Abstract
Familial haemophagocytic lymphohistiocytosis (FHL) is a genetically heterogeneous disorder characterised by constitutive defects in cellular cytotoxicity resulting in fever, hepatosplenomegaly and cytopenia, and the outcome is fatal unless treated by chemoimmunotherapy followed by haematopoietic stem‐cell transplantation. Since 1999, mutations in the perforin gene giving rise to this disease have been identified; however, these account only for 40% of cases. Lack of a genetic marker hampers the diagnosis, suitability for transplantation, selection of familial donors, identification of carriers, genetic counselling and prenatal diagnosis. Mutations in the Munc13–4 gene have recently been described in patients with FHL. We sequenced the Munc13–4 gene in all patients with haemophagocytic lymphohistiocytosis not due to PRF1 mutations.
In 15 of the 30 families studied, 12 novel and 4 known Munc13–4 mutations were found, spread throughout the gene. Among novel mutations, 2650C→T introduced a stop codon; 441del A, 532del C, 3082del C and 3226ins G caused a frameshift, and seven were mis sense mutations. Median age of diagnosis was 4 months, but six patients developed the disease after 5 years of age and one as a young adult of 18 years. Involvement of central nervous system was present in 9 of 15 patients, activity of natural killer cells was markedly reduced or absent in 13 of 13 tested patients. Chemo‐immunotherapy was effective in all patients.
Munc13–4 mutations were found in 15 of 30 patients with FHL without PRF1 mutations. Because these patients may develop the disease during adolescence or even later, haematologists should include FHL2 and FHL3 in the differential diagnosis of young adults with fever, cytopenia, splenomegaly and hypercytokinaemia.
Haemophagocytic lymphohistiocytosis (HLH) is a genetically heterogeneous disorder characterised by a hyperinflammatory syndrome with fever, hepatosplenomegaly, cytopenia and, less often, central nervous system (CNS) involvement. Frequent changes include low levels of fibrinogen, high levels of ferritin, triglycerides and the α‐chain of the soluble interleukin 2 receptor (sCD25).1,2,3 As symptoms may resemble those of leukaemia or lymphoma, bone‐marrow aspiration is usually carried out early during the diagnostic investigation, showing—at first or at repeated evaluations during the disease course—haemophagocytosis by activated macrophages.
The natural course of HLH is rapidly fatal within a few weeks in most cases unless appropriate treatment, including corticosteroids, ciclosporin, etoposide and anti‐thymocite globulin result in transient disease control.3,4,5,6 So far, only patients who underwent haematopoietic stem‐cell transplantation have been cured.7,8,9
Key points
We investigated the role of Munc13–4 mutations in patients with haemophagocytic lymphohistiocytosis, in whom PRF1 mutations had been excluded.
The prevalence of families in which Munc13–4 mutations were found (FHL3) was 50%. Mutations were spread throughout the gene, but interestingly they occur in the protein functional domains. Only one quarter of the mutations found had been previously reported. Among the novel mutations, 2650C→T introduced a stop codon; 441del A, 532del C, 3082del C and 3226ins caused a frameshift; and seven were mis‐sense mutations.
Although patients with FHL3 do not have a particular phenotype, 6 of 15 patients developed the disease after 5 years of age and one as a young adult; central nervous system involvement seems to be a major feature of FHL3.
Differential diagnosis of HLH may be difficult.10 For this purpose, diagnostic guidelines for HLH have been established by the Histiocyte Society.11 In particular, demonstration of frequent association with common pathogens and evidence of impaired natural killer cell cytotoxic activity provided the rationale for considering HLH as a selective immune deficiency.12,13,14 Starting from the original report by Farquhar et al1, autosomal recessive inheritance was proposed as the basis for the familial form of HLH (FHL).
