU/l, leading to a tentative diagnosis of BMD. Consent for this study was obtained from the patient's parents. The ethics committee of the Kobe University Graduate School of Medicine, Kobe, Japan, approved the study.
Mutational analysis
For mutational analysis of the dystrophin gene, genomic DNA was isolated from the lymphocytes of this index case and a control patient using a Wizard genomic DNA extraction kit (Promega Corporation, Madison, Wisconsin, USA). Nineteen deletion‐prone exons were amplified by polymerase chain reaction (PCR) to look for a deletion mutation.15,16 Southern blot analysis was carried out using HindIII restriction‐enzyme‐digested DNA as a template and a dystrophin cDNA fragment as a probe. This technique allows for the full extent of any deletions or duplications to be determined. To look for small mutations, all 79 exons and the flanking introns were amplified17 and each amplified product was directly sequenced using a BigDye terminator v1.1 cycle sequencing kit (Applied Biosystems, Foster City, California, USA) with an automatic DNA sequencer (ABI PRISM 310; Applied Biosystems).
The dystrophin mRNA expressed in lymphocytes was examined by nested PCR. Total RNA was extracted from peripheral lymphocytes and cDNA was synthesised as described previously.11 A fragment extending from exon 36 to exon 41 was amplified using two sets of primers: an inner set with a forward primer corresponding to a segment of exon 36 (3E, 5′‐TTT GAC CAG AAT GTG GAC CA‐3′) and a reverse primer complementary to a segment of exon 41 (c41r, 5′‐TGC GGC CCC ATC CTC AGA CAA‐3′), and an outer set with a forward primer (3A, 5′‐GCT TGA ACA GAG CAT CCA GTC‐3′) and a reverse primer (3B, 5′‐ACT GGC ATC TGT TTT TGA GGA T‐3′). The PCR‐amplified product was electrophoresed on an agarose gel. The purified PCR product was sequenced either directly or after it was subcloned into vector pT7 Blue (Novagen, Madison, Wisconsin, USA).
In vivo splicing assay
Hybrid minigenes
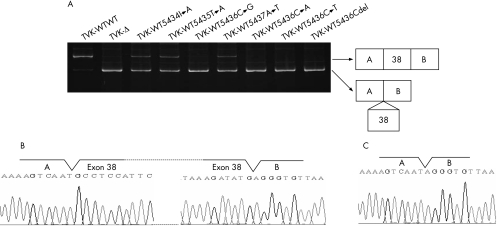
To study the splicing pattern, a minigene vector (H492) that contained two exons (exons A and B) and one intervening intron encoding a multicloning site was constructed from the pcDNA 3.0 mammalian expression vector (Invitrogen, Carlsbad, California, USA).18 A hybrid minigene was created by inserting a test sequence consisting of exon 38 and its flanking introns into the multicloning site (fig 1). The fragment encompassing exon 38 was amplified from both the control sample and the patient's genomic DNA by the PCR with primers that corresponded to introns 37 and 38, and included recognition sites for restriction enzymes NheI and BamHI (In37F‐Nhe, 5′‐GCC GCT AGC GAT TAG TTT AGC AAC AGG AGG‐3′ and In38R‐Bam, 5′‐CGG GAT CCG TGC TCT GAA AAT TCA GTT GGA G‐3′). Amplified products were digested with NheI and BamHI (New England Biolabs, Hertfordshire, UK), and inserted into the minigene that had been digested with the same restriction enzymes. In this way, both wild‐type (TVK‐WT) and mutant (TVK‐Δ) hybrid minigenes, which carried wild‐type exon 38 and exon 38 with the 4‐bp deletion, respectively, were constructed. Site‐specific mutagenesis by overlap extension was carried out to introduce mutations by PCR‐based mutagenesis into the TVK‐WT minigene.19 The four nucleotides at positions 5434, 5435, 5436 and 5437, which were missing in the deletion construct, were each replaced with their complementary nucleotides to make the TVK‐WT5434T→A, TVK‐WT5435T→A, TVK‐WT5436C→G, and TVK‐WT5437A→T minigenes. Furthermore, additional mutations were introduced into position 5436 to yield the TVK‐WT5436C→A, TVK‐WT5436C→T and TVK‐WT5436Cdel minigenes (fig 1). After checking their sequences, these hybrid minigenes were transfected into HeLa cells for a splicing assay.
