Abstract
Prolidase deficiency (PD) is a rare autosomal recessive connective tissue disorder caused by mutations in the prolidase gene. The PD patients show a wide range of clinical outcomes characterised mainly by intractable skin ulcers, mental retardation and recurrent respiratory infections. Here we describe five different PEPD mutations in six European patients. We identified two new PEPD mutant alleles: a 13 bp duplication in exon 8, which is the first reported duplication in the prolidase gene and a point mutation resulting in a change in amino acid E412, a highly conserved residue among different species. The E412K substitution is responsible for the first reported phenotypic variability within a family with severe and asymptomatic outcomes.
Prolidase deficiency (OMIM 170100) is a rare autosomal recessive disorder of the connective tissue caused by mutations in the prolidase (PEPD) gene, located on chromosome 19 and coding for the prolidase enzyme (EC 3.4.13.9).1 Prolidase is a ubiquitous metalloenzyme requiring Mn2+ as cofactor.2 It is found in mammals1,3 as well as in bacteria and archaeon.4,5,6,7,8 Prolidase is the only enzyme able to hydrolyse the peptide bond in iminodipeptides with a C‐terminal proline or hydroxyproline. Thus, it is involved in the final stage of degradation of endogenous and dietary proteins, in particular in collagen catabolism.9
The phenotypic spectrum of patients with prolidase deficiency is very wide. The clinical outcome includes dermatological manifestations with chronic, recurrent, slowly healing ulcerations mainly located on the legs and feet, although lesions of the upper limbs and face have also been described. The intractable ulcers are often preceded by other dermatological manifestations that may occur anywhere and include erythematous papular eruptions, telangiectasias with pruritus and photosensitivity, impetigo‐like eruptions, pruritic eczematous lesions and necrotic papules. Patients with prolidase deficiency may have facial dysmorphism. Features include low hairline and facial hirsutism, a saddle nose, ocular hypertelorism, ptosis, micrognathia, a high arched palate, mandibular protrusion and exophthalamus. Mild to severe mental retardation is also reported in many cases. Other common clinical features are splenomegaly, recurrent infections of the respiratory tract, hypotonia, skeletal anomalies and in two cases systemic lupus erythematosus.1,10,11
The clinical manifestations are usually detectable after birth or in early childhood, but late‐onset cases have also been reported.12 In a small number of cases no clinical symptoms were reported, but the disease evolves constantly and two patients, asymptomatic until age 6 and 8 years, developed skin lesions around puberty.13,14,15,16,17,18 Two other cases were asymptomatic at 26 years and 4 months, but follow‐up was not possible.19,20
Key points
Prolidase deficiency is an autosomal recessive connective tissue disorder characterised mainly by intractable skin lesions, mild to severe mental retardation, and recurrent infections of the respiratory tract. Its severity varies widely, but no relationship between genotype and phenotype has been established yet due to the limited number of described mutations.
Molecular analysis of prolidase deficiency cases identified 13 different mutations in the prolidase gene (PEPD): 6 missense and 4 exon skipping mutations, 2 amino acid deletions and a large genomic deletion.
Here, we describe five different PEPD mutations in six patients from three European countries: Turkey, Denmark and Italy. We identified two new PEPD mutant alleles in homozygotic patients from two unrelated Turkish families: a 13 bp duplication in exon 8, which is the first reported duplication in the prolidase gene, and a point mutation resulting in a change in amino acid E412, a highly conserved residue among different species such as human, mouse, fungi, bacteria and archeon. The E412K substitution is responsible for the first reported phenotypic variability within a family with severe and asymptomatic outcomes.
The metabolic hallmarks of prolidase deficiency are iminodipeptiduria and lack of or reduced prolidase activity in erythrocytes, leukocytes or cultured fibroblasts.21
No definitive cures are available so far, but oral supplementation with manganese, a cofactor of prolidase, and vitamin C, acting on collagen synthesis, have been attempted. Also, blood transfusions and aphaeresis, corticosteroid treatment, oral supplementation with antioxidants and topical antibiotics for the skin lesions have been tested.1
So far, around 60 cases of confirmed prolidase deficiency have been described in the literature, but only 13 mutant alleles have been characterised (table 1).