Linkage analysis led to the identification of an association between FHL and the genomic region 9q21.3–22 (FHL1, Mendelian Inheritance in Man (MIM) 267700),15 where the gene responsible for the disease was not then identified. A simultaneous report established a linkage with another region, 10q21–22,16 in which the perforin (PRF1) gene was identified as responsible for a large proportion of cases of FHL (FHL2, MIM 603553).17 In patients with FHL2, PRF1 mutations induce a complete or partial reduction in the synthesis of the perforin protein. As a result, the cytotoxic machinery of natural killer cells is markedly impaired.18,19 As a formal genotype–phenotype study is not yet available, the exact contribution of the different mutations reported in the literature is not yet clear, although some linkage between a few individual mutations and particular ethnic groups seem to be preliminarily defined.20,21,22,23,24,25
In 2003, a third locus, 17q25, was reported in linkage with FHL (FHL3, MIM 608898).26 The product of the involved gene, the Munc13–4 protein, is thought to contribute to the priming of the secretary granules before they fuse into the plasma cell membrane. Mutations in this gene impair the delivery of the effector proteins, perforin and granzymes into the target cells, resulting in defective cellular cytotoxicity and a clinical picture that seems identical to that associated with PRF1 mutations.26
Recently, on the basis of a genomewide screening, a fourth chromosomal region (6q24) has been reported in linkage with FHL in a subset of Kurdish patients (FHL4, MIM 603552).27 Mutations of the syntaxin 11 gene, mapped in this region, are thought to alter intracellular vesicle trafficking of the phagocytic system.
We have previously reported a series of mutations in the PRF1 gene observed in Italian patients.20 Here, we report the result of the screening of Munc13–4 gene in 30 patients with FHL not associated with PRF1 mutations.
Patients and methods
Patient selection
Patients with HLH diagnosed according to current diagnostic criteria underwent a clinical evaluation, determination of natural killer cell activity and a genetic study.10 In this analysis, we focus on those patients who lacked PRF1 mutations and thus were possibly affected by other genetic subtypes of FHL.
Munc13–4 gene sequencing
Genomic and mRNA sequences of the Munc13–4 gene were retrieved from the National Center for Biotechnology Information (LOC201294 of the genome annotation program, genomic contig NT_010641, model mRNA XM_113950). Genomic DNA was prepared from the peripheral blood samples obtained from the patients and their family members. To analyse the Munc13–4 gene, exons and adjacent intronic regions were amplified, obtaining 21 fragments directly sequenced, in both directions, with the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA, USA). Sequence primers used for amplification are available on request. Sequences obtained using an ABI Prism 3130 Sequence Detection System (Applied Biosystems) were analysed and compared with the reported gene structure using the dedicated software SeqScape (Applied Biosystems).
Human CTL and natural killer cell derivation
Human cytotoxic T lymphocyte (CTL) clones were derived from peripheral blood from patients 7, 9 and 10. The mononuclear cells were purified by separation on Ficoll gradient (Pharmacia) after centrifugation at 2000 rpm for 20 min (peripheral blood mononuclear cells (PBMC); 6×106) were then stimulated with 6×106 irradiated PBMC from a healthy donor in RPMI, 5% human serum, 2 mM glutamine, 100 U/ml penicillin, 100 U/ml streptomycin, 100 U/ml interleukin 2, 0.1 mM 2‐mercaptoethanol and 1 μg/ml PHA. After 20 days in culture, cells were cloned in 96‐well plates at 0.3 cells/well with 1×105 irradiated PBMC and 1 μg/ml PHA. CD8 CTL clones were selected and grown in culture.
Natural killer cells were purified from peripheral blood using the RosetteSep method (StemCell Technologies, Vancouver, Canada) following the manufacturer's instructions. Natural killer cells were cultured on irradiated feeder cells (allogeneic PBMC and 721.221 lymphoblastoid cell line) in the presence of 100 U/ml rIL2 (Proleukin, Chiron Italia, Milan, Italy) and 2 μg/ml PHA to obtain polyclonal natural killer cell populations.