Figure 1 In vivo splicing analysis. (A) Schematic representation of the hybrid minigene. A minigene vector (H492) was constructed to encode two cassette exons (A and B) and an intervening sequence containing a multicloning site. The polymerase chain reaction (PCR)‐amplified regions encompassing exon 38 were inserted into the multicloning site after digestion with NheI and BamHI. The minigene vector contained a cytomegalovirus enhancer–promoter and a polyadenylation signal (BGH; dark shaded boxes) for complete synthesis of mRNA. Box and lines indicate exon and its flanking introns, respectively. The primers used in the reverse transcriptase (RT)‐PCR assay are represented by arrows. (B) Sequences of the 3′ ends of exon 38 in the hybrid minigenes. TVK‐WT and TVK‐Δ denote the sequences of wild‐type exon 38 and exon 38 with the 4‐bp deletion, respectively. Single nucleotide changes were introduced into TVK‐WT by mutagenesis to produce seven other hybrid minigenes. Boxes highlight the mutated nucleotides and open boxes indicate deletions. Bold letters correspond to the deleted nucleotides identified in the index case.
Transfection
HeLa cells were grown in six‐well plates to approximately 70% confluency in Dulbecco's modified Eagle's medium containing 5% fetal bovine serum (Trace Biosciences, Castle Hill, Australia) at 37°C under 5% carbon dioxide. Hybrid minigenes (1.5 μg each) were transfected into the cells using Plus Reagent and Lipofectamine (Invitrogen) according to the manufacturer's protocol. Cells were harvested 24 h after transfection and total RNA was extracted using ISOGENE (Nippon Gene, Toyama, Japan).
Analysis of splicing products
Five μg of total RNA was subjected to reverse transcription (RT) using random hexamer primers as described previously.18 PCR was carried out using a forward primer corresponding to a segment of the upstream exon A (YH303, 5′‐GGT ACC ACA GCT GGA TTA CTC GCT C‐3′) and a reverse primer complementary to a segment of the downstream exon B (YH304, 5′‐CTC GAG CAG CCA GTT AAG TCT CTC AC‐3′; fig 1). Amplification was carried out in a total volume of 20 µl containing 4 µl cDNA, 2 µl 10× Ex Taq buffer, 2 µl 2.5 mM deoxyribonucleotide triphosphates, 10 pmol of each primer and 1 U Ex Taq Polymerase (Takara Bio, Kyoto, Japan). The PCR cycling conditions were as follows: an initial denaturation at 94°C for 2 min, 30 cycles of denaturation at 94°C for 1 min, annealing at 58°C for 1 min, extension at 72°C for 2 min and a final extension at 72°C for 5 min. PCR products were analysed by electrophoresis on an 8% polyacrylamide gel.
To identify the differences between the splicing products, semiquantitative PCR was used. PCR was carried out as described earlier, except that the number of PCR cycles was reduced from 30 to 18. To quantify the amplified products, 1 µl of each reaction mixture mixed with 5 µl of the loading buffer solution containing size markers (15 and 1500 bp) was analysed by capillary electrophoresis (Agilent 2100 Bioanalyzer with DNA 1000 LabChip; Agilent Technologies, Palo Alto, California, USA). The amount of each product was quantified by measuring fluorescence of the peak areas (Agilent 2100 Bioanalyzer).
Analysis of the exonic splicing enhancer
Two programs designed to find exonic splicing enhancers were used to examine exon 38: (1) ESE finder and (2) RESCUE‐ESE. ESEfinder (http://rulai.cshl.edu/ESE/) is a web‐based resource that facilitates rapid analysis of exon sequences to identify binding motifs for four serine/arginine‐rich (SR) proteins: SF2/ASF, SC35, SRp40 and SRp55.20 The RESCUE‐ESE program (http://genes.mit.edu/burgelab/rescue‐ese/) uses a computational method that identifies exonic splicing enhancers in human genomic sequences by searching for specific hexanucleotides.21
Results
The dystrophin gene of an index patient who had been clinically diagnosed with dystrophinopathy was analysed for mutations. Conventional mutational analysis of deletion‐prone exons by PCR amplification and Southern blotting did not disclose any mutations. Next, all 79 exons of the dystrophin gene were examined by direct sequencing of PCR‐amplified products. Although two single‐nucleotide changes were found (ca 7096C→A in exon 48 and ca 9804A→G in exon 67), these changes were variants leading to an amino acid change.22 In the amplified region encompassing exon 38, four nucleotides (TTCA) 11 bp upstream and 108 bp downstream from the 3′ and 5′ ends of exon 38, respectively, were found to be absent (ca 5434–5437del TTCA; fig 2A). We did not find this deletion in more than 100 other patients with DMD or those with BMD, and this deletion has not been previously reported in the literature. This novel deletion shifted the translational reading frame of the dystrophin mRNA, which resulted in a premature stop codon after the fifth amino acid residue of the wild‐type protein. A deletion leading to a translational frame shift suggested a severe DMD phenotype.
