Abstract
Objective
To describe the clinical features of and genetic locus associated with autosomal‐dominant macular dystrophy (MCDR5) in a large Greek family.
Methods
26 members of a single family underwent clinical examinations and venepuncture. A genomewide linkage scan using 400 microsatellite markers distributed with an average spacing of 10 cM throughout the human genome.
Results
14 members of the study family exhibited clinical features of the disease including decreased central vision and macular abnormalities in the posterior pole of the retina. Analysis of loci known to be associated with macular dystrophy did not show positive linkage. A genomewide linkage scan showed linkage to chromosome 19q, with a two‐point maximum LOD score of 5.809 at θ = 0 between the disease and marker locus D19S412. On the basis of recombination events, the disease interval was localised between markers D19S420 and D19S540 on chromosome 19q, at a span of about 3.8 cM, in an area known to contain 120 known genes/transcripts. Eleven of these genes/transcripts were sequenced, and no disease‐causing mutation was identified.
Conclusions
This study describes a new locus on 19q associated with autosomal‐dominant macular dystrophy, designated as MCDR5. Additional study of other family members will be necessary to further narrow the interval and identify the responsible gene. The study of MCDR5 will aid in elucidation of the underlying pathogenic mechanisms for this and other macular diseases, including age‐related macular degeneration.
Macular dystrophies can present with autosomal dominant, autosomal recessive, X‐linked recessive and mitochondrial inheritance. The autosomal dominant form of macular dystrophy is a heterogeneous group of disorders that typically present within the first two decades of life with progressive central visual loss and macular atrophy. Ophthalmoscopic findings include bilateral atrophic macular lesions with or without subretinal deposits at the level of the retinal pigment epithelium (RPE). The resulting central visual loss is variable and can be severe with visual acuities ranging between 20/20 and 20/400. So far, 12 loci for autosomal dominant forms of macular dystrophy have been localised to particular regions of the human genome.1,2 They include Stargardt‐like macular dystrophy (STGD3 and STGD4).3,4 Best macular dystrophy,5 adult vitelliform dystrophy and pattern dystrophy,1,2,6,7 Doyne honeycomb retinal dystrophy,8 progressive bifocal chorioretinal atrophy,9 Sorsby's fundus dystrophy,10 central areolar choroidal dystrophy,11,12 dominant cystoid macular dystrophy,13 North Carolina macular dystrophy (MCDR1),14,15 autosomal dominant “bull's eye” macular dystrophy (MCDR2),16 autosomal dominant macular dystrophy resembling MCDR1 (MCDR3)17 and North Carolina‐like macular dystrophy associated with deafness (MCDR4).18 Five genes have been identified.6,7,8,11,19,20,21
In this report, we describe a large Greek family with autosomal‐dominant macular dystrophy that maps to a new disease locus on chromosome 19. Linkage was established with a two‐point maximum LOD score of 5.809 at θ = 0 between the disease and marker locus D19S412. We designated this locus MCDR5.
Methods
Study subjects
This project was approved by the Institutional Review Board of the Aghia Sophia Children's Hospital, Athens, Greece and the Institutional Review Board of the University of Utah Health Sciences Center, Salt Lake City, Utah, USA. Twenty six members of the study family, 11 females and 15 males, were included. Informed consent was obtained from all members. In all, 14 affected and 12 unaffected members underwent ophthalmic examination including best corrected Snellen visual acuity determination and fundus examination. Members were designated as affected on the basis of decreased visual acuity and the presence of flecks with or without atrophic macular lesions. Fluorescein angiography was carried out on five members.
Genotyping and linkage analysis
Blood was collected by venepuncture and genomic DNA was isolated from the samples with the Puregene genomic DNA purification kit (Gentra Systems, Minneapolis, Massachusetts, USA) according to the manufacturer's instructions. A genomewide linkage scan was carried out using 400 microsatellite markers distributed with an average spacing of 10 cM throughout the human genome (Proligo LLC, Boulder, Colorado, USA). Forward primers were labelled with fluorescent dye D2, D3 or D4 and samples were amplified using the polymerase chain reaction (PCR) and a standard cycling programme of 30 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s. A 1 μl sample of PCR product and 0.5 μl internal size standard (Size Standard Kit 400, Beckman‐Coulter, Fullerton, California, USA) were combined in a total volume of 40 μl sample loading solution (SLS, Beckman‐Coulter) and loaded on to the CEQ 8000 Genetic Analysis System (Beckman‐Coulter).