Table 1 Summary of all known mutant alleles of the prolidase gene causing prolidase deficiency.
| Exon | Intron | Mutation | Effect | Reference |
|---|---|---|---|---|
| 4 | IVS4‐1G→C | delex5 | Ledoux et al22 | |
| 6 | IVS6‐2A→G | delex7 | Ledoux et al22 | |
| 7 | IVS7‐1G→A | Alternative splicing | Forlino et al23 | |
| 8 | 611duplAGGCCCACCGTGA | Fs and premature Stop | * | |
| 8 | 551G→A | R184Q | Ledoux et al24 | |
| 8 | 551C→T | R184X | Kikuchi et al25 | |
| 10 | 691delTAC | 231delY | Lupi et al26 | |
| 11 | IVS11+1G→C | delex11 | Forlino et al23 | |
| 11 | 793C→T | R265X | Wang et al27 | |
| 12 | 826G→A | D276N | Endo et al15 and * | |
| 12 | 833G→A | G278D | Ledoux et al24 and * | |
| 14 | 1234G→A | E412K | * | |
| 14 | 1342G→A | G448R | Ledoux et al,22 Forlino et al23 and * | |
| 14 | del774bp | delex14 | Tanoue et al28 | |
| 15 | 1354delGAG | 452delE | Ledoux et al22 |
*Mutant alleles characterised in the present paper.
The mutations are scattered in the last two thirds of the gene but due to the limited number of known mutations, mutational hot spots have not been identified.
We describe the molecular characterisation of six patients with prolidase deficiency, two of whom are siblings, of Turkish, Danish and Italian origin.
Of the two novel mutant PEPD alleles causing prolidase deficiency (table 1), one is the first reported small duplication in the PEPD gene and the other is a missense mutation, E412K, changing a highly conserved residue in PEPD; this mutation was found in a family with a wide range of phenotypic manifestations. Three of our patients carried in heterozygotic (patients C and D) or homozygotic (patient D) conditions the G448R substitution, previously reported in four other cases,22,23 which seems to be the most frequent in different populations.
Patients and methods
Patients
Patients A and B
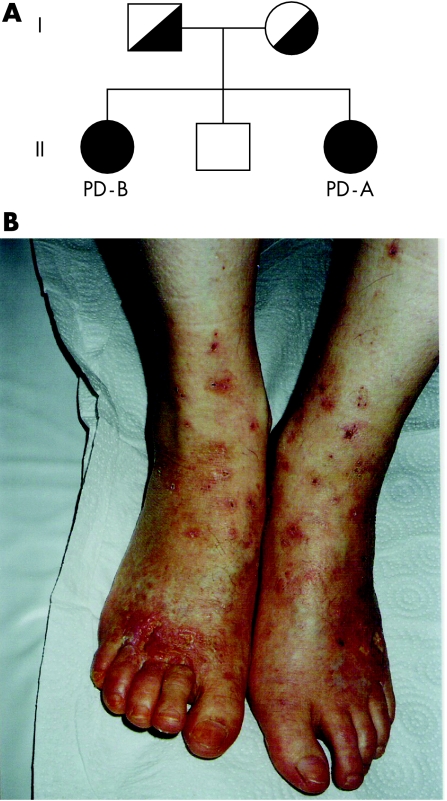
A detailed clinical description of patient A will soon be published by Aytug et al.23a Figure 1 shows the pedigree; all the family members are molecularly characterised. Patient A is the third child from unrelated healthy Turkish parents. She is now 21 years old. She was born after a normal pregnancy and delivery. Her medical history was unremarkable apart from eczema‐like lesions on the face during childhood and recurrent leg ulcers, which started following a trauma after puberty.
Figure 1 (A) Pedigree of the Turkish family carrying the 1234G→A mutant allele causing the E412K substitution in the prolidase. The parents were heterozygotic for this mutation, the two sisters were homozygotic, patient A was symptomatic, patient B essentially asymptomatic; the healthy brother did not carry the mutant allele. (B) Severe skin ulcers located at the lower legs and feet of patient A.
She presented multiple deep irregular and tender ulcers covered with fibrin exudates and depressed atrophic scars on the dorsal surface of the feet and ankle (fig 1B). She had inherited keratosis pilaris from her maternal family; her grandmother, aunt, mother and elder sister were affected as well. Her skin was particularly dry, especially on the legs which had a scaly appearance during the winter. Her facial skin was also dry, with tender scars. She was tall and mentally normal. Since age 16 years, she had been treated with various topical agents, wound dressing, systemic steroids, pentoxyphylline and psoralen phototherapy with ultraviolet A (PUVA), with no effect on her leg ulcers. She had anaemia, haemoglobin O trait and raised IgE. In the urine she had no free hydroxyproline, but increased total hydroxyproline. There was no prolidase activity in erythrocytes or serum. Family screening showed haemoglobin O trait in her father and elder two siblings. Both siblings were clinically normal.