Reverse transcriptase‐PCR and sequencing of Munc13–4 cDNA
Total RNA was extracted from 107 CTL clones after 10 days of stimulation with the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). Munc13–4 cDNA was amplified by reverse transcriptase‐PCR performed on the first‐strand cDNA synthesised with SuperScript II Reverse Transcriptase Kit (Invitrogen Corporation, Carlsbad, California, USA). Munc13–4 cDNA was amplified with the following primers: 5′‐ATGGCGACACTCCTCTCCCATCCGCAG‐3′, annealing at the 5′ end of the cDNA and 5′‐CTACGGTGCCGGCCGCAAGGCATGCTG‐3′, annealing at the 3′ end of the cDNA. The PCR product was cloned into pCR‐TOPO‐Blunt II‐TOPO vectors. Fifteen clones of Munc13–4 cloned into TOPO vector were sequenced to investigate whether the mutations affect Munc13–4 mRNA splicing.
Immunoblotting
Natural killer cells were washed once in phosphate‐buffered saline (PBS) and lysed at 2×107 cells/ml in lysis buffer (50 mM Tris‐HCl (pH 8), 150 mM NaCl, 1 mM MgCl2, 1% Triton X‐100) with complete protease inhibitor (Roche Diagnostics, Welwyn, Garden City, UK) for 15 min on ice, vortexing every 5 min. Nuclei and membranes were spun out at 13 000 rpm for 15 min at 4°C. Lysates were resolved by SDS gel electrophoresis (SDS‐PAGE) on NuPAGE 4–12% Bis‐Tris gels (Invitrogen, Paisley, UK) under reducing conditions. Proteins were transferred to nitrocellulose membranes (Invitrogen) using a XCellII blot module (Invitrogen) in 25 mM Tris (pH 8.3), 192 mM glycine and 20% methanol. Membranes were then blocked for 1 h at room temperature in blocking buffer (PBS plus 5% milk powder and 0.1% Tween20), incubated for 16 h at 4°C with rabbit anti‐Munc13–4 antibody (a gift from H Horiuchi).28 Membranes were washed three times in PBS and 0.1% Tween20 for 10 min each and incubated for 1 h with horseradish peroxidase (HRP)‐labelled anti‐rabbit immunoglobulin secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) diluted in blocking buffer. Excess HRP was removed by washing three times in PBS/0.1% Tween20 for 10 min each and developed for 5 min in Supersignal (Perbio Science UK, Cramlington, UK), exposed for 1 min to 1 h using Biomax Film (Kodak, Chalon‐sur‐Saine, France). SeeBlue Plus2 (Invitrogen) was loaded on each gel as a molecular weight standard. The membranes were normalised using a rabbit anti‐actin antibody (Sigma‐Aldrich).
Immunofluorescence and electron microscopy
CTL cells from patients and controls and P815 target cells were washed in serum‐free medium (RPMI‐1640) and resuspended at 5×106 cells/ml in RPMI‐1640 with no added serum for adherence to glass slides. CTLs and P815 were conjugated at 37°C for 15–20 min in the presence of 1 μg/ml PHA (Sigma‐Aldrich). Samples were fixed for 5 min in ice‐cold methanol, washed three times with PBS, once in PBS plus 1% goat serum, and blocked in PBS plus 1% goat serum for 15 min at room temperature. Primary antibodies were diluted in PBS plus 1% bovine serum albumin (BSA; Sigma‐Aldrich), incubated at room temperature for 40 min and washed three times in PBS. The samples were blocked in PBS plus 1% BSA for 5 min. Secondary antibodies were diluted in PBS plus 1% BSA and incubated with samples for 40 min, washed three times in PBS plus 1% BSA, six times in PBS, and mounted in mounting media (PBS plus 90% glycerol and 2.5% 1,4‐diazabicyclooctane; Sigma‐Aldrich). Images were obtained using a Zeiss Axioplan 2 microscope (Carl Zeiss, Hertfordshire, UK) mounted with a CoolSnap HQ camera (Universal Imaging, Marlow, UK) and processed using Metamorph software (Molecular Devices, Downington, PA, USA) and AutoDeblur+AutoVisualize software (AutoQuant Imaging, Waterwliet, New York, USA).