Figure 2 Analysis of the dystrophin gene and mRNA. (A) Genomic DNA analysis. Sequences of the 3′ end of exon 38 and the 5′ end of intron 38 of the control (upper panel) and the patient (lower panel) are shown. Four nucleotides (TTCA; boxed) located 11 bp upstream from the 3′ end of exon 38 were missing from the patient. The nucleotide numbers correspond to the position in the wild‐type dystrophin cDNA. The 4‐bp deletion is predicted to introduce a stop codon in the sixth codon after the deletion. The deletion does not change Shapiro's probability score for splicing. The sequence AAAGA is found on both sides of the deletion (underlined). A predicted SC35‐binding site (GACTTCAA) is marked by asterisks. (B) Schematic representation of the dystrophin exon 38 and the flanking exons. Splicing events in the control (dotted lines) and the patient (solid lines) are indicated. The arrowhead indicates the position of the four‐nucleotide deletion. The figure is not drawn to scale. (C) The dystrophin mRNA. Dystrophin cDNA from exon 36 to 41 was amplified by polymerase chain reaction (PCR). Although the control (C) and the patient (P) both produced one PCR product, the size of the products clearly differed. The patient produced a smaller PCR product, and sequencing of this product showed that exon 38 was missing. On the right, the exon structure of each band is schematically presented. Boxes and the numbers in the boxes indicate the exon and exon number, respectively. The sequence at the junction of the exons is shown.
Despite the genetic diagnosis of DMD for the patient, his serum creatine kinase level was not constantly raised (the lowest measured value was 1074 IU/l), a phenotype that did not completely match with that of DMD. To examine this discrepancy, the peripheral lymphocytes of the patient were examined for the production of aberrant dystrophin mRNA transcripts. The region spanning from exon 36 to 41 was amplified by a nested PCR. Interestingly, the amplified product from the index case was found to be smaller than the expected product (fig 2C), suggesting that an aberrant splicing product was being amplified. In fact, sequencing the product disclosed that the dystrophin mRNA no longer contained the 123‐bp exon 38 (an exon 38− transcript). Subcloning and sequencing of the PCR‐amplified product did not produce any clone that retained exon 38. We concluded that the ca 5434–5437delTTCA genomic deletion induced complete exon 38 skipping in vivo. The exon 38− transcript was expected to produce a truncated but semifunctional dystrophin protein, thereby resulting in the observed mild phenotype.
As in other dystrophinopathies,23 the exon 38− transcript was expected to be produced by disruption of one of the strictly conserved splicing consensus sequences. No mutations, however, were identified in the flanking introns. Moreover, the ca 5434–5437delTTCA deletion did not influence Shapiro's probability score for the splicing donor site.24 It was therefore likely that the deletion interrupted an exonic splicing enhancer. To examine this, the mutant exon was subjected to in vivo splicing analysis. A hybrid minigene containing exon 38 and its flanking introns between two cassette exons was constructed (fig 1) and transfected into HeLa cells. The resulting splicing products were amplified by RT‐PCR amplified. In the control sample (TVK‐WT), a mature hybrid mRNA consisting of exons A, 38 and B was produced (fig 3). When a mutant exon 38 with the 4‐bp deletion replaced the wild‐type exon 38 (TVK‐Δ), a smaller PCR product was exclusively obtained (fig 3). Sequencing of the product disclosed the absence of exon 38, indicating that this exon was skipped. Therefore, the four nucleotides were concluded to be part of an exonic splicing enhancer that was required for normal splicing.
