Abstract
Previous structural and biochemical studies have revealed that the inner arm dynein I1 is targeted and anchored to a unique site located proximal to the first radial spoke in each 96-nm axoneme repeat on flagellar doublet microtubules. To determine whether intermediate chains mediate the positioning and docking of dynein complexes, we cloned and characterized the 140-kDa intermediate chain (IC140) of the I1 complex. Sequence and secondary structural analysis, with particular emphasis on β-sheet organization, predicted that IC140 contains seven WD repeats. Reexamination of other members of the dynein intermediate chain family of WD proteins indicated that these polypeptides also bear seven WD/β-sheet repeats arranged in the same pattern along each intermediate chain protein. A polyclonal antibody was raised against a 53-kDa fusion protein derived from the C-terminal third of IC140. The antibody is highly specific for IC140 and does not bind to other dynein intermediate chains or proteins in Chlamydomonas flagella. Immunofluorescent microscopy of Chlamydomonas cells confirmed that IC140 is distributed along the length of both flagellar axonemes. In vitro reconstitution experiments demonstrated that the 53-kDa C-terminal fusion protein binds specifically to axonemes lacking the I1 complex. Chemical cross-linking indicated that IC140 is closely associated with a second intermediate chain in the I1 complex. These data suggest that IC140 contains domains responsible for the assembly and docking of the I1 complex to the doublet microtubule cargo.
INTRODUCTION
Dyneins constitute a family of molecular motors responsible for several different functions, including ciliary and flagellar motility, minus end–directed transport of membrane-bound organelles, assembly of the Golgi apparatus, and formation and function of the mitotic apparatus. To perform these transport functions each dynein must be correctly anchored to distinctive organelles, or “cargoes” (Mitchell, 1994; Witman et al., 1994; Porter, 1996). Although we know much about the composition and activity of dynein (reviewed in Holzbaur and Vallee, 1994; Mitchell, 1994; Porter, 1996), we know little about the protein–protein interactions involved in the assembly and binding of dynein to its cargo. It has been proposed that the dynein intermediate chain (IC) subunits play a central role in the docking mechanism (King and Witman, 1990; King et al., 1991, 1995).
The ICs were first identified as proteins that copurify with heavy chains of the flagellar outer dynein arms (Mitchell, 1994; Witman et al., 1994; Porter, 1996). Subsequent biochemical analysis has revealed that IC subunits are also found in cytoplasmic (Holzbaur and Vallee, 1994) and flagellar inner arm dyneins (Piperno et al., 1990; Smith and Sale, 1991, 1992; Kagami and Kamiya, 1992; Porter et al., 1992). Based on the structure of isolated flagellar outer arm dyneins and subfractionated dynein complexes, the ICs were predicted to be located at the base of the dynein structure, at the site of attachment with the doublet microtubule cargo (Goodenough and Heuser, 1983; Sale et al, 1985). Immunoelectron microscopy of isolated dynein complexes supports this predicted localization (King and Witman, 1990: Steffen et al., 1996). Moreover, chemical cross-linking demonstrated that IC78 from the outer dynein arm of Chlamydomonas flagella is closely associated with α-tubulin of the doublet microtubule cargo (King et al., 1991). Analysis of mutant cells indicates that the ICs are required for dynein assembly (Mitchell and Kang, 1991; Wilkerson et al., 1995; Perrone et al., 1998). Sequence analysis revealed that many of the ICs form a family of WD-repeat proteins, possibly indicating a common functional substructure (Ogawa et al., 1995; Wilkerson et al., 1995). Finally, indirect evidence also indicates that cytoplasmic dynein is linked to organelles by ICs (Karki and Holzbaur, 1995; Steffen et al., 1997). Together, these data support the hypothesis that IC subunits play a role in the positioning and docking of different dyneins to specific cargoes. However, despite these data, we know little about the precise functions and protein interactions of the dynein ICs.
The inner row of dynein arms in the flagellar axoneme presents an ideal system for studying the mechanism of cargo–dynein interaction. Structural and biochemical analyses of wild-type and mutant axonemes have established that the inner arm dyneins are heterogeneous in composition and location along each doublet microtubule (reviewed in Porter, 1996). Current models suggest that the inner arms are organized in precise groups, which repeat in a 96-nm pattern, in exact register with the paired radial spokes (Goodenough and Heuser, 1984; Piperno et al., 1990; Burgess et al., 1991; Mastronarde et al., 1992; Porter et al., 1992; Gardner et al., 1994; Woolley, 1997). Notably, in vitro reconstitution of axonemes with isolated dyneins revealed that each inner arm dynein is targeted and anchored to a unique position within the 96-nm axoneme repeat (Smith and Sale, 1992).
The I1 inner arm dynein complex offers several advantages for study of dynein assembly. First, I1, like outer arm and cytoplasmic dynein, can be isolated as a complex, composed of two heavy chains, three ICs (IC140, IC138, and IC97) (IC97 is also referred as IC110 [Perrone et al., 1998, accompanying article]), and three light chains (Goodenough et al., 1987; Piperno et al., 1990; Porter et al., 1992; Harrison et al., 1998). Second, the I1 complex is defined by mutations in four loci (Brokaw and Kamiya, 1987; Piperno et al., 1990; Kamiya et al., 1991; Porter et al., 1992; Myster et al., 1997; Perrone et al., 1998). Defects in any of these loci result in the failure to assemble the I1 complex in the axoneme. Structural analysis of I1 mutant axonemes revealed that the complex is located in a proximal position in each 96-nm repeat unit (Goodenough and Heuser, 1984; Piperno et al., 1990; Mastronarde et al., 1992; Smith and Sale, 1992; Porter et al., 1992). Notably, the resulting gaps located at 96-nm intervals indicate that each inner arm dynein can be assembled and targeted independent of neighboring dyneins (Piperno et al., 1990). Finally, as described above, I1 can be reconstituted in vitro, precisely restoring I1 structure and function to the axoneme (Smith and Sale, 1992).
Based on the hypothesis that the IC subunits play a role in targeted assembly of dynein, we focused on studying IC140, one of the IC subunits of the I1 complex. To determine whether IC140 plays a role in I1 assembly and anchoring, we began by cloning IC140 with the dual goals of characterizing its molecular structure and producing specific reagents such as fusion proteins and antibodies. Sequence analysis, with focus on β-sheet structure, revealed that IC140 contains seven WD repeats arranged in an order similar to that of the seven WD/β-sheet repeats predicted in other dynein ICs. We also report that IC140 is located along the length of axonemes in both Chlamydomonas flagella, consistent with conclusions based on structural analysis of axonemes lacking the I1 structure (Piperno et al., 1990; Piperno and Ramanis, 1991; Porter et al., 1992). Finally, based on in vitro reconstitution and chemical cross-linking, we suggest that IC140 plays a central role in the assembly and anchoring of the I1 complex. These predictions are supported by the observations of Perrone et al. (1998) that mutations in the IC140 gene block assembly of the I1 complex and that transformation of these mutants with a wild-type copy of the IC140 gene will rescue the assembly defect.
MATERIALS AND METHODS
Cell Strains
Cell strains were obtained from the Chlamydomonas Genetics Center (Dr. E.H. Harris, Duke University, Durham, NC). Wild-type CC-125 was used for preparation of DNA and RNA. Mutant strains pf28 (Mitchell and Rosenbaum, 1985) and pf28pf30 (Piperno et al., 1990) were used for biochemical and cell biological experiments. The cell wall defective mutant cw-15 was used for immunofluorescent staining. All cells were grown in Tris-acetate-phosphate medium with aeration over a 14/10-h light/dark cycle (Witman, 1986).
Purification of IC140
All reagents, unless stated otherwise, were from Sigma Chemical (St. Louis, MO). For cloning IC140, our strategy was first to obtain partial amino acid sequence from the isolated protein and then to design degenerate primers for reverse transcription (RT)-PCR. Axonemes were isolated from pf28 cells, and dynein extraction and sucrose gradient fractionation were carried out as described previously (Smith and Sale, 1992). For sucrose gradients, the dynein-containing extracts were first dialyzed into buffer A (10 mM HEPES, 5 mM MgSO4, 1 mM DTT, 0.5 mM EDTA, 30 mM NaCl, 0.1 mM PMSF, 0.6 trypsin inhibitor units aprotinin, pH 7.4), followed by sedimentation through 5–20% sucrose gradients prepared in buffer A. The I1 dynein fractions sedimenting between 18 and 21S were identified by SDS-PAGE and silver staining and concentrated by centrifugation in an Ultrafree-CL Polysulfone filter unit (Millipore, Bedford, MA). The concentrated sample was resolved by 7% SDS-PAGE and then transferred onto a polyvinylidene difluoride membrane (Applied Biosystems, Foster City, CA) in 10 mM 3-[cyclohexylamino]-1-propane sulfonic acid/10% methanol, pH 11. The IC140 band was detected on the membrane by Amido Black staining (0.1% in 1% acetic acid/40% methanol), excised, and shipped to Dr. J. Leszyk (University of Massachusetts, Worcester, MA; formerly the Worcester Foundation for Experimental Biology) for peptide microsequencing. Briefly, the membrane bearing the IC140 protein was exposed to endoproteinase lys-C, and the resulting peptides were eluted and fractionated by HPLC. Three peak fractions were selected and sequenced with an Applied Biosystems Procise Sequencer (see Table 1).