Patient B is now 29 years old and is a sister of patient A. She was diagnosed only because her symptomatic sister was investigated for prolidase deficiency due to her severe skin lesions. Patient B had no prolidase activity in serum and erythrocytes, and also does not have ulcers or any other typical symptoms of prolidase deficiency. However, according to her family members, her skin heals slowly after injury compared with her healthy brother.
No marked differences in physical activity were reported between the two sisters.
Patient C
This boy was born to Danish parents after a normal pregnancy and delivery in gestational week 36. APGAR (Activity, Pulse, Grimace, Appearance, Respiration) scores were normal. He developed hypoglycaemia during the first day, which was quickly corrected by intravenous glucose. Hyperbilirubinaemia was noted on day 2 and treated with phototherapy for 2 days. After discharge, he developed normally until age 18 months when he was referred for hypotonia and retarded gross motor development. His skin was normal and he never had skin ulcers or eczema except for a perianal eczema. At follow‐up at age 3 years his skin remains normal. He has no splenomegaly and growth is normal. His gross motor development remains about 8 months behind his chronological age, whereas his intellectual development is normal. He is now 4 years old.
Urine amino acid analysis showed a high level of prolidase‐hydrolysable dipeptides. This finding was confirmed in vitro; prolidase activity in fibroblasts was reduced at 2.7 nmol/min/mg protein (reference range 196–274 nmol/min/mg). He was prescribed ascorbic acid and extra skin care was instituted.
Patient D
This girl was born to Danish parents after a normal pregnancy and delivery. She is now 12 years old. She was well until age 11 months when she presented with developmental delay, splenomegaly and transfusion‐dependent anaemia. Skin ulcers were already the major problem at that age and she also developed secondary infections and septicaemia. She had been treated with ascorbic acid since diagnosis, with no relevant clinical effect. Because of the increasing number of infections and severe neutropenia, neupogen was started at age 10 years. She had no septic episodes on this treatment and her skin healed; she had always had severe and very painful skin ulcers on her feet, making the use of a wheelchair for extended periods necessary. She is now capable of walking long distances. The size of her spleen increased over the years and was removed at age 11 years. She is attending a special‐needs school and remains developmentally delayed, but this delay has been non‐progressive. Urine iminodipeptiduria was detected and prolidase activity in fibroblasts was reduced to 5 nmol/min/mg protein.
Patient E
This woman is 30 years old. Her clinical outcome was described previously.29 She was born after a normal pregnancy and delivery to Turkish consanguineous parents. She presented within the first year of life with developmental delay, skin ulcers and failure to thrive. She was diagnosed with prolidase deficiency at age 4 years after analysis of urine amino acids showed increased excretion of prolidase hydrolysable dipeptides. Prolidase acitivity was reduced to 12.8 nmol/min/mg protein. Treatment with ascorbic acid, l‐proline and manganese was attempted, with initially good, but later questionable effect on her ulcers. Her developmental delay has been mild and skin ulcers severe, although they also vary with time. Splenomegaly was noted at age 4 years during several examinations but was not present when she was evaluated from age 15 years onwards; she never had an ultrasound of the abdomen. At age 14 years she was involved in a car accident leading to further intellectual damage. At the last follow‐up at age 29 years, her skin ulcers and skin infections remain her main complaint. Her nails are severely dystrophic. She has had a skin transplant on some areas of her lower legs with good effect. She constantly wears bandages and is treated with ascorbic acid and topical creams. She lives in a house for mentally retarded people.
Patient F
This boy was born to southern Italian parents 6 years ago. At age 4 years he was treated for splenomegaly and presented mild mental retardation and severe skin lesions at clinical examination. He was diagnosed with systemic lupus erythematosus and treated for 2 years with steroids and azathioprine, with amelioration of the immunological abnormalities but worsening of the skin lesions. Iminodipeptiduria was detected, with essentially no prolidase activity in erythrocytes. Topical care of skin ulcers and antibiotic treatment was adopted, with temporary improvement of the skin lesions. Later, one of his toes needed amputation.