For electron microscopy, CTLs were labelled for 16 h in the presence of 1–2 mg/ml HRP (Boehringer Mannheim, Mannheim, Germany) added directly to the growth medium to load the lytic granules, and washed three times by pelleting and resuspension in RPMI to remove free HRP and serum, then the final CTL pellet was resuspended at 5×106 cells/ml. CTLs were either taken alone (unconjugated) or mixed 1:1 with P815 target cells in the presence of 1 μg/ml PHA to form conjugates. Cells were left in suspension at room temperature for 5 min, plated in individual wells of 12‐well plastic tissue culture plates (Nunc, Denmark) and transferred to 37°C for a further 60 min. Samples were then fixed and processed for DAB cytochemistry post‐fixation, with reduced osmium and EPON embedding as previously described.29 Sections (100–150‐ nm thick) were stained with lead citrate and viewed using a Phillips Technai G2 Electron Microscope. Images were captured using Kodak photographic film, and the negatives subsequently scanned and digitally recorded.
Results
Study population
We investigated a total of 30 families in which HLH had been diagnosed in one or more members and PRF1 mutations had been excluded by sequence analysis. These patients had been diagnosed between 1983 and 2005: 26 were of Italian origin, 2 were from Slovenia, 1 from the USA and 1 from Morocco. In 14 Italian families and 1 from Slovenia, we did not find any Munc13–4 mutation in the 32 coding exons. In the remaining 15 families (50%), we identified Munc13–4 mutations.
Munc13–4 mutations
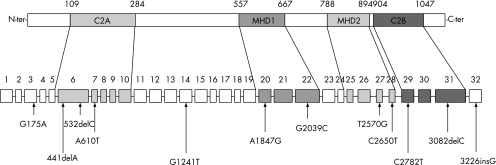
We identified 12 novel mutations (table 1; fig 1). One mutation, in patient 9, introduces a nucleotide change, 2650C→T, giving rise to a predicted protein product of 884 amino acids. Three mutations cause a frameshift, giving rise to premature termination and predicted proteins that would be truncated. The 441delA in patient 13 results in a stop codon at residue 161; nucleotide deletions at 532delC in patient 8 and 3082delC in patient 5 cause frameshifts resulting in stop codons, and would truncate the proteins at residues 248 and 1028, respectively, while a fourth mutation, a single nucleotide insertion (3226insG in patient 9), causes a frameshift in which the stop codon occurs in the 3′ untranslated region, giving rise to a predicted protein product of increased size.
Table 1 Details of mutations in Munc13–4 gene observed in 15 families with haemophagocytic lymphohistiocytosis.
| Patient | UPN | Origin | Nucleotide numbering | Exon | Nucleotide change | Predicted effect on protein level |
|---|---|---|---|---|---|---|
| 1 | 237 | Italy | 1241 | 14 | G→T | R414L |
| 2782 | 29 | C→T | R928C | |||
| 1847 | 20 | A→G | E616G (splice error) | |||
| 2 | 196 | Italy | 753 | 9 | +1G→T | Splice error |
| 2570 | 27 | T→G | F857C | |||
| 3 | 180 | Italy | 175 | 3 | G→A | A59T |
| 753* | 9 | +1G→T | Splice error | |||
| 4 | 253 | Slovenia | 2346 | 24 | del GGAG | R782 FsX12 |
| 5 | 277 | Italy | 1241 | 14 | G→T | R414L |
| 2782 | 29 | C→T | R928C | |||
| 3082 | 31 | del C | L1028 FsX | |||
| 6 | 205 | USA | 2346 | 24 | del GGAG | R782 FsX12 |
| 7 | 249 | Italy | 1847* | 20 | A→G | E616G (splice error) |
| 8 | 3 | Italy | 532* | 6 | del C | Q178 FsX70 |
| 9 | 289 | Italy | 610 | 7 | A→G | M204V |
| 2650 | 28 | C→T | Q884X | |||
| 3226 | 32 | ins G | H1076 FsX51 | |||
| 10 | 225 | Italy | 753 | 9 | +1G→T | Splice error |
| 1847 | 20 | A→G | E616G (splice error) | |||
| 11 | 293 | Morocco | 1822* | 20 | del 12bp | Del V608‐A611 |
| 12 | 198 | Italy | 1847* | 20 | A→G | E616G (splice error) |
| 13 | 336 | Italy | 441* | 6 | del A | P147 FsX14 |
| 175* | 3 | G→A | A59T | |||
| 14 | 159 | Italy | 817* | 10 | C→T | R273X |
| 15 | 132 | Italy | 2039* | 22 | G→C | R680P |
Fs, frameshift.