Table 1.
Amino acid sequences of peptides obtained by direct microsequencing of band-purified IC140
| Peptide 1 | HRFMTDTQFSHHQVVTTLQ |
| Peptide 2 | AAAAAAAGMPLPTANERK |
| Peptide 3 | LEQGMFYAGSFDGELVYADF |
Molecular Biology
For cloning, total RNA was derived from cells before (control) or 45 min after deflagellation, a process that up-regulates the transcription of many flagellar genes (Lefebvre and Rosenbaum, 1986). Genomic DNA was prepared as described (Wilkerson et al., 1994). Poly(A) RNA was further purified with oligo(dT) cellulose (Collaborative Biochemical Products, Bedford, MA) following the methods of Schnell and Lefebvre (1993). Primers for RT-PCR used for various cloning steps (Figure 1) include P1S (CGCGGAATTCATGACIGACACICARTTY), P3A (CGCGGGATCCAISWICCIGCRTARAACAT), RT1 (TTGCCCTTCTTCAGCAGCTTCTCA), Cr5 (GCGTGTCCGTCATGAATCGGTGCTT), FUS (CGCTACCAGAGGACTACGTG), FUSrev (CGTAGTCCTCTGGTAGCG), and T3 (CTCCTTGCTAGGGATCTG).
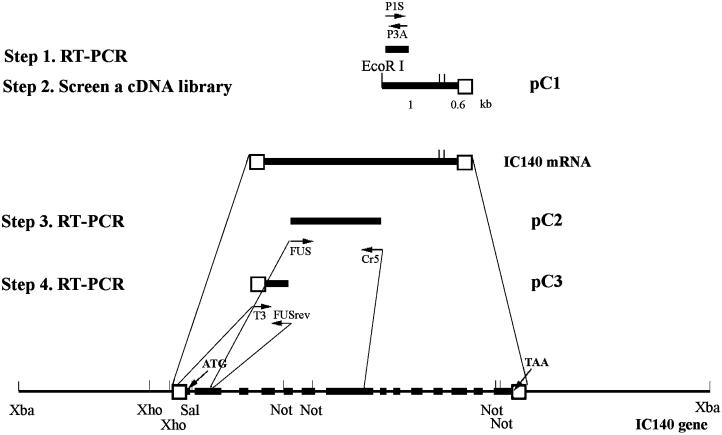
Figure 1.
Schematic picture illustrates the strategy and steps used to clone Chlamydomonas IC140 gene and message. The 11.5-kb XbaI genomic subclone contains the complete IC140 gene consisting of 14 exons (solid bars) and 13 introns. Open boxes indicate 5′ and 3′ untranslated regions. Arrows indicate the position and the direction of primers for RT-PCR (see MATERIALS AND METHODS). The pC1 clone was recovered from screening a cDNA library. PC2 and pC3 resulted from RT-PCR using the gene-specific primer RT1 for reverse transcription and primer pairs FUS/Cr5 and T3/FUSrev, respectively (see RESULTS for details).
RT was primed either with oligo(dT)12–18 (Pharmacia Biotech, Piscataway, NJ) or with the gene-specific primers (RT1) using the reverse transcriptase Superscript II (Life Technologies, Gaithersburg, MD) according to the manufacturer’s instructions. Each 50-μl PCR reaction contained 0.5 μl RT mixture, 0.2 mM dNTP, a 0.2 μM concentration of each primer (2 μM for degenerate primers), 2.5 U Taq or 1.25 U Pfu DNA polymerase (Strategene, La Jolla, CA) and 1× manufacturer-provided buffer. The PCR mixture was initially denatured at 95°C for 3 min, followed by 35 cycles of 95°C, 1.5 min, 48–50°C, 1.5 min, and 72°C, 3 min. The reaction mixture was maintained at 72°C for an additional 10 min, followed by separation of products in 0.8–1% agarose (Fisher Scientific, Pittsburgh, PA). Notably, after extensive experimentation and numerous primers, we found that RT-PCR, using the gene-specific primer RT1, was optimal when dGTP was replaced with 7-deaza-2′-dGTP/dGTP (3:1; see Innis, 1990) and that the sample was treated with RNAse H after reverse transcription. These steps resulted in the 1.55-kb RT-PCR product known as pC2. The bands of interest were excised and spun through siliconized glass wool. The DNA in the flow through was extracted with phenol-chloroform and precipitated with ethanol. Most purified PCR products were then blunt end cloned into the SmaI site of pBluescript II KS(−). The 350-bp product primed with P1S/P3A was cloned into the EcoRI and BamHI sites.
Library Screening
To obtain cDNA clones containing IC140 sequence, we screened a λzapII cDNA library, made from RNA derived from cells regenerating flagella (kindly provided by K. Wilkerson and G. Witman, University of Massachusetts; Wilkerson et al., 1995). To obtain genomic clones containing IC140 sequence, a λfixII genomic library, constructed from strain A54-e18, was screened (kindly provided by E. Smith and P. Lefebvre, University of Minnesota, Minneapolis, MN; Smith and Lefebvre, 1997). For screening, libraries were plated and transferred to Hybond N filters (Amersham International, Buckinghamshire, UK), followed by UV cross-linking with Strata-linker (Strategene). Positive clones were identified by hybridizing the filters with the [P32]dCTP-labeled, random-primed probes using the 350-bp PCR product as a template and were incubated at 65°C overnight in buffer (1% BSA, 1 mM. EDTA, 0.5 M sodium phosphate buffer, pH 7.5, 7% SDS). The membranes were then washed once at 65°C with 2× SSC/0.1% SDS and three times with 0.2× SSC/0.1% SDS for 20 min each. The cDNA clones from purified plaques were excised with helper phage (Wilkerson et al., 1995). The 1.6-kb insert of cDNA clone pC1 (Figure 1) was released with EcoRI and XhoI and further digested to a 1-kb 5′ fragment, containing the coding sequence, and a 0.6-kb 3′ fragment. The 1-kb fragment was subcloned and used as a probe for Northern and Southern blots. The inserts from genomic clones were released from phage vector by XbaI digestion and ligated into pBluescript II KS(−). The XbaI genomic clone was further digested with SalI and NotI and subcloned into pBluescript II KS(−) to further facilitate sequencing.
Southern and Northern Blots
For Southern analysis, genomic DNA (5 μg) was digested with restriction endonucleases, fractionated in 0.8% agarose gels, and transferred to a Nytran membrane following the manufacturer’s instructions (Schleicher & Schuell, Keene, NH). For Northern analysis, poly(A) RNA (3 μg) was fractionated in 1.2% formamide-containing gels and transferred to Hybond N+ filters (Amersham). Membranes were probed with the 1-kb EcoRI–NotI fragment of pC1 (Figure 1) using the procedures described for library membrane lifts.
Fusion Protein and Antibody Production
The 1.6-kb insert from the pC1 cDNA clone was released by EcoRI and XhoI and ligated, in frame, into pET28(a) expression vector (Novagen, Madison, WI). The expression construct was transformed into Escherichia coli strain BL21(DE3) (Novagen). Production of the N-terminal His-tagged fusion protein was induced by 100 mM isopropyl-1-thio-β-d-galactopyranoside, and proteins contained within the inclusion body were solublized in the 6 M urea/imidazole buffer designed for Ni+ column affinity purification and according to the manufacturer’s instructions (Novagen). The eluent was slowly dialyzed against PBS (without pressure or stirring) for 20 h. Most of the purified fusion protein was soluble and used as an antigen to produce polyclonal antibodies in rabbits (Spring Valley Laboratories, Sykesville, MD). For Western blots, samples were separated in SDS-PAGE followed by Coomassie blue staining or transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA) for immunoblot. The membrane was blocked with 5% nonfat dry milk followed by incubation first with anti-IC140 antibody (1:3000–1:6000 dilution) and then with HRP-conjugated goat anti-rabbit secondary antibody (1:5000, Bio-Rad). The antibody reactivity was detected with 4-chloro-1-naphthal and hydrogen peroxide or with enhanced chemiluminescent reagents (Amersham). M. Porter and S. Myster (University of Minnesota) generously provided the antibody to the 1-α heavy chain of inner arm dynein I1 (Myster et al., 1997).
DNA Sequencing and Sequence Analysis
Sequencing of both strands was carried out manually using dideoxy methods with a Sequenase II kit (United States Biochemicals, Cleveland, OH) or in most cases by automated cycle sequencer (ABI 377, Applied Biosystems). Sequence was analyzed using programs from Wisconsin Sequence Analysis Package Incremental Release 8.1 (Genetics Computer Group, Madison, WI). Programs used include CodonPreference to reveal regions of codon bias in genomic sequences, Findpattern to identify the consensus sequence for exon–intron junctions (/GAXXGX . . . . . . . CAG/, kindly provided by C. Silflow, University of Minnesota), COMPARE for comparing pairs of dynein IC sequences (window, 30; stringency, 18), PileUp for plotting dendrograms, BLAST to search for homologous molecules (Altschul et al., 1990), Motifs to identify WD repeats, and PeptideStructure for predicting the secondary structure. Coiled-coil structures were predicted using the Coils program (Lupas et al., 1991) (with the MTIDK matrix and a window of 21) from Pedros Biomolecular Research Tools (www.public.iastate.edu/∼pedro/research_ tools.html). For analysis, default parameters were used unless stated otherwise.