Electrophoresis was carried out using the following programme: (a) 0.5 μl sample injection; (b) 5 min DNA strand denaturation at 93°C; (c) separation at 6000 V at 50°C; (d) signal detection with calibrated D2, D3 or D4 emission spectra. Fragment size determinations were carried out using the default fragment analysis parameters of the CEQ 8000 software. Two‐point LOD scores were calculated using the subroutine MLINK of the LINKAGE program (V.5.1; http://www.hgmp.mrc.ac.uk/; Human Genome Mapping Project Resources Center, Cambridge, UK).22,23 An autosomal‐dominant mode of inheritance with full penetrance and a disease allele frequency of 0.0001 were assumed in the computations. Additional microsatellite markers for fine mapping on chromosome 19 were chosen from the Marshfield database, amplified using PCR (30 cycles at 94°C for 30 s, 55°C for 30 s and 72°C for 45 s), labelled with α[32P]‐2′‐deoxycytidine 5′‐triphosphate, and separated by electrophoresis on a 6.6% denaturing polyacrylamide gel.24,25
DNA sequence analysis
Direct sequencing of GNG8, SIX5, ZNF224, XTP7, GPR4, FKRP, ZNF45, ZNF342, PLAUR, CCDC8, RTN2,RDS,ELOVL4 and CRX was carried out using the Taq Dyedeoxy Terminator Cycle Sequencing Kit (Beckman‐Coulter). For RDS, ELOVL4 and CRX, the primers used were described previously.19,26,27 For the other genes, primers were designed to amplify the complete coding regions and intron splice sites. Amplified products were purified using the QIAquik Gel Extraction Kit (Qiagen, Valencia, California, USA) and sequenced with forward and reverse primers using the Taq Dyedeoxy Terminator Cycle Sequencing Kit (Beckman‐Coulter) according to the manufacturer's instructions.
Results
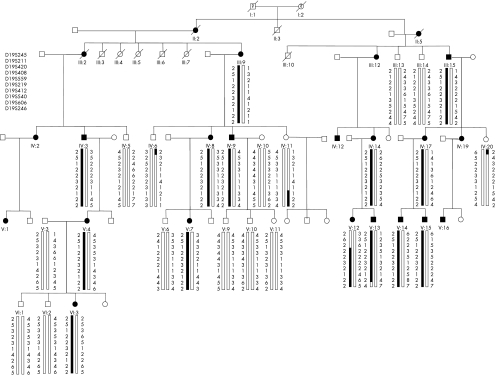
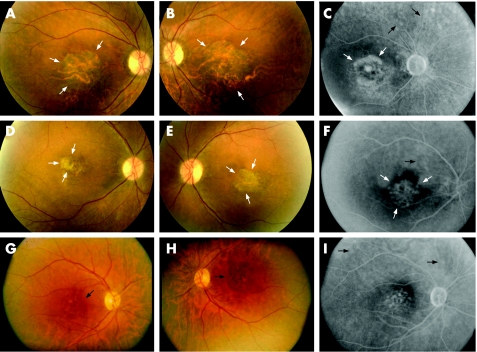
Twenty six members of a large Greek family spanning four generations were included in this study. In all, 14 members were affected and 12 were unaffected (fig 1). All 26 family members underwent clinical examination and ophthalmoscopy. Ages of examined members ranged from 14 to 88 years. Of the 26 individuals examined, 11 were females and 15 were males. Affected patients experienced decreased central vision in the second decade of life and exhibited fundus changes including central macular atrophy with or without subretinal deposit surrounding the macula (fig 2). No dark choroid phenomena were seen on fluorescein angiography (fig 2). Colour vision and night vision were not affected.
Figure 1 Pedigree of a large Greek family with autosomal‐dominant macular dystrophy. Haplotypes of microsatellite markers spanning the linked region on 19q13.2–q13.3 are shown. Affected members are identified by solid squares (males) or solid circles (women). Unaffected members are identified by open symbols; deceased members are indicated by a slash (/). The clinical status of deceased grandparents (I‐1 and I‐2) is not known. Solid bars denote the haplotype that segregates with the disease phenotype.