Figure 3 Hybrid minigene splicing. Hybrid minigenes containing the indicated variants were tested in an in vivo splicing assay and the resulting mRNA was amplified by reverse transcriptase‐polymerase chain reaction (RT‐PCR). (A) Products electrophoresed on a polyacrylamide gel is shown. A 319‐bp full‐length transcript was generated from a minigene carrying the wildtype sequence of exon 38 (TVK‐WT), whereas a 196‐bp transcript was generated from a minigene carrying the mutant exon 38 (TVK‐Δ). TVK‐WT and TVK‐Δ contain the wild‐type exon 38 and the patient's genomic sequence with the 4‐bp deletion, respectively. Each of the four nucleotides of the deleted region in the TVK‐WT minigene was replaced with respective complementary nucleotide to produce the TVK‐WT5434T→A, TVK‐WT5435T→A, TVK‐WT5436C→G and TVK‐WT5437A→T minigenes. To analyse the position effect, 5436C was deleted (TVK‐WT5436Cdel), or replaced with A (TVK‐WT5436G→A) or T (TVK‐WT5436G→T). A schematic description of the RT‐PCR products is shown on the right. The wild‐type product (top) consists of exons A, 38 and B, whereas the smaller product did not contain exon 38, with exon A joining directly to exon B (bottom). (B,C) Nucleotide sequences at the junctions between exons are shown.
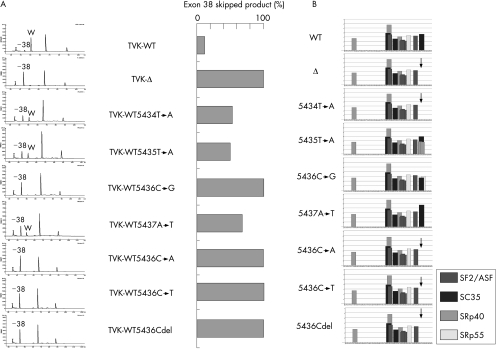
To explore the role of each of the four nucleotides in the regulation of exon 38 splicing, site‐directed mutagenesis was used. Four hybrid minigenes in which each of the four nucleotides was replaced with its complementary nucleotide were prepared (TVK‐WT5434T→A, TVK‐WT5435T→A, TVK‐WT5436C→G and TVK‐WT5437A→T) and transfected into HeLa cells. RT‐PCR analysis of hybrid mRNA from cells expressing TVK‐WT5436C→G produced a single band corresponding to a transcript lacking exon 38, indicating complete exclusion of exon 38; the other three constructs resulted in two amplified bands that corresponded to hybrid mRNAs with and without exon 38 skipping, indicating partial exon 38 skipping (fig 3). Quantification of the splicing products showed that TVK‐Δ and TVK‐WT5436C→G induced complete exon 38 skipping, whereas the production of mRNA without exon 38 was not absolute in cells expressing TVK‐WT5434T→A, TVK‐WT5435T→A and TVK‐WT5437A→T (fig 4).
Figure 4 Splicing enhancer. (A) Semiquantitative measurement of in vivo splicing products. Capillary electrophoretic patterns from semiquantitative reverse transcriptase‐polymerase chain reaction (RT‐PCR) analysis of minigene transcripts are shown (left). A fragment encompassing exon A to exon B was amplified. From their size, two peaks were identified: the 319‐bp wild‐type (WT) and the 196‐bp exon 38‐skipped product (−38). The area under the peak of each amplified product was quantified and the percentage of the total product, which was the exon 38‐skipped product, was calculated and is graphically shown (right panel). Five minigene products (TVK‐Δ, TVK‐WT5436C→G, TVK‐WT5436G→A, TVK‐WTG5436→T and TVK‐WT5436Gdel) completely lacked exon 38. (B) ESEfinder predictions of exonic splicing enhancers. ESEfinder predictions of exonic splicing enhancers in the wild‐type exon 38 (WT) and a variety of exon 38 mutants including the 4‐bp deletion (Δ) are shown. The arrows indicate the SC35 motif that disappeared as a result of the 4‐bp deletion and some of the single nucleotide mutations. Exonic splicing enhancer motifs that had scores above the threshold for each SR protein (SF2/ASF, SC35, SRp40 and SRp55) are indicated with differently shaded bars. The heights of the bar reflect the scores of the motifs.