Immunolocalization
Immunofluorescent staining was carried out as described (Johnson and Rosenbaum, 1992) with modification. The cell wall–deficient cells were allowed to attach to the Nalge-Nunc Permanox chamber slide (Fisher Scientific). Immobilized cells were fixed by immersion in −20°C methanol for 10 min, followed by immersion in a blocking buffer composed of 3% BSA in PBS at room temperature. Slides were incubated with either preimmune serum or the anti-IC140 polyclonal antibody, diluted 1:1000 in 1% BSA/PBS, overnight at 4°C, and FITC-conjugated goat anti-rabbit antibody 1:500 (Cappel/INC Pharmaceuticals, Costa Mesa, CA) in 1% BSA/PBS for 2 h. The images were captured by a Zeiss (Thornwood, NY) epifluorescent microscope equipped with an air-cooled charge-coupled device camera (CCD300T; Dage-MTI, Michigan City, IN).
Fusion Protein–Doublet Microtubule Binding Assay
Axonemes from pf28 and pf28pf30 were prepared as described (Smith and Sale, 1992). Axonemal and fusion protein concentration was determined with the Bradford assay (Bio-Rad). The mixture of 2 mg/ml axonemes in buffer A and 15–150 ng fusion protein in a total volume of 100 μl was incubated on ice or at 22°C for 30 min. After dilution with 1.5 ml buffer A, axonemes were sedimented, and the pellet was resuspended with 100 μl buffer A for SDS-PAGE and Western blots.
For cross-linking, axonemes (2–5 mg/ml) and dynein extract were prepared in buffer A but in the absence of DTT and protease inhibitors. Cross-linking was carried out as described (King et al., 1991) by incubating axonemes or dynein extract with 1–10 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC; Pierce, Rockford, IL) for 1 h at room temperature and was terminated with at least a 10-fold molar excess of β-mercaptoethanol. For sucrose gradient sedimentation, the cross-linked axonemes were further extracted with 0.6 M NaCl/buffer A, and the extract was sedimented through a 5–20% sucrose gradient made with buffer A. Fractions were analyzed by SDS-PAGE and Western blots.
RESULTS
Cloning of IC140
To clone and analyze the IC140 gene, our strategy was to obtain partial protein sequences from purified IC140 (Table 1), design oligonucleotide primers for RT-PCR (Figure 1), and use the product for screening libraries and for Southern and Northern analysis (Figure 2). IC140 was purified by a combination of velocity sedimentation fractionation of the dynein extract on sucrose gradients followed by SDS-PAGE and transfer to polyvinylidene difluoride membranes. We found that the N terminus of the band-purified IC140 was naturally or experimentally blocked to the Edman degradation reaction. Therefore, internal amino acid sequence was obtained by digestion of IC140 with endoproteinase lys-C, followed by fractionation of peptides by HPLC. The sequences of three peptides are listed in Table 1.
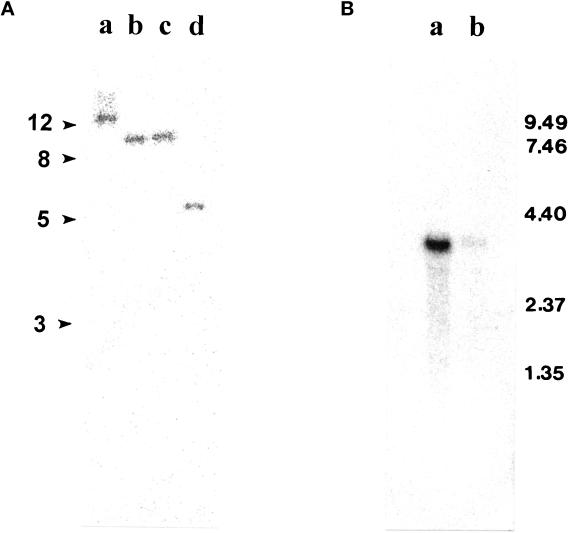
Figure 2.
(A) Southern blot analysis of 5 μg genomic DNA digested with EcoRI, XhoI, SalI, and SacI (lanes a–d, respectively). (B) Northern blot analysis of 3 μg poly(A) RNA prepared from Chlamydomonas regenerating half-length of flagella (lane a) and from control cells (no deflagellation) (lane b). Both membranes were probed with the EcoRI–NotI fragment of pC1. The Northern blot was reprobed with the CRY1 gene to confirm the equal loading.
Sense and antisense degenerate oligonucleotides P1S and P3A, listed in MATERIALS AND METHODS, were designed based on the amino acids underlined in Table 1. RT-PCR with this primer pair resulted in a single 350-bp band (Figures 1 and 3, nucleotides 4048–4962). The 350-bp PCR product was cloned and sequenced. The predicted amino acid sequence, flanking the primers, was identical to the amino acid sequence obtained from microsequencing, with the sole exception that VVTT in peptide 1 (Table 1) was later confirmed to be VVTD (Figure 3, amino acids 638–641) based on the cDNA and genomic sequence. The predicted peptide sequence (Figure 3, amino acids 628–741) is novel but homologous to the “C”-WD repeat described in other dynein ICs (Ogawa, et al., 1995), suggesting that the 350-bp clone derived from IC140 represented a new member of the dynein IC family.
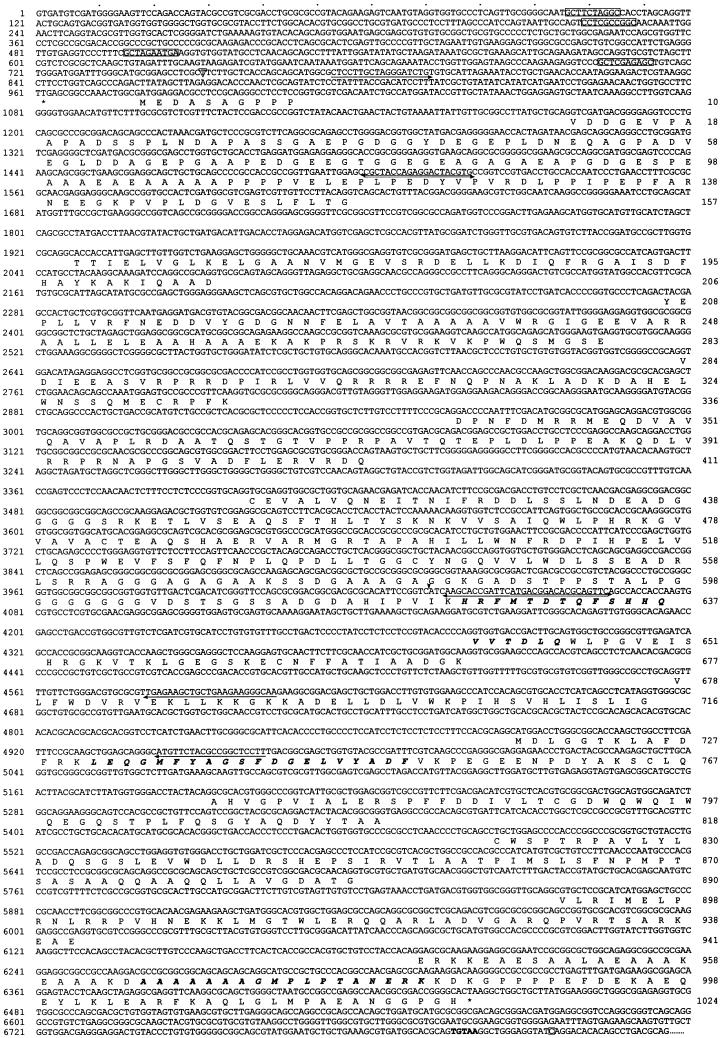
Figure 3.
The IC140 genomic and protein sequence. The amino acid sequence predicted from cDNA sequence aligned with the exons of the genomic sequence. Bold italic letters, aa sequences from peptide microsequencing; box, predicted tub boxes; asterisk, stop codons preceding the predicted first ATG and for terminating IC140 translation; arrow, primer for RT-PCR; arrowhead, 5′ end of the first partial cDNA clone from cDNA library; letter in an open triangle, transcription initiation site; letter in an open square, polyadenylation site. The bold TGTAA is probably the polyadenylation signal.
The 350-bp PCR product was used to screen a λzapII cDNA library constructed from RNA obtained from Chlamydomonas cells after deflagellation (Wilkerson et al., 1995). Three identical 1.6-kb cDNA clones were recovered and are represented as pC1 in Figure 1 and nucleotides 4027–6814 in Figure 3. The pC1 sequence extended 9 bp beyond the PCR product in the 5′ direction, contained a poly(A) tail, and, based on the predicted amino acid sequence, contained peptide 2 near the 3′ end of the cDNA clone (Figure 3, amino acids 965–982). Based on these results, the 1.6-kb cDNA pC1 represents part of the IC140 message.