Molecular studies
Dermal fibroblast cultures of patients A, C, D and E were established from skin punch biopsies after informed consent and grown in Dulbecco's modified Eagle medium at 37°C in the presence of 5% CO2.
Total RNA was extracted from subconfluent cultured fibroblasts using TRI (Sigma, Milan, Italy) Reagent and 1 μg was reverse transcribed for 1 h at 42°C using the cDNA synthesis kit (Roche, Milan, Italy) for reverse transcriptase‐polymerase chain reaction (PCR) according to the manufacturer's specifications. The entire prolidase transcript was amplified in seven overlapping fragments and sequenced as described by Forlino et al.23
For patients C and D, the mutations were confirmed by sequencing two independent reverse transcriptase‐PCR fragments from two independent RNA extractions. For patients A, B, E and F, genomic confirmation of the mutations was undertaken. The presence of the mutation in all members of the families of patients A and B was investigated at the genomic level.
Genomic DNA from patient E was extracted from cultured fibroblasts. For patients A and F, as well as for the family members of patients A and B, DNA was obtained from peripheral blood by standard techniques.
The PCR conditions used for genomic DNA amplification and sequence analysis were 94°C for 1 min; 35 cycles of the following three steps: 94°C for 2 min, 55–62°C for 1 min, 72°C for 1 min, and a final cycle at 72°C for 10 min. Table 2 shows the specific primers and annealing temperatures.
Table 2 Primer sets used for genomic sequences.
| Patient | Primer orientation | Primer sequence (5′ to 3′) | Primer location | Annealing temperature (°C) |
|---|---|---|---|---|
| A, B | Sense | CGTGGAGCGCATCGACGAGCCC | Exon14 (1155–76) | |
| and their family members | Antisense | CCGCGAAAGCGCTGCAGGACC | Exon14 (1314–34) | 62 |
| E | Sense | CCAGTGCCTCCTGAAAGTCACTG | IVS7 | |
| Antisense | CTCTCGCCACACAGCAACACTGC | IVS8 | 58 | |
| F | Sense | AGGTCCTGCAGCGCTTTCGCG | Exon14 (1313–33) | |
| Antisense | CGCAGGTCAGCAGCTCTATGC | Exon15 (1382–02) | 55 |
The PCR products were run on 1.8% agarose gels, gel purified and directly sequenced using an ABI PRISM 310 and the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems).
The sequences obtained were compared with the reported PEPD gene sequence (NM_000285) using the BLASTN program.
This study was approved by the ethics committee of the University of Pavia (2 August 2005).
Results and discussion
Prolidase deficiency is a rare autosomal recessive connective tissue disorder caused by mutation in the PEPD gene. Its incidence of 1–2 in 1 000 000 is probably underestimated due to doctors' unfamiliarity with this condition. Moreover, the frequency might be dependent on the population considered, as is often the case for recessive disorders. Our laboratory is an international centre for the molecular diagnosis of prolidase deficiency (www.orphan.net).
Only 13 mutant alleles have so far been described in 22 patients (table 3). In the past 2 years, we have collected six new patients with prolidase deficiency (table 3) and carried out molecular diagnosis by full sequencing of the prolidase transcript according to the screening method established in our laboratory.23
Table 3 Summary of the patients who underwent molecular characterisation.