*Homozygotic mutation.
We identified 16 different mutations in the Munc13–4 gene, of which four had been reported previously: 753+1G→T, causing a change in the splice donor site, initially reported by Feldmann et al26 and subsequently by zur Stadt et al22; 817C→T causing R273X22; 2346–9delGGAG (R782FsX12)22; 1822del 12 bp (del V608‐A611).26
Figure 1 Sites of novel mutations described in this paper relative to exon structure and protein domains: C2 (C2A and C2B) and Munc homology domains (MHD1 and MHD2).
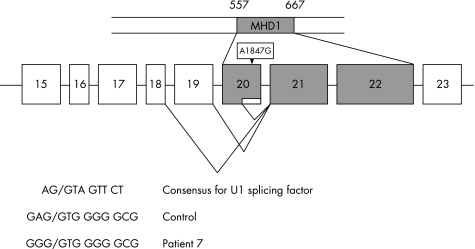
Two mutations occurred at splice recognition sites. One, 1847A→G (E616G) in patients 1, 7, 10 and 12, is located two nucleotides from the end of exon 20, where it is predicted to disrupt splice site recognition. To verify this, we cloned and sequenced cDNA from patients 7 and 10. Fifteen cDNA clones from patient 7 showed abnormal splicing of exon 20, with four causing a deletion of nine amino acids, which remained in frame (fig 2). In patient 10, cDNA analysis showed that both exons 9 and 20 were aberrantly spliced. These results indicate that 1847A→G (E616G) disrupts splicing and results in several RNA products, some of which may encode proteins. The second mutation, 610A→G, (M204V, in patient 9) is located five nucleotides from the end of exon 7. Although it could be expected to affect splicing, we have no experimental data to further support this concept.
Figure 2 Representative cDNA products of Munc13–4 from patient 7. Full‐length cDNAs were isolated with deletions of last 27 or 49 nucleotides of exon 20, deletion of exon 20 or exons 19 and 20. Both the consensus U1 recognition splice site and the genomic mutation in patient 7 are shown.
Five novel missense mutations causing single amino acid changes were also identified in this study. 175G→A (A59T) in patients 3 and 13 occurred in the N‐terminal region of Munc 13–4; 1241G→T (R414L) in patients 1 and 5 between the C2A and the MHD1 domains of the gene; 2039G→C (R680P) in patient 15 in MHD1; 2570T→G (F857C) in patient 2 in MHD2; and 2782C→T (R928C) in patients 1 and 5 between MHD2 and C2B.
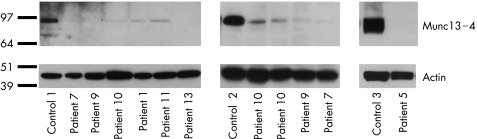
To understand the effects of the mutations, we analysed Munc 13–4 protein expression from CTL or natural killer cells by western blot analysis using an antibody raised to amino acids 1–262 as described by Shirakawa et al.28 Figure 3 shows western blot analysis of lysates from patients 1, 5, 7, 9–11 and 13, all normalised against actin. In addition, in patient 3 (data not shown) no protein was detected. Although nucleotide sequencing suggests that some protein might be produced from all of these cases, trace amounts of protein could be detected in patients only 1, 7 and 9–11, showing that the protein products arising from these mutations are present at markedly reduced levels.
Figure 3 Munc13–4 protein expression is greatly reduced or absent in many cells. Western blot of natural killer cells (left and right) or CD8+CTL (middle panels) lysates from healthy donors (controls 1–3) and patients as indicated, probed with anti‐Munc 13–4 and re‐probed with anti‐actin. Molecular weight markers (kD) are shown on the left.