Southern blots of genomic DNA, digested with four different restriction enzymes (indicated in Figure 2A) and probed with the 1-kb fragment of pC1, revealed only a single band of similar intensity for each enzyme. Given that there were no restriction sites for these enzymes within the 1-kb probe, these data suggest that there is a single gene coding for IC140. Northern blot analysis of poly(A) RNA from Chlamydomonas revealed a single 3.7-kb transcript coding for IC140 (Figure 2B), which was up-regulated 30–45 min after deflagellation (Figure 2B, lane a), as expected for a message that encodes a protein with a flagellar-specific function (Silflow and Rosenbaum, 1981; Le-febvre and Rosenbaum, 1986).
Rescreening the cDNA library and 5′ rapid amplification of cDNA ends both failed to yield additional clones at the 5′ end of IC140. Therefore, we chose to obtain the genomic clone of IC140 to complete the sequence. Screening a λfixII genomic library (Smith and Lefebvre, 1997) with the 1-kb fragment of pC1 as a probe resulted in the recovery of 10 overlapping clones (17–22 kb each) covering a total of 30 kb of genomic sequence. Seven of the clones were distinct, and, after restriction mapping and analysis, a detailed map of the genomic clones was constructed. Southern blot analysis of the genomic clones using the 1-kb 5′ and 0.6-kb 3′ fragments of pC1 (Figure 1) as probes identified the 5′ end of the IC140 gene and resulted in recovery of an 11.5-kb XbaI clone (Figure 1). The 11.5-kb XbaI clone contains pC1, and, as described in the accompanying article, the clone rescues a Chlamydomonas mutant strain, which fails to assemble inner arm I1 because of a deletion mutation in the IC140 gene (Perrone et al., 1998). Both strands of the XbaI subclone were therefore sequenced. As predicted, the entire pC1 sequence was located close to the 3′ terminus of the XbaI genomic subclone (Figure 3).
To identify the coding sequence, we took advantage of two properties of Chlamydomonas nuclear genes. First, most protein-coding sequences in nuclear genes are strongly biased toward G/C, particularly at the third position of each codon (LeDizet and Piperno, 1995). Therefore, the three reading frames of genomic and pC1 sequences were analyzed with CodonPreference using a codon frequency table based on the Chlamydomonas outer arm γ-heavy chain coding sequence as a standard (Mitchell and Brown, 1994; Wilkerson et al., 1994). In theory, the exons of the gene showing strong codon bias would be located at the codon bias peak. As predicted, the coding sequence of pC1 showed strong codon bias and corresponded to the codon-biased peaks of the genomic sequence. In contrast, the 3′ untranslated region and introns are not biased. Second, a consensus sequence (5′-/GTXXGX … CAG/-3′) for Chlamydomonas exon–intron–exon junctions has been compiled (Silflow, University of Minnesota, unpublished data). As predicted, each codon-biased peak within the region coding for the pC1 sequence was flanked by the exon–intron consensus sequence. Therefore the codon-biased peaks flanked by the consensus sequence using Findpattern were identified as potential exons throughout the gene.
Several sense primers located within the hypothetical exons were designed, and after two additional RT-PCR steps (illustrated in Figure 1), the 1.55-kb clone pC2 and the 550-bp clone pC3 were obtained. The pC2 clone contained the second to the seventh exons, and the pC3 clone contained the first two exons, resulting in recovery of the 5′ untranslated region and the first ATG, which was preceded by a stop codon TGA 18 bp upstream (Figure 3, 962) in the first exon. The genomic sequence along with the predicted exons and amino acid sequence for IC140 are shown in Figure 3.
Sequence Analysis
The IC140 gene consists of 14 exons interrupted by 13 introns illustrated in Figures 1 and 3. The sequences at the exon–intron junction (Figure 3) completely conform to those from most known Chlamydomonas nuclear genes. In addition to several stretches of AT-rich segments, a number of “tub box” sequences were found upstream of the transcription initiation site (Figure 3). It was proposed that the tub box (GCTC[G/C]AAGGC) is involved in the regulation of tubulin transcript during flagellar regeneration and during the cell cycle (Davis and Grossman, 1994). The presence of tub boxes is consistent with the up-regulation of the IC140 transcript after deflagellation as shown by Northern blot (Perrone et al., 1998).
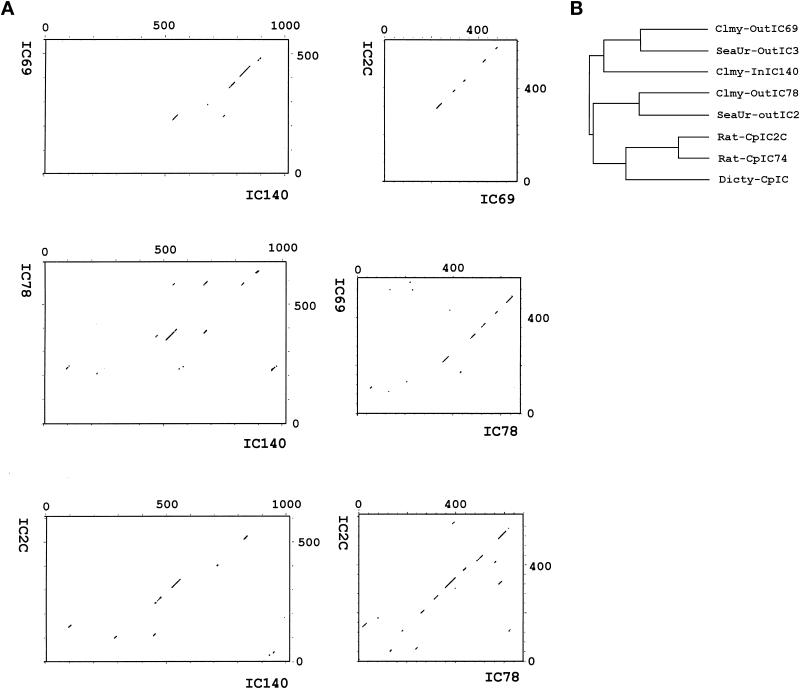
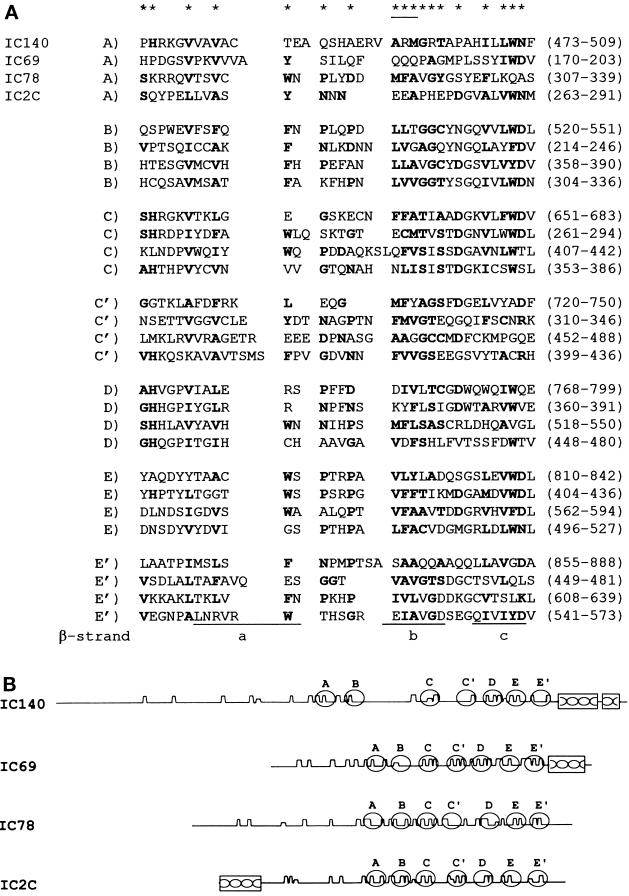
The 3072-bp open reading frame predicts a protein of 1024 amino acids with a mass 110 kDa and a pI of 4.68 (Figure 3). Similar differences between predicted and measured size were also reported for other axonemal proteins (LeDizet and Piperno, 1995; Ogawa et al., 1995; Koutoulis et al., 1997). However, this difference is probably not due to phosphorylation (Habermacher and Sale, 1997). The N-terminal 125 amino acids of IC140 are particularly rich in G and A (44%), D and E (28%), and P (16%). No basic residues are found in this region. As expected, the top of the list of the homologous proteins revealed by BLAST search includes dynein IC family members (smallest sum probability ranging from 4.4 × 10−31 to 1.2 × 10−10). The homologous regions between the ICs, summarized as a dot plot in Figure 4A, are primarily located around and within a set of WD-repeat motifs common to the ICs studied to date (Ogawa et al., 1995; Wilkerson et al., 1995). Analysis from BLAST search, Compare (Figure 4A, dot plot), and PileUp (Figure 4B, dendrogram) suggests that IC140 is most closely related to sea urchin IC3 and Chlamydomonas IC69 of the outer dynein arm (Mitchell and Kang, 1991; Ogawa et al., 1995). Motif search and initial visual evaluation revealed five WD repeats in the middle of IC140 (Figure 3, amino acids 461–842). However, closer examination revealed two additional WD repeats, C′ and E′ (Figure 5, A and B). This assignment of WD repeats is based on the predicted β-sheet position, on the consensus sequence for the WD repeat, and on homologous sequences within the IC family. Based on this new analysis it is possible to assign seven WD/β-sheet repeats in each dynein IC family member, as summarized in Figure 5 (see DISCUSSION). (IC140, IC69, IC78, and IC2 were selected because these four ICs represent distinct classes of IC.) Notably, the WD repeats of the ICs shown in Figure 5B are located in relatively similar positions. One exception is an unusually long variable region of 100 amino acid (aa) residues between the B and C repeats of IC140 (Figure 5B). The variable regions of WD-repeat proteins identified so far consist of 6–94 residues (Neer et al., 1994). Several observations indicate that the long stretch of aa is not a cloning artifact. First, the seventh exon, which contains A and B WD repeats and an 87-aa variable region, is predicted as a continuous strong condon-biased peak. Second, there are no consensus sequences for intron–exon junctions between the B repeat and the end of the seventh exon. Furthermore, the independently recovered pC1 and pC2 overlap and are identical at the 3′ end of the seventh exon. The functional significance of this region remains to be tested.