As the 5436C→G mutation produced the same result as the complete disruption of the splicing enhancer sequence, the role of nucleotide 5436 in the splicing reaction was further examined by deleting this nucleotide or substituting the nucleotide with A or T. Three hybrid minigenes (TVK‐WT5436C→A, TVK‐WT5436C→T and TVK‐WT5436Cdel) were constructed and their transcripts were analysed. As all three mutants showed the complete exon 38‐skipping phenotype (fig 4A), we believe that nucleotide 5436C is determinant for proper exon 38 splicing.
The deletion identified in the index case is located in a purine‐rich sequence (14 of 20 nucleotides are purines). Purine‐rich sequences are known to serve as recognition motifs for SR proteins, which are required to define exons and consequently for splicing.25 When exon 38 was analysed for candidate splicing enhancer motifs using the ESE‐finder program,21 many SR protein‐binding motifs were identified (fig 4B). In particular, an SC35‐binding site was found to be located at the deletion site (GACTTCAA, the underlined sequence was deleted in the index case). These findings suggested that disruption of a predicted SC35‐binding site caused the skipping of exon 38 in the index case. Similarly, introduction of the C5436A, C5436T and C5436del mutations into exon 38 disrupted the predicted SC35‐binding site (fig 4). This agrees with our experimental results (fig 4). Four results, however, indicated that the identified motifs were not absolutely responsible for the in vivo splicing reaction (fig 4). Firstly, C5436G induced complete exon skipping but maintained a predicted SC35‐binding site and introduced an additional SRp55‐binding site. Secondly, T5434A induced partial enhancement of exon skipping in vivo and disrupted a predicted SC35‐binding site. Thirdly, T5435A improved exon skipping in vivo, but produced two additional SR‐protein‐binding sites while maintaining a predicted SC35‐binding site. Finally, A5437T induced partial exon skipping, but maintained the same binding site as the wild‐type sequence.
The RESCUE‐ESE program identified exonic splicing enhancers in several regions of exon 38. Two predicted enhancers, AAGACT and ACTTCA (the underlined sequences were deleted in the index case), were affected by the deleted region (data not shown). This again indicated that the disruption of an exonic splicing enhancer by the deletion caused the exon skipping observed in the index case.
Discussion
Genomic DNA analyses led to the identification of a novel molecular basis for dystrophinopathy: a 4‐bp deletion (ca 5434–5437delTTCA) in exon 38 of the dystrophin gene. The 4‐bp deletion mutation created a premature stop codon and was expected to cause DMD. The patient's moderately raised serum creatine kinase level, however, was more indicative of BMD. Interestingly, the exon 38− transcript was found to be the sole product of the mutant dystrophin gene (fig 2C). The exon 38− transcript contained an uninterrupted translational reading frame, leading to the production of an internally deleted, semifunctional dystrophin protein. Careful follow‐up with the index case will be necessary to make a conclusion about his phenotype.
Although 4‐bp deletions have been found in some exons of the dystrophin gene, none has been reported in exon 38. The genomic region surrounding the deletion was searched for sequences that could have predisposed the area to a deletion. Intrastrand complementarity, which is known to cause small intraexonic deletions,26 was ruled out because the exon 38 sequence is unlikely to form the required hairpin structure. Additionally, deletion motifs were not found in the genomic sequences.27 The sequence AAAGA, however, was identified both 7 bp upstream and 2 bp downstream from the deletion (fig 2). Although the slipped‐mispairing model28 suggests that this repeat may have had a role in the generation of the 4‐bp deletion, the exact mechanism that led to the TTCA deletion remains unclear.