| Patient ID | Mutation | Prolidase activity (%)* | Clinical phenotype | Ethnic origin |
|---|---|---|---|---|
| Previously described patients | ||||
| 1 (WG1298) | IVS4‐1G→C/null allele | ≈8† | Skin ulcers, borderline mental retardation, recurrent infections | USA |
| 2 (WG1625) | IVS6‐2A→G/IVS6‐2A→G | <1‡ | Skin ulcers, mild mental retardation, systemic lupus erythematosus | Canada |
| 3§ | IVS7‐1G→A/IVS7‐1G→A | <9† | Skin ulcers, mental retardation | Italy |
| 4§ | IVS7‐1G→A/IVS7‐1G→A | <9† | Skin ulcers | Italy |
| 5 (WG1077) | 551G→A/833G→A | ≈8† | Asymptomatic at birth, no data available later | Canada |
| 6 | 551C→T/551C→T | None†,‡ | Skin ulcers, mental retardation, recurrent infections, dysmorphic facies, dislocations of joints, partial deafness | Japan |
| 7¶ | 691delTAC/691delTAC | ≈5† | Skin ulcers, abnormal behaviour | Portugal |
| 8¶ | 691delTAC/691delTAC | ≈5† | Skin ulcers, anaemia, increased IgE | Portugal |
| 9 | IVS11‐1G→A/IVS11‐1G→A | <9† | Skin ulcers, mental retardation, dysmorphic facies, telangiectasia, photosensitivity, pigmented skin | Italy |
| 10** | 793C→T/793C→T | <1‡ | Skin ulcers, recurrent infections, dysmorphic facies, hepatomegaly | Ohio |
| 11** | 793C→T/793C→T | <1‡ | Skin ulcers, recurrent infections, dysmorphic facies, hepatomegaly | Ohio |
| 12** | 793C→T/793C→T | <1‡ | Skin ulcers, recurrent infections, dysmorphic facies, hepatomegaly | Ohio |
| 13** | 793C→T/793C→T | <1‡ | Skin ulcers, recurrent infections, dysmorphic facies, hepatomegaly | Ohio |
| 14†† | 826G→A/826G→A | <5† | Moderate skin ulcers, abnormalities of the bone and joints | Middle East |
| 15†† | 826G→A/826G→A | <5† | Moderate skin ulcers, splenomegaly | Middle East |
| 16 (WG1343) | 1342G→A/null allele | <2† | Mild skin ulcers, mild mental retardation, recurrent infections | Canada |
| 17 (WG1194) | 1342G→A/1342G→A | <7† | Skin ulcers, mental retardation, recurrent infections, dysmorphic facies, splenomegaly | UK |
| 18‡‡ | 1342G→A/1342G→A | <10† | Severe skin ulcers, mental retardation, recurrent infections | Italy |
| 19‡‡ | 1342G→A/1342G→A | <10† | Mild skin ulcers | Italy |
| 20§§ | del774bp | None† | Skin ulcers, mental retardation | Japan |
| 21§§ | del774bp | None† | Skin ulcers | Japan |
| 22 (WG1082) | 1354delGAG/null allele | <5† | Mild skin ulcers, borderline mental retardation, chronic liver disease | Australia |
| Patients described in this paper | ||||
| A¶¶ | 1234G→A/1234G→A | None‡ | Skin ulcers, increased IgE | Turkey |
| B¶¶ | 1234G→A/1234G→A | None‡ | Asymptomatic | Turkey |
| C | 826G→A/1342G→A | <2† | Perianal eczema, gross motor delay | Denmark |
| D | 833G→A/1342G→A | <3† | Skin ulcers, recurrent infections, splenomegaly | Denmark |
| E | 611duplAGGCCCACCGTGA/611duplAGGCCCACCGTGA | <10† | Skin ulcers, mental retardation, recurrent infections, splenomegaly, dysmorphic facies | Turkey |
| F | 1342G→A/1342→A | None† | Skin lesions, mild mental retardation, splenomegaly, systemic lupus erythematosus | Italy |
Names of the original cell lines are given in parentheses, where available.
*The prolidase activity is expressed as a percentage with respect to normal values; †prolidase activity measured in fibroblasts; ‡prolidase activity measured in serum; §unrelated; ¶unrelated; **family related; ††unrelated; ‡‡brothers; §§sisters; ¶¶sisters.
The patients characterised in this paper have different ethnic origins: the two sisters (patients A and B) are Turkish, patient F is Turkish as well, although referred to us from a Danish doctor, patients C and D are Danish and patient E is from southern Italy.
Molecular analysis of patient A showed homozygosity for a 1234G→A mutation causing the change E412K. Patient A and her asymptomatic sister patient B, who were diagnosed only on the basis of biochemical and molecular analysis, were both homozygotic for the same mutation.
Such phenotypic variability in the presence of an identical mutation in siblings is relatively frequent in genetic disorders and its molecular basis needs further investigation. The healthy brother of patients A and B did not carry this mutation, whereas the parents were, as expected, heterozygotic carriers.
We detected the mutation 1342G→A, causing the amino acid change G448R, in three different patients: C, D and F. The same molecular defect has already been reported in four other patients with prolidase deficiency, one of whom was heterozygotic, with a null mutation on the other allele22: the other three, two of whom were brothers, were homozygotic for this mutation.22,23
The phenotype associated with the 1342G→A mutation is similar in all described patients, all of whom presented with mild to severe skin manifestation and mental retardation.