Figure 4.
Dot plot (A) and dendrogram (B) reveal the homology among the ICs: IC140 of the Chlamydomonas inner arm dynein I1, IC69 and IC78 of Chlamydomonas outer arm dynein, and IC2C of rat cytoplasmic dynein. The dot plots were generated by the program Compare with a stringency 18 to find 18 similarity matching values in a comparison window size of 30 (Maizel and Lenk, 1981). The dendrogram was compiled by the PileUp program using default parameters.
Figure 5.
(A) Sequence alignment of the seven predicted WD/β-sheet repeats in dynein ICs. A–E WD repeats were based on the WD consensus structure discussed by Wilkerson et al. (1995). The C′ and E′ repeats were assigned based on β-sheet location discussed in RESULTS and DISCUSSION. The asterisks indicate the consensus amino acid residues for WD repeats (Neer et al., 1994). The underlined asterisk indicates that two of the three residues were required to match the consensus sequence. The residues in the ICs matching the consensus sequence are shown in bold characters. Underlined regions (a–c) correspond to the first three β-strands as seen in the structure of Gβb. The fourth strand (d) is not illustrated, because it is located in the variable region between WD repeats. (B) Schematic depicting the predicted Chou-Fasman β-sheets (sharp sawtooth wave), WD repeats (open oval), and the coiled coils (box) of the IC proteins listed in Figure 4A.
The regions outside of the WD repeats are mainly composed of α-helices and turns. Analysis with the Coils program predicts two juxtaposed coiled coils (99 and 60% propensity with a window size of 28) in the C terminus (Figure 3, approximately amino acids 920-1020). With the exceptions of IC78 and IC2, the coiled-coil structure is also found in the other IC members, including the C terminus of IC69, IC3, and the N-terminus of IC2C, a cytoplasmic dynein homologue (Figure 5B; see Mitchell and Kang, 1991; Paschal et al., 1992; Ogawa et al., 1995; Wilkerson et al., 1995).
Binding of IC140 to the Axoneme
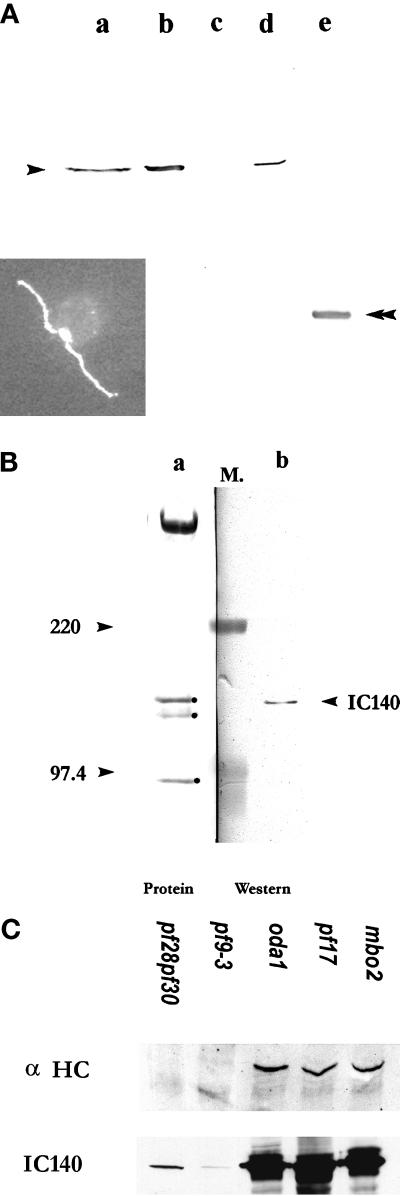
To explore the function of IC140, fusion protein and antibody probes were produced. A N-terminally 6-His tagged 53-kDa fusion protein was overexpressed in E. coli strain BL21(DE3). The cells were transformed with a pET28 (a) expression vector containing the 1.6-kb cDNA from pC1 (Figure 1), inserted in frame. This construct contains the C–E′ WD repeats and the C terminus (Figure 5B). The fusion protein was purified on an Ni+ column and used to raise polyclonal antibodies and for the microtubule binding assays described below. A rabbit polyclonal antibody specifically recognized IC140 on Western blots of isolated dynein I1 (Figure 6A) or isolated flagella and axonemes (Figure 6, A and B). In contrast, the antibody failed to recognize IC138 from the I1 complex (Figure 6B), confirming that IC138 and IC140 are distinct proteins (Habermacher and Sale, 1997; King and Dutcher, 1997). Immunofluorescent staining, using the IC140 antibody, revealed an even distribution of IC140 along the length of both flagellar axonemes in wild-type Chlamydomonas (Figure 6A, inset). The preimmune serum failed to stain IC140 on immunoblots or by immunoflorescence. Western blots of flagella or axonemes from mutant cells lacking the outer dynein arms, radial spoke heads, central pair apparatus, or the mbo2 gene product were indistinguishable from wild type (Figure 6C, oda1, pf17, and mbo2). In contrast, immunoreactivity is detectable but greatly reduced in Western blots of whole axonemes from two strains that fail to assemble the I1 complex (Figure 6C, pf9-3 or pf28pf30) using either colorimetric or enhanced chemiluminescence developing reagents (see MATERIALS AND METHODS). In identical blots, immunoreactivity is completely absent in axonemes from ida7, an I1 mutant defective in the gene for IC140 (see Perrone et al., 1998). The low level of IC140 in pf9-3 and pf28pf30, mutants shown to be defective in an HC gene of I1 (Myster et al., 1997), indicates that IC140 can partially assemble independent of other I1 subunits, consistent with a role for IC140 in anchoring the I1 complex to the doublet microtubule cargo.
Figure 6.
Polyclonal antibody raised against the 53-kDa fusion protein of IC140 specifically recognized IC140. Western blots for Figure 6, A and B, were probed with the anti-IC140 serum followed by HRP-conjugated secondary antibody and colorimetric development. (A) Lane a, wild-type high-salt extract; lane b, pf28 axonemes; lane c, pf28pf30 axonemes; lane d, I1 fraction from sucrose gradient of pf17 dynein extract; lane e, 53-kDa fusion protein. Inset, immunofluorecent microscopy illustrating that IC140 is located along both flagellar axonemes in wild-type Chlamydomonas cells. (B) Lane a, Amido Black staining revealing the proteins of the I1 fraction frompf28 axonemes including 3 ICs: 140, 138, and 97 kDa (dot); lane b, corresponding Western blot of the same 21S fraction. Molecular weight markers (M.) between lanes a and b were used for alignment. (C) Western blots and protein stains of overloaded gels revealed that IC140 is present in minor but detectable quantity in axonemes from pf28pf30 and pf9-3. For comparison, axonemes from control strains oda1, pf17, and mbo2 bear wild-type amounts of IC140. Axonemes (50 μg/lane) were separated in SDS-PAGE (4.5% for α HC, 7% for IC140 and Tubulin). Top panel, Western blot probed with affinity-purified anti-α HC (Myster, et al., 1997). Middle panel, Western blot probed with anti-IC140 serum. Bottom panel, tubulin (25 μg/lane) was revealed by Amido Black staining, showing equal loading of samples.
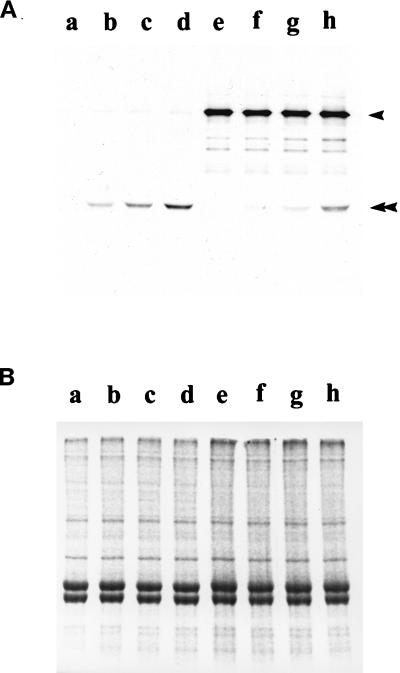
To further test this hypothesis, we examined whether the purified IC140 fusion protein would bind axonemes. The prediction was that the fusion protein would bind doublet microtubules but only in the absence of endogenous I1 complex, when the presumed I1 anchor would be exposed. Axonemes were isolated from either control cells (pf28, lacking the outer dynein arms but bearing the full compliment of inner dynein arms) or experimental, double mutant cells (pf28pf30, lacking both the outer dynein arms as well as the I1 complex: see Smith and Sale, 1992) and mixed with the 53-kDa fusion protein. After reconstitution, the fusion protein specifically sediments with axonemes derived from pf28pf30 (Figure 7). In contrast, little fusion protein binds to axonemes derived from pf28 (Figure 7). Rebinding is not affected by the addition of 1 mM ATP. This result suggests that IC140 is capable of binding directly to the microtubule cargo and may be involved in anchoring the I1 complex to the doublet microtubule. The result also indicates that at least one IC140 domain, responsible for binding, is located within the C-terminal 53 kDa (see Perrone et al., 1998).