Figure 2 Fundus photographs and fluorescein angiograms of three affected members: IV:9 (A–C), 49 years old, visual acuity (VA): 20/800 OD, 20/800 OS; IV:8 (D–F), 53 years old, VA: 20/400 OD, 20/400 OS; V:4 (G–I), 49 years old, VA: 20/50 OD, 20/70 OS. Fundus photographs and angiograms exhibit central macular atrophy (white arrows) and subretinal deposits (black arrows).
Polymorphic STR markers surrounding loci previously known to be associated with autosomal dominant macular dystrophy, cone dystrophy (COD), central areolar choroidal dystrophy (CACD) and cone‐rod dystrophy (CORD) were examined first. These included RDS/Peripherin on 6p21.1‐cen, MCDR1 on 6q14‐q16.2, MCDR2 and STGD4 on 4p, MCDR3 on 5p15.33‐p13.1, MCDR4 on 14p, EFEMP1 on 2p16, autosomal dominant butterfly‐shaped macular dystrophy on 5q21.2‐q33, CORD7 on 6cen‐q14, STGD3 on 6q14, VMD2 on 11q13, CACD on 17p, CORD5 on 17p13‐p12 and TIMP3 on 22p12.1‐q13.1,2,3,4,11,12,13,14,15,19,21,27,28,29,30 No marked linkage was found to any of these loci. Additionally, direct sequencing of the coding regions of RDS/Peripherin, ELOVL4 and CRX for affected member III:9 did not show any mutations. We then carried out a whole genome scan using 400 microsatellite markers spaced at 10 cM intervals throughout the human genome.
Results of the genome scan showed that the disease phenotype was linked to marker D19S559. Negative or non‐significant two‐point LOD scores were obtained for all other test loci. This locus was refined using the following markers: D19S245, D19S211, D19S420, D19S408, D19S559, D19S219, D19S412, D19S540, D19S606 and D19S246. Table 1 shows the LOD scores. A maximum two‐point LOD score of 5.809 was obtained at θ = 0 with marker D19S412. The LOD scores for loci D19S559, D19S219 and D19S412 were all >4.
Table 1 Two‐point LOD score between macular dystrophy and STR markers on 19q.
| Markers | LOD scores at different recombination fractions (θ) | ||||
|---|---|---|---|---|---|
| 0 | 0.1 | 0.2 | 0.3 | 0.4 | |
| D19S245 | −5.5743 | 0.49704 | 0.82841 | 0.7649 | 0.46702 |
| D19S211 | −4.2199 | 2.0171 | 1.6731 | 1.103 | 0.4586 |
| D19S420 | −6.8152 | 2.3554 | 2.0798 | 1.4075 | 0.6123 |
| D19S408 | 2.8386 | 4.6271 | 3.6037 | 2.3543 | 1.0499 |
| D19S559 | 5.4475 | 4.4733 | 3.4061 | 2.2386 | 1.0182 |
| D19S219 | 4.4364 | 3.6128 | 2.7689 | 1.8829 | 0.9381 |
| D19S412 | 5.8086 | 4.7384 | 3.5628 | 2.2856 | 0.9876 |
| D19S540 | −1.6092 | 3.9727 | 3.2824 | 2.2841 | 1.1271 |
| D19S606 | −3.1395 | 4.8823 | 3.8078 | 2.5003 | 1.1291 |
| D19S246 | −5.3555 | 3.3266 | 2.8207 | 1.9723 | 0.9725 |
Extended haplotypes were constructed using markers in the following order: D19S420–D19S408–D19S559–D19S219–D19S412–D19S540–D19S606–D19S246. The disease‐associated haplotype was determined as a common extended haplotype for all affected members. On the basis of recombination events, the disease interval was localised between markers D19S420 and D19S540 on chromosome 19q, at a span of about 3.8 cM. No recombination was detected at loci D19S408, D19S559, D19S219 or D19S412 (fig 1). There are about 120 known genes/transcripts within this interval. Among these, 11 (GNG8, SIX5, ZNF224, XTP7, GPR4, FKRP, ZNF45, ZNF342, PLAUR, CCDC8, RTN2) were sequenced, and no disease‐causing mutation was identified.