Our analysis also identified polymorphisms 279C→T and 3198A→G. The 2599A→G transition in exon 27 was found in six unrelated families, including one homozygotic parent. To assess the role of this previously unreported frequent amino acid change, we studied the incidence of the 2599A→G transition in a series of 50 consecutive newborns, of which, 65% were found to be carriers of this amino acid change.
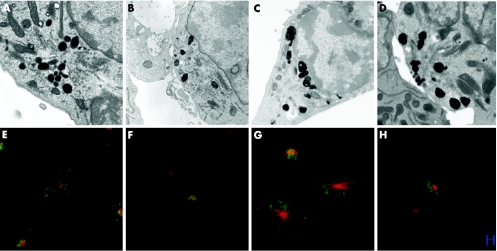
We analysed granule polarisation in FHL3 CTL from patients 6 and 9, in which trace amounts of the Munc13–4 protein could be detected, to see if the typical profile of docked and unsecreted granules previously reported26 could be observed. Figure 4 shows granules distributed throughout CTL before polarisation in both control and FHL3 CTL, with granules polarising at the immunological synapse formed between CTL and target, with many more docked granules visible in patients with FHL3 than in controls (B, D, F, H), consistent with a block in granule secretion in these patients in whom low levels of the Munc 13–4 protein are expressed.
Figure 4 Granule polarisation in control and patients with FHL3. Electron microscopy (A–D) and confocal microscopy (E–H) of control (A, B, E, F) and FHL3 (C, D, G, H) cytotoxic T lymphocyte (CTL). Single CTL (A, E, C, G) shows granules distributed throughout the cells and CTL conjugated to target cells (B, F, D, H). FHL3 cells from patients 7 (C, D) and 9 (G, H). Scale bar = 10μm. Tubulin (red), lamp (green).
Clinical findings
Twelve of our FHL3 families were of Italian origin; among these, no geographical cluster could be identified.
Parental consanguinity was documented in four families (7, 11–12, 15) and was likely in three (patients 8, 13, 14), and also in light of documented close geographical origin of the parents or even of the same family name. Among 321 patients who enrolled in the International HLH Registry (Aricò et al2; Aricò, unpublished data) and evaluated for this parameter, 61 (19%) had related parents.
Among the 15 families with Munc13–4 mutations, 13 had one or more siblings. Of a total of 25 siblings, 8 were affected; 1 additional sibling was found to be genetically identical to the index case and thus considered potentially affected, although in a pre‐symptomatic phase. For this reason, despite being matched for HLA, she could not serve as a stem‐cell donor for her affected brother.
As two families (patients 1 and 5) shared the same couple of heterozygotic mutations in one parent, we investigated their geographical origin, which turned out to be common.
The clinical and laboratory features of the 15 patients with Munc13–4 mutations fit the diagnostic criteria for HLH (fever, splenomegaly, cytopenia, hypertriglyceridaemia or hypofibrinogenaemia and haemophagocytosis).11 Table 2 summarises the main presenting features. The natural killer cell activity in these patients was variably impaired ranging between no activity and 8% killing at erythrocyte: thyrocyte ratio of 20%. Among them, the median age at onset of the disease was 4 months, which is not different from that of our additional 15 patients in whom no Munc13–4 or PRF1 mutations were found (5 months; range 1–10.7 years), yet, remarkably, one of our patients was diagnosed with HLH at the age of 18 years.