Figure 7.
Selective binding of the C1 fusion protein of IC140 to axonemes lacking the I1 complex. Increasing amounts of purified fusion protein were mixed and incubated with isolated axonemes from either pf28pf30, lacking the I1 complex (lanes a–d), or pf28 (lanes e–h) (see MATERIALS AND METHODS). (A) Western blots using anti-IC140 serum, which reveals the 53-kDa fusion protein (double arrowhead) and IC140 (arrowhead) and using HRP secondary antibodies and developed with chemiluminescent reagents. (B) Duplicate gel stained with Commasie brilliant blue, confirming equal loads for each sample.
Cross-linking of IC140 to a Second IC of I1
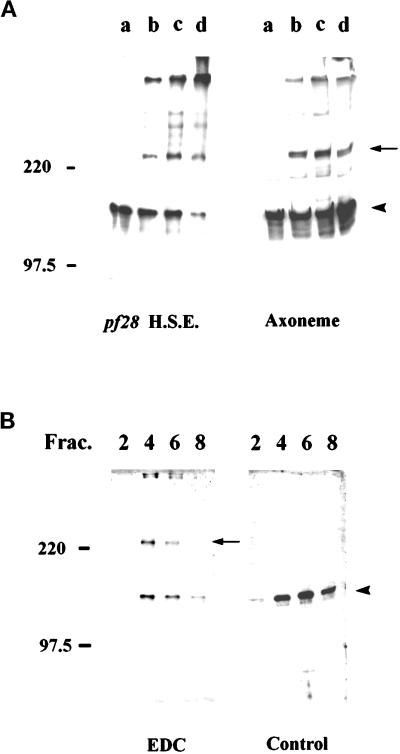
Cross-linking experiments using the zero length cross-linker EDC were performed in an attempt to identify the putative axonemal docking protein to which IC140 binds and to define interacting proteins within the I1 complex (see King et al., 1991; Koutoulis et al., 1997). Western blot analysis of both axonemes and dynein extract after exposure to increasing concentration of EDC revealed a new band migrating at 240 kDa, suggesting that IC140 interacts with a salt-extractable axonemal protein of ∼100 kDa (Figure 8A). For these experiments, axonemes were derived from pf28, lacking the outer dynein arms. However, similar results were obtained using wild-type, pf14, and pf2 cells. We hypothesized the ∼100 kDa protein cross-linked to IC140 is IC97 of the I1 complex. We therefore predicted the 240-kDa cross-linked protein complex would be found in the 19S I1 fraction after salt extraction. To test this, EDC-treated pf28 axonemes were extracted with 0.6 M NaCl/buffer A, and the extract was fractionated by zonal centrifugation using 5–20% sucrose gradients. As predicted, the 240-kDa protein in the cross-linked axoneme was salt extractable and present solely in the I1 fraction (Figure 8A), just as IC140 is present solely in the I1 fraction in the control samples (Figure 8B). The results are consistent with a model in which IC140 is closely associated with one of the other ICs in the I1 complex.
Figure 8.
Western blot analysis, using anti-IC140 antibodies, of EDC cross-linked axonemal proteins. (A) Axonemes (right) or dynein-containing high-salt extracts (left), derived from pf28, were treated with 0, 1.5, 3.5, or 8.5 mM EDC (A, lanes a–d). Notably a novel 240-kDa band (arrow) in addition to IC140 (arrow) appears after exposure to EDC. (B) The same 240-kDa immunoreactive band appears in the 19S-I1 fraction (left, EDC gradient fractions 2, 4, 6, and 8 shown). In contrast, the same gradient fractions of the control, nontreated axonemes do not reveal the 240-kDa band (right, Control, fractions 2, 4, 6, and 8). In each case samples were separated by 5% SDS-PAGE followed by Western blot using anti-IC140 serum and the enhanced chemiluminescent reagents.
DISCUSSION
Isolation of the Gene for IC140
We have recovered the complete genomic and cDNA sequence for IC140 (Figure 3). The basis for this conclusion is founded on a number of independent experiments. First, using DNA-mediated transformation, an 11.5-kb genomic clone rescues a Chlamydomonas mutant lacking the gene for IC140 (see Perrone et al., 1998). Second, the net combined sequences of pC1, pC2, and pC3 (Figure 1) total 3.7 kb, the size predicted from the Northern blots (Figure 2). Because pC1 was derived from a cDNA library, and the RT-PCR clones pC3 and pC2 contain multiple predicted exons, the clones represent the complete mRNA of IC140. Third, through a combination of primer extension and sequencing, the transcription initiation site (only 24 bp upstream of the pC3) was identified (Figure 3, nucleotide 750; see Perrone et al., 1998). Consistent with this result, probes made from the 290-bp XhoI fragment (nucleotides 415–710), which is 5′ of the transcription initiation site, failed to reveal any message. Fourth, the open reading frame predicts a protein of 1024 aa that contains the three peptide sequences obtained by direct protein sequencing (compare sequences in Table 1 and bold letters Figure 3). Finally, a monospecific antibody against a 53-kDa fusion protein constructed with the pC1 insert specifically recognizes IC140 by Western blot analysis.
Structural Domains in IC140: IC140 Is a WD-Repeat Protein
The structural basis for the assembly and docking of dynein is not known. However, sequence and secondary structural predictions for the IC proteins have shown that each is a member of the WD-repeat family of proteins, implying functional homology among the ICs (Paschal et al., 1992; Ogawa et al., 1995; Wilkerson et al., 1995). WD-repeat proteins were originally defined based on a repeating module of approximately defined size that begins with a GH and ends with WD, or respective equivalent residues (van der Voorn and Ploegh, 1992; Neer et al., 1994). In each case the module repeats four to eight times, suggesting that multiple adjacent WD modules are required for protein stability and function. The structural predictions for WD-repeat proteins have been confirmed by the crystal structure of the β subunit of G protein (Wall et al., 1995; Lambright et al., 1996; Sondek et al., 1996). Crystallography reveals that Gβ folds into a circular sevenfold β-sheet “propeller.” Each blade consists of four antiparallel β-strands. However, similar β-propeller structures have also been discovered from non-WD-repeat proteins consisting of repeated β-sheet structures (Faber et al., 1995; Neer and Smith, 1996), suggesting that the consensus expression for defining a WD-repeat protein is insufficient to identify the less conserved repeats predicted to have the similar β-sheet structure.
Based on a WD-repeat consensus sequence and homology among the ICs, Wilkerson et al., (1995) and Ogawa et al., (1995) clearly established that the ICs are WD-repeat proteins. However, many repeats vary considerably from the consensus expression (for example, A and C repeats of IC78; Figure 5A). Therefore, to assign the WD repeats in IC140, we compared WD-repeat regions with the predicted β-sheet structure and the homologous regions of the ICs. We discovered that on this basis each dynein IC contains seven repeated structures, including newly identified C′ and E′ repeats. The C′ and E′ repeats are located in homologous positions among the ICs. Thus, repeating WD/β-sheet units at equivalent positions in different proteins are more similar to each other than to other repeating units within a protein (Figure 5A). For example, the C repeat is more similar to the C repeat between ICs than to other repeats in the same protein, suggesting each repeat serves a conserved function. The simplest model is that each dynein IC contains seven WD/β-sheet repeats, which are likely to be arranged in a sevenfold β-sheet propeller, similar to that described in Gβ.
Another region of conserved structure is the predicted coiled coil present in the C termini of IC140 and IC69 (Figure 5), suggesting a common role. For example, deletion of the C-terminal coiled coil in IC69 resulted in failure to rescue Oda6 (Mitchell, personal communication). We demonstrate here that the fusion protein containing the last five WD repeats and the coiled coil of IC140 specifically bound the axonemes missing I1, suggesting at least part of the targeting domain for I1 is located in the C-terminal region of IC140. Moreover, the N-terminal coiled coils of IC74 are critical for interaction between dynein and dynactin (Vaughan and Vallee, 1995; Burkhardt et al., 1997; Steffen et al., 1997). Because the sheets within the propeller are not necessarily accessible, one plausible region for anchoring could be the coiled coil and/or the loops and variable regions between the WD repeats. The structure outside the WD/β-sheet repeats is predicted to primarily form α-helices and turns. Thus, BLAST searches have identified homology between IC140 and filamentous proteins such as collagen, fibroin, and intermediate filament protein. There may be little significance to these homologies.