Three models have been proposed to explain exon skipping caused by an exonic mutation. The first model is a nuclear scanning model in which a translation‐like machinery in the nucleus preserves the integrity of the reading frame by surveying the sequence and inducing the skipping of exons containing premature termination codons during pre‐mRNA splicing.29 This model, however, does not explain our results, because the transcripts produced in the in vivo splicing assay did not contain a continuous open‐reading frame. The second model is a secondary structure model in which a change in the pre‐mRNA secondary structure leads to exon skipping.30 The results from the in vivo splicing assay using minigenes that did not contain the full wild‐type introns make it unlikely that this model underlies the skipping of exon 38. The third model, which is supported by our results, is a cis‐element model in which disruption or creation of exonic splicing enhancers or silencers causes mutation‐associated exon skipping.31,32,33
A growing number of disease‐causing mutations have been shown to inactivate exonic splicing enhancers, and thereby cause exon skipping.25,31 For the dystrophin gene, the disruption of exonic splicing enhancers has been reported in exons 19 and 27.6,11,12 Our results showed that exon 38 contains an exonic splicing enhancer sequence. It has been reported that the strength of the splice sites and the position of the exonic splicing enhancer along the exon have major roles in the activity of the splicing enhancer.32,34 Among the 79 dystrophin exons, exon 38 is classified as a normal exon according to the exon definition model35; weak exons of the dystrophin gene are reported to need an exonic splicing enhancer sequence for proper recognition of the exon. In fact, exonic splicing enhancers have been identified in several dystrophin exons in experiments that searched for antisense oligonucleotides inducing exon skipping.13,14
The hybrid minigene proved to be a good model for the exon skipping induced by the deletion mutation (fig 3). This hybrid minigene system will facilitate the molecular understanding of mutations that are not classified as disease‐causing mutations.18 Although tissue‐specific splicing patterns are well documented, our results suggest that lymphocytes and HeLa cells contain the same splicing machinery for dystrophin pre‐mRNA splicing. As the deleted region was shown to be essential for proper splicing in two different cell types, the deleted sequence probably contributes to the constitutive splicing of exon 38 instead of alternative or tissue‐specific splicing.36
Purine‐rich exonic splicing enhancers activate splicing by binding SR proteins, which recruit the splicing machinery to adjacent splice sites.25 Although individual SR proteins are known to exhibit distinct substrate specificities, the degeneracy of the RNA recognition sequences for the different SR proteins makes the prediction of SR‐protein‐dependent enhancers uncertain. The ESE‐finder program showed that the deletion removed a predicted SC35‐binding motif (GACTTCAA) from exon 38. Moreover, our in vivo splicing study (fig 4) suggests that a single nucleotide (C5436) is essential for proper splicing of exon 38. Although splicing error due to abnormal SC35 binding has been shown to cause genetic diseases,37 a single nucleotide in a predicted SC35‐binding site has never been shown to be determinant for proper splicing. Our finding will facilitate the understanding of splicing regulation by SR proteins.
Presently, there is no effective way to treat DMD. Recent treatments for DMD have focused on converting the DMD phenotype to a BMD phenotype by changing dystrophin mRNAs from out of frame to in frame. We have shown that induction of exon 19 skipping in a patient with DMD carrying a deletion in exon 20 led to the production of in‐frame dystrophin mRNA and dystrophin‐positive skeletal muscle cells.38 Therefore, artificial induction of exon skipping using antisense oligonucleotides is now being extensively studied.14,39,40,41,42 Our findings indicated that the 4‐bp sequence TTCA is a good target for treatment with antisense oligonucleotides to induce exon 38 skipping. Seven nonsense mutations have been identified in exon 38 of patients with DMD. If these patients are treated with antisense oligonucleotides that are able to induce exon 38 skipping, the resulting dystrophin mRNA is expected to produce a truncated and semifunctional dystrophin protein. This would shed new light on designing antisense oligonucleotides used for treatment of DMD.
Our results provide important insights for the understanding of the molecular basis of splicing regulation and the clinical application of exon skipping treatment for patients with DMD.
Acknowledgements
We thank Ms A Hosoda for her secretarial help.
Abbreviations
BMD - Becker's muscular dystrophy
DMD - Duchenne's muscular dystrophy
PCR - polymerase chain reaction
RT - reverse transcription
TVK‐Δ - mutant hybrid minigene
TVK‐WT - wild‐type hybrid minigene
Footnotes
Funding: This work was supported by a Grant‐in‐Aid for Scientific Research from the Japan Society for the Promotion of Science; Health and Labour Sciences Research Grants from the Ministry of Health, Labour, and Welfare for research on psychiatric and neurological diseases and mental health; a grant for nervous and mental disorders from the Ministry of Health, Labour, and Welfare; and the Mitsubishi Foundation.
Competing interests: None declared.