Patient F was homozygotic for the G448R mutation. He originates from the same southern Italian region (Puglia) as two other patients harbouring the same defect, and his phenotype is similar to that of these two other patients with prolidase deficiency.23
Patient C was compound heterozygotic for the G448R and the D276N mutations and patient D was compound heterozygotic for the same G448R substitution and a G278D mutation.
The occurrence of the same change at a conservative amino acid in six unrelated families favours its relevance for protein structure or function.
The 826G→A transition responsible for the D276N change has already been described in the homozygotic state in two unrelated patients,30 who presented the typical skin ulcers, suggesting that the phenotype caused by this mutation is consistent with the classical prolidase deficiency skin manifestation. Our patient did not have any ulcers yet, but this is probably due to his young age and a follow‐up is needed for a definitive answer.
The G278D mutation was reported previously in heterozygosity with R184Q in an asymptomatic patient.24
In patient D the presence of the G448R allele could be responsible for the more severe outcome.
A phylogenetic comparison shows that R184, D276, G278, E412 and G448 are highly conserved in the prolidase sequence in organisms high up in the phylogenetic tree, such as Mus musculus (BBA11685, 93% homology with Homo sapiens prolidase, NP_000276), Emericella nidulans (CAC39600, 49% homology), Suberites domuncula (CAA75231, 80% homology), Lactobacillus delbrueckii (CAB07978, 44% homology), Pseudoalteromonas haloplanktis (AAA99824, 44% homology) and Pyrococcus furiosus (AAC61259, 43% homology; fig 2).
Figure 2 Alignment of amino acid sequences for prolidase enzyme from: Homo sapiens, Mus musculus, Emericella nidulans, Suberites domuncula, Lactobacillus delbrueckii, Pseudoalteromonas haloplanktis and Pyrococcus furiosus. The asterisks indicate the amino acids conserved among the species; the residues changed by point mutations in patients with prolidase deficiency are shown in red.
Patient E carried a homozygotic 13‐bp duplication in exon 8, generating a premature stop codon after 18 amino acids from the insertion site and resulting in the absence of prolidase.
Two other mutations described in the literature, R184X and R265X, generate a premature stop codon.25,27 Both were found in homozygotic subjects and caused the synthesis of a truncated protein. In both cases the phenotype was particularly severe, suggesting that nonsense mutations are associated with more severe clinical outcome.
The structure of human prolidase, as well as the composition of its active site, is still unknown.
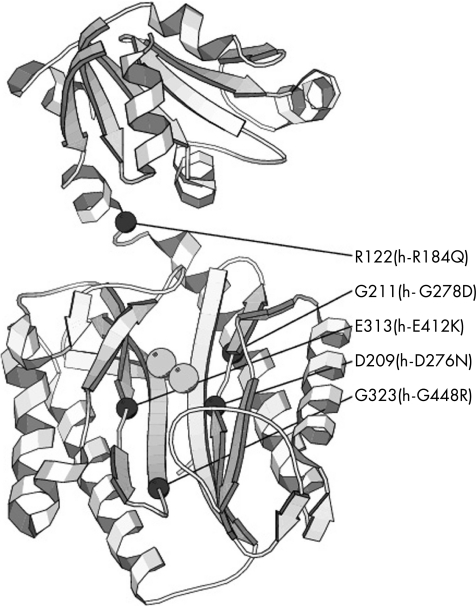
Recently, the complete structure of the P furiosus prolidase (Pfprol) and the organisation of its active site have been described.7,31 On the basis of homology observations, four causative mutations for prolidase deficiency are judged to occur in highly conserved amino acids that have been shown to be relevant for structure or function of Pfprol: R184Q (R122 in Pfprol), G278D (G211 in Pfprol) and G448R (G323 in Pfprol) with structural functions and D276N (D209 in Pfprol) relevant for the cofactor metal binding, which in Pyrococcus is Co32 (fig 3).
Figure 3 Position of the pathogenic mutations in the structure of Pyrococcus furiosus prolidase. The figure shows the ribbon representation of Pfprol (PDB entry 1PV9) together with the two metal ions bound to the active site (grey spheres). The positions of the C‐α atoms of the residues corresponding to the mutation sites in the human enzyme are shown as black spheres.
Using the same type of comparison, E412 corresponds to the amino acid E313 in Pfprol, which has been identified as a member of the dinuclear metal centre‐active site.