Discussion
Here we describe a new locus, MCDR5, on 19q associated with autosomal dominant macular dystrophy. This locus shows variable expressivity evidenced by variable fundus appearance and visual acuities in affected patients. In addition, there is no dark choroid on fluorescein angiography. Several loci and genes have been identified for an autosomal dominant form of macular dystrophy. We now add to this list a new locus, on the basis of the description of a large Greek family with autosomal‐dominant macular dystrophy that links to a locus on chromosome 19q13.2–13.3. Recombination and haplotype analyses have narrowed the interval containing the gene responsible for this disease to a region spanning 3.8 cM between markers D19S420 and D19S540. Additional study of other family members will be necessary to further narrow the interval.
The identification of this locus will hopefully lead to further understanding and treatment of this disabling disease. The study of MCDR5 and other similar hereditary macular disorders will also potentially help elucidate the complex mechanisms behind age‐related macular degeneration, the most common cause of irreversible blindness in older people in the developed world. Through investigation of early‐onset macular dystrophy associated with single‐gene mutations that share clinical and histopathological features with age‐related macular degeneration, it is possible to identify pathways that are common to these diseases. In this manner, a better understanding of the pathogenesis of retinal degeneration and macular dystrophies can be attained. This knowledge will hopefully lead to earlier diagnosis and new strategies for prevention and treatment.
Abbreviations
CORD - cone‐rod dystrophy
PCR - polymerase chain reaction
RPE - retinal pigment epithelium
Footnotes
Funding: K Zhang was supported by grants from the U.S. National Institutes of Health (NIH) NIH RO1 EY14428, NIH RO1 EY14448; the American Health Assistance Foundation; the Karl Kirchgessner Foundation; the Ruth and Milton Steinbach Fund; Ronald McDonald House Charities; Val and Edith Green Foundation and an unrestricted grant to the Department of Ophthalmology and Visual Sciences from Research to Prevent Blindness, New York, NY: Z Yang was supported by grants from the Department of Science and Technology of Sichuan Province, Sichuan, People's Republic of China.
Competing interests: None declared.
References
- 1.Michaelides M, Hunt D M, Moore A T. The genetics of inherited macular dystrophies. J Med Genet 200340641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.den Hollander A I, van Lith‐Verhoeven J J, Kersten F F.et al Identification of novel locus for autosomal dominant butterfly shaped macular dystrophy on 5q21.2–q33.2. J Med Genet 200441699–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stone E M, Nichols B E, Kimura A E.et al Clinical features of a Stargardt‐like dominant progressive macular dystrophy with genetic linkage to chromosome 6q. Arch Ophthalmol 1994112765–772. [DOI] [PubMed] [Google Scholar]
- 4.Kniazeva M, Chiang M F, Morgan B.et al A new locus for autosomal dominant stargardt‐like disease maps to chromosome 4. Am J Hum Genet 1999641394–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forsman K, Graff C, Nordstrom S.et al The gene for Best's macular dystrophy is located at 11q13 in a Swedish family. Clin Genet 199242156–159. [DOI] [PubMed] [Google Scholar]
- 6.Felbor U, Schilling H, Weber B H. Adult vitelliform macular dystrophy is frequently associated with mutations in the peripherin/RDS gene. Hum Mutat 199710301–309. [DOI] [PubMed] [Google Scholar]
- 7.Nichols B E, Sheffield V C, Vandenburgh K.et al Butterfly‐shaped pigment dystrophy of the fovea caused by a point mutation in codon 167 of the RDS gene. Nat Genet 19933202–207. [DOI] [PubMed] [Google Scholar]
- 8.Stone E M, Lotery A J, Munier F L.et al A single EFEMP1 mutation associated with both Malattia Leventinese and Doyne honeycomb retinal dystrophy. Nat Genet 199922199–202. [DOI] [PubMed] [Google Scholar]
- 9.Kelsell R E, Godley B F, Evans K.et al Localization of the gene for progressive bifocal chorioretinal atrophy (PBCRA) to chromosome 6q. Hum Mol Genet 199541653–1656. [DOI] [PubMed] [Google Scholar]