References
- 1.Tanaka K, Watakabe A, Shimura Y. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol Cell Biol 1994141347–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pagani F, Stuani C, Tzetis M.et al New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum Mol Genet 2003121111–1120. [DOI] [PubMed] [Google Scholar]
- 3.Monaco A P, Bertelson C J, Liechti‐Gallati S, Moser H, Kunkel L M. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 1988290–95. [DOI] [PubMed] [Google Scholar]
- 4.Chelly J, Gilgenkrantz H, Lambert M.et al Effect of dystrophin gene deletions on mRNA levels and processing in Duchenne and Becker muscular dystrophies. Cell 1990631239–1248. [DOI] [PubMed] [Google Scholar]
- 5.Barbieri A M, Soriani N, Ferlini A.et al Seven novel additional small mutations and a new alternative splicing in the human dystrophin gene detected by heteroduplex analysis and restricted RT‐PCR heteroduplex analysis of illegitimate transcripts. Eur J Hum Genet 19964183–187. [DOI] [PubMed] [Google Scholar]
- 6.Shiga N, Takeshima Y, Sakamoto H.et al Disruption of the splicing enhancer sequence within exon 27 of the dystrophin gene by a nonsense mutation induces partial skipping of the exon and is responsible for Becker muscular dystrophy. J Clin Invest 19971002204–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melis M A, Muntoni F, Cau M.et al Novel nonsense mutation (C→A nt 10512) in exon 72 of dystrophin gene leading to exon skipping in a patient with a mild dystrophinopathy. Hum Mutat 1998(Suppl 1)S137–S138. [DOI] [PubMed]
- 8.Ginjaar I B, Kneppers A L, vd Meulen J D.et al Dystrophin nonsense mutation induces different levels of exon 29 skipping and leads to variable phenotypes within one BMD family. Eur J Hum Genet 20008793–796. [DOI] [PubMed] [Google Scholar]
- 9.Fajkusova L, Lukas Z, Tvrdikova M.et al Novel dystrophin mutations revealed by analysis of dystrophin mRNA: alternative splicing suppresses the phenotypic effect of a nonsense mutation. Neuromuscul Disord 200111133–138. [DOI] [PubMed] [Google Scholar]
- 10.Hamed S, Sutherland‐Smith A, Gorospe J.et al DNA sequence analysis for structure/function and mutation studies in Becker muscular dystrophy. Clin Genet 20056869–79. [DOI] [PubMed] [Google Scholar]
- 11.Matsuo M, Masumura T, Nishio H.et al Exon skipping during splicing of dystrophin mRNA precursor due to an intraexon deletion in the dystrophin gene of Duchenne muscular dystrophy Kobe. J Clin Invest 1991872127–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeshima Y, Nishio H, Sakamoto H.et al Modulation of in vitro splicing of the upstream intron by modifying an intra‐exon sequence which is deleted from the dystrophin gene in dystrophin Kobe. J Clin Invest 199595515–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Surono A, Tran V K, Takshima Y.et al Chimeric RNA/ethylene bridged nucleic acids promote dystrophin expression in myocytes of Duchenne muscular dystrophy by inducing skipping of the nonsense‐mutation‐encoding exon. Hum Gene Ther 200415749–757. [DOI] [PubMed] [Google Scholar]
- 14.Aartsma‐Rus A, De Winter C L, Janson A A.et al Functional analysis of 114 exon—internal AONs for targeted DMD exon skipping: indication for steric hindrance of SR protein binding sites. Oligonucleotides 200515284–297. [DOI] [PubMed] [Google Scholar]
- 15.Chamberlain J S, Gibbs R A, Ranier J E.et al Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res 19881611141–11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beggs A H, Koenig M, Boyce F M.et al Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction. Hum Genet 19908645–48. [DOI] [PubMed] [Google Scholar]
- 17.Roberts R G, Gardner R J, Bobrow M. Searching for the 1 in 2,400,000: a review of dystrophin gene point mutations. Hum Mutat 199441–11. [DOI] [PubMed] [Google Scholar]
- 18.Thi Tran H T, Takeshima Y, Surono A.et al A G‐to‐A transition at the fifth position of intron 32 of the dystrophin gene inactivates a splice donor site both in vivo and in vitro. Mol Genet Metab 200585213–219. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Russell D W. eds. Site‐specific mutagenesis by overlap extension. In: Molecular cloning. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 200113.36–13.39.