Function of IC140
Our primary goal was to test the hypothesis that IC140 anchors the I1 complex to the doublet microtubule, targeting I1 proximal to the first radial spoke. Evidence from two independent experiments is consistent with this hypothesis. First, using an in vitro binding assay we demonstrated that the purified IC140 fusion protein specifically binds to pf28pf30 axonemes, which lack the I1 complex (Figure 7). In contrast, pf28 axonemes, which contain the I1 complex, bind greatly reduced amounts of the fusion protein. The simplest interpretation is that I1 is anchored through interaction of IC140 with the doublet microtubule. Second, low levels of IC140 are assembled, in vivo, onto axonemes derived from other I1 mutants defective in the heavy-chain genes (Figure 6C; also see Myster et al., 1997). The interpretation is that IC140 can be transported and anchored to the axoneme in the absence of assembly of other I1 components. This result is also consistent with a model that IC140 anchors I1 to the microtubule cargo. We therefore predicted that in using in vitro binding experiments the IC140 fusion protein would compete with the isolated I1 complex for binding to pf28pf30 axonemes. However, in preliminary reconstitution experiments, the IC140 fusion protein failed to block the rebinding of the I1 complex. Evidently isolated I1 has a higher affinity than the fusion protein. However the significance of this negative result is unknown.
The observation that IC140 preferentially binds to axonemes derived from pf28pf30 cells supports the hypothesis that IC140 binds to a docking complex, which specifically anchors and positions the I1 complex to the doublet microtubules. Unlike the outer dynein arms, which can bind both axonemal and brain microtubules (Haimo et al., 1979) and can be cross-linked to tubulin (King et al., 1991), the I1 complex does not appear to be directly associated with tubulin. For example, in vitro reconstitution studies demonstrated that isolated I1 would bind to purified, single microtubules, but that binding was prevented in the presence of MgATP (Smith and Sale, 1991). Evidently, the isolated I1 complex does not bind the microtubule by an ATP-insensitive anchoring mechanism. In contrast, purified I1 bound specifically to axonemes, and ATP did not interrupt binding (Smith and Sale, 1992). Sequence analysis demonstrates that IC140 contains no homology to the microtubule binding domains of structural MAPs such as tau, MAP2, and MAP4 (reviewed in Chapin and Bulinski, 1992). Finally, unlike IC78 of outer arm dynein, we did not find that IC140 could be cross-linked to tubulin (Yang and Sale, unpublished results). Therefore, it is not likely that IC140 binds directly to tubulin.
To attempt to directly identify the axonemal protein that anchors I1, we adopted a chemical cross-linking approach. This approach has been used successfully to identify protein–protein interactions in outer arm dynein and radial spokes (King et al., 1991; Diener et al., 1993). However, despite working with two cross-linkers, applying multiple conditions, and taking advantage of a number of Chlamydomonas flagellar mutants, we have failed to identify candidate anchor proteins interacting with IC140. However, we demonstrated that treatment of isolated axonemes with the zero-length cross-linker EDC resulted in a new 240-kDa band that contained IC140 (Figure 8). The simplest explanation for this result is that IC140 was cross-linked to an axonemal protein of ∼100 kDa, and obvious candidates included the other ICs of the I1 complex. To test this, the I1 complex was purified after treatment of isolated axonemes with EDC, and as predicted, the 240-kDa cross-link product is located in the I1 complex only (Figure 8B). Based solely on mass, measured by SDS-PAGE, the simplest interpretation of these data is that IC140 is cross-linked to IC97. However, we cannot yet rule out the possibility that the 240-kDa product is a product of cross-linking IC140 and IC138.
Assuming IC140 and IC97 are closely associated in the I1 complex, it is possible they play a role similar to that of IC69 and IC78 in the outer dynein arm in Chlamydomonas flagella. The basis for predicting that IC140 and IC69 serve related roles includes sequence homology between IC140 and IC69 and C-terminal coiled-coil and related cross-linking studies of the outer dynein arm (King et al., 1991). Our model is that IC140, like IC69, bears a scaffold of multiple domains responsible for interactions with the docking complex and other subunits of the dynein complex. With the discovery of a null mutant for IC140 (Perrone et al., 1998), it is possible to test this model by DNA-mediated transformation using modified IC140 constructs.
ACKNOWLEDGMENTS
We are grateful to Drs. M. Porter and C. Perrone (University of Minnesota) for helpful suggestions and for our collaboration. We are also thankful to Drs. C. Wilkerson and G. Witman (Worcester Foundation/University of Massachusetts) and E. Smith and P. Lefebvre (University of Minnesota) for providing the cDNA and genomic libraries and advice for cloning. Drs. S. Myster and M. Porter (University of Minnesota) generously contributed the 1-α heavy-chain antiserum. Drs. D. Mitchell (State University New York), S. L’Hernault, L. Quarmby, and G. Benian (Emory University) contributed valuable suggestions. This work was supported by grants from the National Institute of General Medical Sciences, National Institutes of Health.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers E W, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Brokaw CJ, Kamiya R. Bending patterns on Chlamydomonas flagella: IV. Mutants with defects in inner and outer arms indicate differences in dynein arm function. Cell Motil Cytoskeleton. 1987;8:68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- Burgess SA, Carter DA, Dover SD, Woolley DM. The inner dynein arm complex: compatible images from freeze-etch and thin section methods of microscopy. J Cell Sci. 1991;100:319–328. doi: 10.1242/jcs.100.2.319. [DOI] [PubMed] [Google Scholar]
- Burkhardt JK, Echeverri CJ, Nilsson T, Vallee RB. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin S, Bulinski JC. Microtubule stabilization by assembly promoting MAPs: a repeat performance. Cell Motil Cytoskeleton. 1992;23:236–243. doi: 10.1002/cm.970230403. [DOI] [PubMed] [Google Scholar]
- Davis JP, Grossman AR. Sequence controlling transcription of the Chlamydomonas reinhardtii β2-tubulin gene after deflagellation and during the cell cycle. Mol Cell Biol. 1994;14:5165–5174. doi: 10.1128/mcb.14.8.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener DR, Ang LH, Rosenbaum JL. Assembly of flagellar radial spoke proteins in Chlamydomonas: identification of axonemal binding domain of RSP3. J Cell Biol. 1993;123:183–190. doi: 10.1083/jcb.123.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber HR, Groom CR, Baker HM, Morgan WT, Smith A, Baker EN. 1.8 Å crystal structure of the C-terminal domain of rabbit serum haemopexin. Structure. 1995;3:551–559. doi: 10.1016/s0969-2126(01)00189-7. [DOI] [PubMed] [Google Scholar]
- Gardner LC, O’Toole E, Perrone CA, Giddings T, Porter ME. Components of a “Dynein Regulatory Complex” are located at the junction between the radial spokes and the dynein arms in Chlamydomonas flagella. J Cell Biol. 1994;127:1311–1325. doi: 10.1083/jcb.127.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, Gebhart B, Memall V, Mitchell DR, Heuser JE. HPLC fractionation of Chlamydomonas dynein extracts and characterization of inner arm dynein subunits. J Mol Biol. 1987;194:481–494. doi: 10.1016/0022-2836(87)90676-0. [DOI] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE. Substructure of the outer dynein arm. J Cell Biol, 1983;95:798–815. doi: 10.1083/jcb.95.3.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE. Structural comparison of purified proteins with in situ dynein arms. J Mol Biol. 1984;180:1083–1118. doi: 10.1016/0022-2836(84)90272-9. [DOI] [PubMed] [Google Scholar]
- Habermacher G, Sale WS. Regulation of flagellar dynein by phosphorylation of a 138 kDa inner arm dynein intermediate chain. J Cell Biol. 1997;136:167–176. doi: 10.1083/jcb.136.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimo LT, Telzer B, Rosenbaum JL. Dynein binds and crossbridges cytoplasmic microtubules. Proc Natl Acad Sci USA. 1979;76:5759–5763. doi: 10.1073/pnas.76.11.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison A, Olds-Clarke P, King SM. Identification of the t complex-encoded cytoplasmic dynein light chain Tctex1 in inner arm I1 supports the involvement of flagellar dyneins in meiotic drive. J Cell Biol. 1998;140:1137–1147. doi: 10.1083/jcb.140.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzbaur ELF, Vallee RB. Dyneins: molecular structure and cellular function. In: Spudich JA, McKnight SL, Schekman R, editors. Annual Review of Cell Biology. Palo Alto, CA: Annual Reviews; 1994. pp. 339–372. [DOI] [PubMed] [Google Scholar]
- Innis MA. PCR with 7-deaza-2′-deoxyguanosine triphos-phage. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols. San Diego: Academic Press; 1990. pp. 54–59. [Google Scholar]