Table 2 Presenting features and treatment outcome in 15 patients with haemophagocytic lymphohistiocytosis and Munc13–4 gene mutations.
| Patient/UPN * | 1/237 | 2/196 | 3/180 | 4/253 | 5/277 | 6/205 | 7/249 | 8/3 | 9/289 | 10/225 | 11/293 | 12†/198 | 13/336 | 14/159 | 15/132 |
| No of +siblings/affected | 0/0 | 1/0 | 2/1 | 1/0 | 1/1‡ | 4/2 | 2/1 | 4/0 | 1/0 | 2/1 | 0/0 | 2/1 | 1/0 | 2/1 | 2/1 |
| Ethnic origin | Italy | Italy | Italy | Slovenia | Italy | USA | Italy | Italy | Italy | Italy | Morocco | Italy | Italy | Italy | Italy |
| Consanguinity | – | – | – | – | – | – | + | Poss | – | – | + | + | Poss | Poss | + |
| Sex | M | F | F | F | M | M | F | F | M | F | F | M | M | M | F |
| Age at diagnosis | 7.9 y | 1 m | 2.5 m | 1.5 y | 9.8 y | 1 day | 8.2 y | 3 m | 3 m | 12.7 y | 2 m | 18 y | 4 m | 2 m | 5.9 y |
| Fever | + | + | + | + | + | + | + | + | + | + | – | + | + | + | + |
| Splenomegaly | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Haemoglobin (<9 g/dl) | 8.5 | 7.4 | 7.5 | 7.1 | 9.7 | 7.4 | 10 | + | 5.0 | 10.6 | 4.1 | 7.1 | 7.1 | 8.2 | 5.8 |
| Platelets (<100 000/mm3) | 82 | 74 | 30 | 35 | 39 | 35 | 69 | 8 | 44 | 100 | 4 | 40 | 4 | 32 | 59 |
| Neutrophils (<1000/mm3 | 400 | 380 | 1300 | 500 | 450 | 930 | 800 | + | 1580 | 1000 | 170 | 620 | 330 | 250 | 600 |
| Triglycerides (>265 mg/dl) | NT | 750 | 250 | 753 | + | 370 | 157 | 1540 | 141 | 70 | 567 | 430 | 620 | 228 | 810 |
| Fibrinogen (<150 mg/dl) | 120 | 200 | 47 | 74 | + | 135 | 222 | 89 | 117 | 190 | 42 | 180 | 28 | 100 | 85 |
| CNS symptoms | + | Seizure | + | Seizure | + | – | + | – | – | Ataxia | Seizure | – | + | ||
| CSF pleocytosis (<5/mm3) | 22 | 6 | 0 | 0 | + | ND | 10 | ND | – | 40 | – | 20 | 12 | – | 6 |
| NK activity (% lysis)‡ | 3 | 0 | 0 | 0 | 8 | ND | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| BMT, donor | MUD | MSD | – | MUD | MUD | – | MSD | MSD | MUD | MUD | MUD | – | MSD | MSD | MUD |
| Present status | Asy | Asy | DOD | DOT | Asy | DOD | Asy | Asy | DOD | DOT | Asy | DOT | Well | DOT | DOT |
| Age at last follow‐up | 12.8 y | 6 y | 3 y | 2.6 y | 11.5 y | 3 d | 12 y | 17 y | 1.3 y | 13 y | 16 m. | 18.8 y | 8 m. | 4 m. | 7 y |
Asy, asymptomatic; d, day; DOD, dead of disease; DOT, dead of toxicity; F, female; M, male; m, month; MUD, matched unrelated donor; MSD, matched sibling donor; NA, not applicable; ND, not determined; NK, natural killer; y, years.
Items in bold face comprise the diagnostic criteria for haemophagocytic lymphohistiocytosis.
*Normal limit values are indicated in brackets.
†The clinical features of this patient had been previously reported by Ishii et al31 as case 2.
‡Natural killer cell activity was tested against the K562 cell line after 4‐h incubation, effector:target ratio 20:1 (net variance >10%).
CNS involvement was present in 9 of the 15 patients: 3 had seizures (patients 4, 6, 13), 1 had ataxia (patient 12), 4 had recurrent signs of encephalopathy (patients 1, 5, 7, 15) and 1 (patient 9) developed fulminant encephalopathy, which was the cause of his death.
All but one patient received current HLH‐directed treatment4 and all showed a favourable response, which, however, was followed by early reactivation in six (43%).