The limited number of patients who were molecularly characterised does not allow prediction concerning the ethnic origin of the mutant alleles. We can only speculate about the 1342G→A allele, found in seven patients from different countries. It does seem that it has a European origin.
The identification of de novo mutations causing prolidase deficiency will help both better understanding of the relationship between function and structure, in the absence of direct structural data, and to elucidate the relationship between molecular defects and clinical outcome.
In addition, detection of mutations in patients with prolidase deficiency allows appropriate genetic counselling: carriers can easily be detected among relatives of people in whom mutations have been identified, and knowledge about the prolidase deficiency mutations segregating in a family opens possibilities for early prenatal diagnosis.
Acknowledgements
We thank all the families for their participation in the study and for providing us with written consent for publication of their clinical and molecular data. We thank A Gallanti for tissue culture support, Dr AF Aytug, Department of Dermatology, Marmara University Hospital, Istanbul, Turkey, for dermatologic evaluation of patients A and B, and Professor A Mattevi, Dipartimento di Genetica e Microbiologia, University of Pavia, Italy, for structure comparison of human and Pyrococcus furiosus prolidase.
Abbreviations
PCR - polymerase chain reaction
PEPD - prolidase gene
Footnotes
Funding: This work was supported by Fondo di Ateneo per la Ricerca (FAR) and Fondazione Cariplo.
Competing interests: None declared.
References
- 1.Royce P M, Steinmann B. Prolidase deficiency. In: Royce PM, Steinmann B, eds. Connective tissue and its heritable disorders. New York: Wiley‐Liss, 2002727–743.
- 2.Richter A M, Lancaster G L, Choy F Y.et al Purification and characterization of activated human erythrocyte prolidase. Biochem Cell Biol 19896734–41. [DOI] [PubMed] [Google Scholar]
- 3.Myara I, Brosset B, Lemonnier A. Tissue distribution of prolidase and prolinase activity in man and rat. Med Sci Res 198715965–966. [Google Scholar]
- 4.Booth M, Jennings P V, Ni Fhaolain I.et al Endopeptidase activities of Streptococcus cremoris. Biochem Soc Trans 199018339–340. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez‐Espla M D, Martin‐Hernandez M C, Fox P F. Purification and characterization of a prolidase from Lactobacillus casei subsp. casei IFPL 731. Appl Environ Microbiol 199763314–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suga K, Kabashima T, Ito K.et al Prolidase from Xanthomonas maltophilia: purification and characterization of the enzyme. Biosci Biotechnol Biochem 1995592087–2090. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh M, Grunden A M, Dunn D M.et al Characterization of native and recombinant forms of an unusual cobalt‐dependent proline dipeptidase (prolidase) from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol 19981804781–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujii M, Nagaoka Y, Imamura S.et al Purification and characterization of a prolidase from Aureobacterium esteraromaticum. Biosci Biotechnol Biochem 1996601118–1122. [DOI] [PubMed] [Google Scholar]
- 9.Cunningham D F, O'Connor B. Proline specific peptidases. Biochim Biophys Acta 19971343160–186. [DOI] [PubMed] [Google Scholar]
- 10.Bissonnette R, Friedmann D, Giroux J M.et al Prolidase deficiency: a multisystemic hereditary disorder. J Am Acad Dermatol 199329(Pt 2)818–821. [DOI] [PubMed] [Google Scholar]
- 11.Shrinath M, Walter J H, Haeney M.et al Prolidase deficiency and systemic lupus erythematosus. Arch Dis Child 199776441–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyne K, Zanaboni G, Bertazzoni M.et al Mild, late‐onset prolidase deficiency: another Italian case. Br J Dermatol 2001144635–636. [DOI] [PubMed] [Google Scholar]
- 13.Umemura S. Studies on a patient with iminodipeptiduria. II. Lack of prolidase activity in blood cells. Physiol Chem Phys 197810279–283. [PubMed] [Google Scholar]