- Johnson KA, Rosenbaum JL. Polarity of flagellar assembly in Chlamydomonas. J Cell Biol. 1992;119:1605–1611. doi: 10.1083/jcb.119.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami O, Kamiya R. Translocation and rotation of microtubules caused by multiple species of Chlamydomonas inner arm dynein. J Cell Sci. 1992;103:653–664. [Google Scholar]
- Kamiya R, Kurimoto E, Muto E. Two types of Chlamydomonas flagellar mutants missing different components of inner arm dynein. J Cell Biol. 1991;112:441–447. doi: 10.1083/jcb.112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki S, Holzbaur ELF. Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J Biol Chem. 1995;270:28806–28811. doi: 10.1074/jbc.270.48.28806. [DOI] [PubMed] [Google Scholar]
- King SJ, Dutcher SK. Phosphorylation of an inner dynein arm complex in Chlamydomonas reinhardtii is altered in phototactic mutant strains. J Cell Biol. 1997;136:177–191. doi: 10.1083/jcb.136.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Patel-King RS, Wilkerson CG, Witman GB. The 78,000-Mr intermediate chain of Chlamydomonas outer arm dynein is a microtubule-binding protein. J Cell Biol. 1995;131:399–409. doi: 10.1083/jcb.131.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Wilkerson CG, Witman GB. The Mr 78,000 intermediate chain of Chlamydomonas outer arm dynein interacts with α-tubulin in situ. J Biol Chem. 1991;266:8401–8407. [PubMed] [Google Scholar]
- King SM, Witman GB. Localization of an intermediate chain of outer arm dynein by immunoelectron microscopy. J Biol Chem. 1990;265:19807–19811. [PubMed] [Google Scholar]
- Koutoulis A, Pazour GJ, Wilkerson CG, Inaba K, Sheng HS, Takada S, Witman GB. The Chlamydomonas reinhardtii ODA3 gene encodes a protein of the outer arm docking complex. J Cell Biol. 1997;137:1069–1080. doi: 10.1083/jcb.137.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambright DG, Sondek J, Bohm A, Skiba N, Hamm HE, Sigler PB. The 2.0 Å crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- LeDizet M, Piperno G. The light chain p28 associates with a subset of inner dynein arm heavy chains in Chlamydomonas axonemes. Mol Biol Cell. 1995;6:697–711. doi: 10.1091/mbc.6.6.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre PA, Rosenbaum JL. Regulation and assembly of flagellar proteins and flagellar proteins during regeneration. Annu Rev Cell Biol. 1986;2:517–546. doi: 10.1146/annurev.cb.02.110186.002505. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Maizel JV, Lenk RP. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc Natl Acad Sci USA. 1981;78:7665–7669. doi: 10.1073/pnas.78.12.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D, O’Toole E, McDonald K, McIntosh JR, Porter ME. Arrangement on inner dynein arms in wild-type and mutant flagella of Chlamydomonas. J Cell Biol. 1992;118:1145–1162. doi: 10.1083/jcb.118.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR. Cell and molecular biology of flagellar dyneins. In: Jeon KW, Jarvik J, editors. International Reviews of Cytology. San Diego: Academic Press; 1994. pp. 141–175. [DOI] [PubMed] [Google Scholar]
- Mitchell D, Brown K. Sequence analysis of the Chlamydomonas alpha and beta dynein heavy chain genes. J Cell Sci. 1994;107:635–644. doi: 10.1242/jcs.107.3.635. [DOI] [PubMed] [Google Scholar]
- Mitchell DR, Kang Y. Identification of oda6 as a Chlamydomonas dynein mutant by rescue of the wild-type gene. J Cell Biol. 1991;113:835–842. doi: 10.1083/jcb.113.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR, Rosenbaum JL. A motile Chlamydomonas flagellar mutant that lacks outer dynein arms. J Cell Biol. 1985;100:1228–1234. doi: 10.1083/jcb.100.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myster SH, Knott JA, O’Toole E, Porter ME. The Chlamydomonas Dhc1 gene encodes a dynein heavy chain subunit required for assembly of the I1 inner arm complex. Mol Biol Cell. 1997;8:607–620. doi: 10.1091/mbc.8.4.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neer EJ, Schmidt R, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- Neer EJ, Smith TF. G protein heterodimers: new structures propel new questions. Cell. 1996;84:175–178. doi: 10.1016/s0092-8674(00)80969-1. [DOI] [PubMed] [Google Scholar]
- Ogawa K, Kamiya R, Wilkerson CG, Witman GB. Interspecies conservation of outer arm dynein intermediate chain sequences defines two intermediate chain subclasses. Mol Biol Cell. 1995;6:685–696. doi: 10.1091/mbc.6.6.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal BM, Mikami A, Pfister KK, Vallee RB. Homology of the 74 kD cytoplasmic dynein subunit with a flagellar dynein polypeptide suggests an intracellular targeting function. J Cell Biol. 1992;118:1133–1143. doi: 10.1083/jcb.118.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone CA, Yang P, O’Toole E, Sale WS, Porter ME. The Chlamydomonas IDA7 locus encodes a 140-kDa dynein intermediate chain required to assemble the I1 inner arm complex. Mol Biol Cell. 1998;9:3351–3365. doi: 10.1091/mbc.9.12.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Ramanis Z. The proximal portion of the Chlamydomonas flagella contains a distinct set of inner dynein arms. J Cell Biol. 1991;112:701–709. doi: 10.1083/jcb.112.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Ramanis Z, Smith EF, Sale WS. Three distinct inner dynein arms in Chlamydomonas flagella: molecular composition and location in the axoneme. J Cell Biol. 1990;110:379–389. doi: 10.1083/jcb.110.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME. Axonemal dyneins: assembly, organization, and regulation. Curr Opin Cell Biol. 1996;8:10–17. doi: 10.1016/s0955-0674(96)80042-1. [DOI] [PubMed] [Google Scholar]
- Porter ME, Power J, Dutcher SK. Extragenic suppressors of paralyzed flagellar mutations in Chlamydomonas reinhardtii identify loci that alter the inner dynein arms. J Cell Biol. 1992;18:1163–1176. doi: 10.1083/jcb.118.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale WS, Goodenough UW, Heuser JE. The substructure of isolated and in situ outer dynein arms of sea urchin sperm flagella. J Cell Biol. 1985;101:1400–1412. doi: 10.1083/jcb.101.4.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell R, Lefebvre PA. Isolation of the Chlamydomonas regulatory gene NIT2 by transposon tagging. Genetics. 1993;134:737–747. doi: 10.1093/genetics/134.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow CD, Rosenbaum JL. Multiple alpha- and beta-tubulin genes in Chlamydomonas and regulation of tubulin mRNA levels after deflagellation. Cell. 1981;24:81–88. doi: 10.1016/0092-8674(81)90503-1. [DOI] [PubMed] [Google Scholar]
- Smith EF, Lefebvre PA. PF20 gene product contains WD repeats and localizes to the intermicrotubule bridge in Chlamydomonas flagella. Mol Biol Cell. 1997;8:455–467. doi: 10.1091/mbc.8.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EF, Sale WS. Microtubule binding and translocation by inner dynein arm subtype I1. Cell Motil Cytoskeleton. 1991;18:258–268. doi: 10.1002/cm.970180403. [DOI] [PubMed] [Google Scholar]
- Smith EF, Sale WS. Structural and functional reconstitution of inner dynein arms in Chlamydomonas flagellar axonemes. J Cell Biol. 1992;117:573–581. doi: 10.1083/jcb.117.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondek J, Bohm A, Lambright DG, Hamm HE, Sigler PB. Crystal structure of a GA protein βγ dimer at 2.1 Å resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- Steffen W, Hodgkinson JL, Wiche G. Immunogold localization of the intermediate chain within the protein complex of cytoplasmic dynein. J Struct Biol. 1996;117:227–235. doi: 10.1006/jsbi.1996.0087. [DOI] [PubMed] [Google Scholar]
- Steffen W, Karki S, Vaughan KT, Vallee RB, Holzbaur ELF, Weiss D, Kuznetsov S. The involvement of the intermediate chain of cytoplasmic dynein in binding the motor complex to the membranous organelles of Xenopus oocytes. Mol Biol Cell. 1997;8:2077–2088. doi: 10.1091/mbc.8.10.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Voorn L, Ploegh HL. The WD-40 repeat. FEBS Lett. 1992;307:131–134. doi: 10.1016/0014-5793(92)80751-2. [DOI] [PubMed] [Google Scholar]
- Vaughan KT, Vallee RB. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150glued. J Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall MA, Coleman DE, Lee E, Iniguez-Lluhi JA, Gilman PA, Sprang SR. The structure of the G protein heterotrimer Giα1Giαa1β1γ2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- Wilkerson CG, King SM, Koutoulis A, Pazour GJ, Witman GB. The 78,000 Mr intermediate chain of Chlamydomonas outer arm dynein is a WD-repeat protein required for arm assembly. J Cell Biol. 1995;129:169–178. doi: 10.1083/jcb.129.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson CG, King SM, Witman GB. Molecular analysis of the γ heavy chain of Chlamydomonas flagellar outer arm dynein. J Cell Sci. 1994;107:497–506. doi: 10.1242/jcs.107.3.497. [DOI] [PubMed] [Google Scholar]
- Witman GB. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 1986;134:280–290. doi: 10.1016/0076-6879(86)34096-5. [DOI] [PubMed] [Google Scholar]
- Witman GB, Wilkerson CG, King SM. Microtubules. H.S. Hyams and C.W. Lloyd, New York: Wiley-Liss; 1994. The biochemistry, genetics and molecular biology of flagellar dynein; pp. 229–249. [Google Scholar]
- Woolley DM. Studies on the eel sperm flagellum. I. The structure of the inner arm complex. J Cell Sci. 1997;110:85–94. doi: 10.1242/jcs.110.1.85. [DOI] [PubMed] [Google Scholar]