- 10.Weber B H, Vogt G, Wolz W.et al Sorsby's fundus dystrophy is genetically linked to chromosome 22q13‐qter. Nat Genet 19947158–161. [DOI] [PubMed] [Google Scholar]
- 11.Hoyng C B, Heutink P, Testers L.et al Autosomal dominant central areolar choroidal dystrophy caused by a mutation in codon 142 in the peripherin/RDS gene. Am J Ophthalmol 1996121623–629. [DOI] [PubMed] [Google Scholar]
- 12.Lotery A J, Ennis K T, Silvestri G.et al Localisation of a gene for central areolar choroidal dystrophy to chromosome 17p. Hum Mol Genet 19965705–708. [DOI] [PubMed] [Google Scholar]
- 13.Kremer H, Pinckers A, van den Helm B.et al Localization of the gene for dominant cystoid macular dystrophy on chromosome 7p. Hum Mol Genet 19943299–302. [DOI] [PubMed] [Google Scholar]
- 14.Small K W, Weber J L, Roses A.et al North Carolina macular dystrophy is assigned to chromosome 6. Genomics 199213681–685. [DOI] [PubMed] [Google Scholar]
- 15.Reichel M B, Kelsell R E, Fan J.et al Phenotype of a British North Carolina macular dystrophy family linked to chromosome 6q. Br J Ophthalmol 1998821162–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michaelides M, Johnson S, Poulson A.et al An autosomal dominant bull's‐eye macular dystrophy (MCDR2) that maps to the short arm of chromosome 4. Invest Ophthalmol Vis Sci 2003441657–1662. [DOI] [PubMed] [Google Scholar]
- 17.Michaelides M, Johnson S, Tekriwal A K.et al An early‐onset autosomal dominant macular dystrophy (MCDR3) resembling North Carolina macular dystrophy maps to chromosome 5. Invest Ophthalmol Vis Sci 2003442178–2183. [DOI] [PubMed] [Google Scholar]
- 18.Francis P J, Johnson S, Edmunds B.et al Genetic linkage analysis of a novel syndrome comprising North Carolina‐like macular dystrophy and progressive sensorineural hearing loss. Br J Ophthalmol 200387893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang K, Kniazeva M, Han M.et al A 5‐bp deletion in ELOVL4 is associated with two related forms of autosomal dominant macular dystrophy. Nat Genet 20012789–93. [DOI] [PubMed] [Google Scholar]
- 20.Petrukhin K, Koisti M J, Bakall B.et al Identification of the gene responsible for Best macular dystrophy. Nat Genet 199819241–247. [DOI] [PubMed] [Google Scholar]
- 21.Weber B H, Vogt G, Pruett R C.et al Mutations in the tissue inhibitor of metalloproteinases‐3 (TIMP3) in patients with Sorsby's fundus dystrophy. Nat Genet 19948352–356. [DOI] [PubMed] [Google Scholar]
- 22.Lathrop G M, Lalouel J M, Julier C.et al Multilocus linkage analysis in humans: detection of linkage and estimation of recombination. Am J Hum Genet 198537482–498. [PMC free article] [PubMed] [Google Scholar]
- 23.Lathrop G M, Lalouel J M, Julier C.et al Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 1984813443–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dib C, Faure S, Fizames C.et al A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 1996380152–154. [DOI] [PubMed] [Google Scholar]
- 25.Broman K W, Murray J C, Sheffield V C.et al Comprehensive human genetic maps: individual and sex‐specific variation in recombination. Am J Hum Genet 199863861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, Lin W, Moshfeghi D M.et al A novel mutation in the RDS/Peripherin gene causes adult‐onset foveomacular dystrophy. Am J Ophthalmol 2003135213–218. [DOI] [PubMed] [Google Scholar]
- 27.Freund C L, Gregory‐Evans C Y, Furukawa T.et al Cone‐rod dystrophy due to mutations in a novel photoreceptor‐specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell 199791543–553. [DOI] [PubMed] [Google Scholar]
- 28.Travis G H, Christerson L, Danielson P E.et al The human retinal degeneration slow (RDS) gene: chromosome assignment and structure of the mRNA. Genomics 199110733–739. [DOI] [PubMed] [Google Scholar]
- 29.Kelsell R E, Gregory‐Evans K, Gregory‐Evans C Y.et al Localization of a gene (CORD7) for a dominant cone‐rod dystrophy to chromosome 6q. Am J Hum Genet 199863274–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balciuniene J, Johansson K, Sandgren O.et al A gene for autosomal dominant progressive cone dystrophy (CORD5) maps to chromosome 17p12–p13. Genomics 199530281–286. [DOI] [PubMed] [Google Scholar]