- 14.Oono T, Arata J. Characteristics of prolidase and prolinase in prolidase‐deficient patients with some preliminary studies of their role in skin. J Dermatol 198815212–219. [DOI] [PubMed] [Google Scholar]
- 15.Endo F, Tanoue A, Kitano A.et al Biochemical basis of prolidase deficiency. Polypeptide and RNA phenotypes and the relation to clinical phenotypes. J Clin Invest 199085162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arata J, Tada J, Yamada T.et al Angiopathic pathogenesis of clinical manifestations in prolidase deficiency. Arch Dermatol 1991127124–125. [PubMed] [Google Scholar]
- 17.Pasquali Ronchetti I, Quaglino D, Jr, Dyne K M.et al Ultrastructural studies on dermis from prolidase deficient subjects. J Submicrosc Cytol Pathol 199123439–445. [PubMed] [Google Scholar]
- 18.Zanaboni G, Dyne K M, Rossi A.et al Prolidase deficiency: biochemical study of erythrocyte and skin fibroblast prolidase activity in Italian patients. Haematologica 19947913–18. [PubMed] [Google Scholar]
- 19.Isemura M, Hanyu T, Gejyo F.et al Prolidase deficiency with imidodipeptiduria. A familial case with and without clinical symptoms. Clin Chim Acta 197993401–407. [DOI] [PubMed] [Google Scholar]
- 20.Mandel H, Abeling N, Gutman A.et al Prolidase deficiency among an Israeli population: prenatal diagnosis in a genetic disorder with uncertain prognosis. Prenat Diagn 200020927–929. [DOI] [PubMed] [Google Scholar]
- 21.Kurien B T, Patel N C, Porter A C.et al Prolidase deficiency and the biochemical assays used in its diagnosis. Anal Biochem 2006349165–175. [DOI] [PubMed] [Google Scholar]
- 22.Ledoux P, Scriver C, Hechtman P. Four novel PEPD alleles causing prolidase deficiency. Am J Hum Genet 1994541014–1021. [PMC free article] [PubMed] [Google Scholar]
- 23.Forlino A, Lupi A, Vaghi P.et al Mutation analysis of five new patients affected by prolidase deficiency: the lack of enzyme activity causes necrosis‐like cell death in cultured fibroblasts. Hum Genet 2002111314–322. [DOI] [PubMed] [Google Scholar]
- 23a. Aytug A F, Ergun T, Ratip S, Elcioglu N, Gultepe M, Mercan E, Gurbuz O. Prolidase deficiency associated with haemoglobin O trait and microcytic anemia. Int J Dermatol 200645877–878. [DOI] [PubMed] [Google Scholar]
- 24.Ledoux P, Scriver C, Hechtman P. Expression and molecular analysis of mutations in prolidase deficiency. Am J Hum Genet 1996591035–1039. [PMC free article] [PubMed] [Google Scholar]
- 25.Kikuchi S, Tanoue A, Endo F.et al A novel nonsense mutation of the PEPD gene in a Japanese patient with prolidase deficiency. J Hum Genet 200045102–104. [DOI] [PubMed] [Google Scholar]
- 26.Lupi A, De Riso A, Della Torre S.et al Characterization of a new PEPD allele causing prolidase deficiency in two unrelated patients; natural‐occurrent mutations as a tool to investigate structure‐function relationship. J Hum Genet 200449500–506. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Kurien B T, Lundgren D.et al A nonsense mutation of PEPD in four Amish children with prolidase deficiency. Am J Med Genet A 2006140580–585. [DOI] [PubMed] [Google Scholar]
- 28.Tanoue A, Endo F, Akaboshi I.et al Molecular defect in siblings with prolidase deficiency and absence or presence of clinical symptoms. A 0.8 Kb deletion with breakpoints at the short, direct repeat in the PEPD gene and synthesis of abnormal messenger RNA and inactive polypeptide. J Clin Invest 1991871171–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedersen P S, Christensen E, Brandt N J. Prolidase deficiency. Acta Paediatr Scand 198372785–788. [DOI] [PubMed] [Google Scholar]
- 30.Tanoue A, Endo F, Kitano A.et al A single nucleotide change in the prolidase gene in fibroblasts from two patients with polypeptide positive prolidase deficiency. Expression of the mutant enzyme in NIH 3T3 cells. J Clin Invest 199086351–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maher M J, Ghosh M, Grunden A M.et al Structure of the prolidase from Pyrococcus furiosus. Biochemistry 2004432771–2783. [DOI] [PubMed] [Google Scholar]
- 32.Du X, Tove S, Kast‐Hutcheson K.et al Characterization of the dinuclear metal center of Pyrococcus furiosus prolidase by analysis of targeted mutants. FEBS Lett 20055796140–6146. [DOI] [PubMed] [Google Scholar]