- 20.Cartegni L, Wang J, Zhu Z.et al ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 2003313568–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fairbrother W G, Yeo G W, Yeh R.et al RESCUE‐ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res 200432W187–W190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts R G, Bobrow M, Bentley D R. Point mutations in the dystrophin gene. Proc Natl Acad Sci USA 1992892331–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sironi M, Bardoni A, Felisari G.et al Transcriptional activation of the non‐muscle, full‐length dystrophin isoforms in Duchenne muscular dystrophy skeletal muscle. J Neurol Sci 200118651–57. [DOI] [PubMed] [Google Scholar]
- 24.Shapiro M B, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 1987157155–7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blencowe B J. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci 200025106–110. [DOI] [PubMed] [Google Scholar]
- 26.Robinson D O, Bunyan D J, Gabb H A.et al A small intraexonic deletion within the dystrophin gene suggests a possible mechanism of mutagenesis. Hum Genet 199799658–662. [DOI] [PubMed] [Google Scholar]
- 27.Sironi M, Pozzoli U, Cagliani R.et al Relevance of sequence and structure elements for deletion events in the dystrophin gene major hot‐spot. Hum Genet 2003112272–288. [DOI] [PubMed] [Google Scholar]
- 28.Todorova A, Danieli G A. Large majority of single‐nucleotide mutations along the dystrophin gene can be explained by more than one mechanism of mutagenesis. Hum Mutat 19979537–547. [DOI] [PubMed] [Google Scholar]
- 29.Dietz H C, Kendzior R J., Jr Maintenance of an open reading frame as an additional level of scrutiny during splice site selection. Nat Genet 19948183–188. [DOI] [PubMed] [Google Scholar]
- 30.Muro A F, Caputi M, Pariyarath R.et al Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol Cell Biol 1999192657–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zatkova A, Messiaen L, Vandenbroucke I.et al Disruption of exonic splicing enhancer elements is the principal cause of exon skipping associated with seven nonsense or missense alleles of NF1. Hum Mutat 200424491–501. [DOI] [PubMed] [Google Scholar]
- 32.Cartegni L, Chew S L, Krainer A R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 20023285–298. [DOI] [PubMed] [Google Scholar]
- 33.Faustino N A, Cooper T A. Pre‐mRNA splicing and human disease. Genes Dev 200317419–437. [DOI] [PubMed] [Google Scholar]
- 34.Wu Y, Zhang Y, Zhang J. Distribution of exonic splicing enhancer elements in human genes. Genomics 200586329–336. [DOI] [PubMed] [Google Scholar]
- 35.Sironi M, Pozzoli U, Cagliani R.et al Analysis of splicing parameters in the dystrophin gene: relevance for physiological and pathogenetic splicing mechanisms. Hum Genet 200110973–84. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Smith P J, Krainer A R.et al Distribution of SR protein exonic splicing enhancer motifs in human protein‐coding genes. Nucleic Acids Res 2005335053–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gabut M, Mine M, Marsac C.et al The SR protein SC35 is responsible for aberrant splicing of the E1alpha pyruvate dehydrogenase mRNA in a case of mental retardation with lactic acidosis. Mol Cell Biol 2005253286–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeshima Y, Yagi M, Wada H.et al Intravenous infusion of an antisense oligonucleotide results in exon skipping in muscle dystrophin mRNA of Duchenne muscular dystrophy. Pediatr Res. 2006;59;690–4. [DOI] [PubMed]
- 39.Aartsma‐Rus A, Janson A A, Kaman W E.et al Antisense‐induced multiexon skipping for Duchenne muscular dystrophy makes more sense. Am J Hum Genet 20047483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuo M, Takeshima Y. Rescue of dystrophin mRNA of Duchenne muscular dystrophy by inducing exon skipping. Acta Myol 2005XXIV110–114. [PubMed] [Google Scholar]
- 41.Gebski B L, Errington S J, Johnsen R D.et al Terminal antisense oligonucleotide modifications can enhance induced exon skipping. Neuromuscul Disord 200515622–629. [DOI] [PubMed] [Google Scholar]
- 42.Bartoli M, Poupiot J, Goyenvalle A.et al Noninvasive monitoring of therapeutic gene transfer in animal models of muscular dystrophies. Gene Therapy 20061320–28. [DOI] [PubMed] [Google Scholar]