Haematopoietic stem‐cell transplantation, the only recognised curative procedure available to date,7,8,9 was given to 12 of the 15 patients; 3 patients had died of early reactivation and progressive disease before transplantation, after initial response to treatment.
Discussion
In this study, we analysed the Munc13–4 gene in 30 families in which HLH had been diagnosed and PRF1 mutations had been ruled out by sequence analysis. Of these, half—15 families—had genetic mutations confirming the pathogenic role of Munc13–4 in families with FHL. Eight families had homozygotic mutations, whereas seven families had compound heterozygotic mutations.
Only four mutations that we observed had been previously reported: 753+1G→T, causing a splice error22,26; 2346–9delGGAG (R782fsX12)22; 817C→T (R273X)22; and 1822del12bp (del V608‐A611).26
Thus, in this study we contribute 12 novel mutations in the Munc13–4 gene, of which 1 (2650C→T) introduced a stop codon and 4 (441delA, 532delC, 3082delC, 3226insG) caused a frameshift.
The pathogenic potential of missense mutations is often questioned. In our series, we identified seven missense mutations. The 1847A→G (E616G) was found in four families, two of which were homozygotic. The western blot analysis of family 7 clearly documents the absence of the band corresponding to the mature form of the protein. Analysis of cDNA products shows that this mutation, which occurs in the U1 recognition site involved in splicing, leads to alternative splicing and loss of functional protein. Recent studies have also shown that exonic sequences contain regulatory elements of splicing that overlap with coding capacity; thus, mutations in exons can also disrupt splicing.30 The drastically reduced levels of proteins observed in patients 1, 5 and 9 could arise either from decreased protein stability or disrupted splicing. The role of the 610A→G mutation in changing splicing can also be predicted for , according to its location.
In family 1, 1847A→G was associated with 1241G→T (R414L) and 2782C→T (R928C), carried by one parent and found in the same combination in another family (patient 5). These two missense mutations were found in association also in family 5. Thus, we are not able at present to evaluate the individual effect of these two missense mutations. The 2570T→G (F857C) observed in family 2 was associated with a known pathogenic mutation (753+1G→T). The 2039G→C (R680P), a mutation in the MHD1 region domain, was observed in one homozygous family (patient 15). The 175G→A (A59T) mutation was found in families 3 and 13. In all these families they were found on the same allele containing a pathogenic mutation such as splice error (family 3) or frameshift (family 13). Thus their pathogenic potential might be limited but cannot be further assessed at present.
Of the 15 families with FHL3, only two had a single mutation. Interestingly both had the same mutation—2346–9delGGAG (R782FsX12)—, and the same pattern was reported by zur Stadt,22 suggesting a non‐random event.
We analysed eight patients for the functional impact of their Munc13–4 mutations on the protein. In keeping with the findings of Ishii et al31 in two patients, none of our patients had a normal protein level; it was either reduced or absent.
In our analysis we found the polymorphisms 279C→T and 3198A→G. Furthermore, we unexpectedly observed as a frequent finding the 2599A→G (K867G) transition in exon 27. It was found both in the homozygous or heterozygous states, in patients with other mutations or in patients with no other detectable mutations, who were thus considered as non‐FHL3. Although the transition was associated with an amino acid change, its incidence and the finding of homozygosity in an healthy father, prompted us to investigate its incidence among normal controls, who were shown to be carriers at a rate of 65%. Thus, we document 2599A→G as a novel polymorphism of the Munc13–4 gene.
The median age at the onset of the disease in our 15 patients with FHL3 was comparable to that of the 15 remaining patients in whom Munc13–4 and PRF1 mutations were not found, and also of patients with HLH from the International registry (data not shown).2 Among 113 patients with HLH enrolled in the HLH‐94 study, the mean age at the disease onset was 19 months (range 0–145).4 Nevertheless, it is remarkable that late onset beyond 5 years was found in 6 of 15 (40%) of our patients with FHL3 compared with only 17% of all Registry patients (data not shown).2 Whether the delayed onset may depend on the presence of mutations allowing residual protein activity remains to be clarified in a genotype–phenotype study when a larger number of cases of FHL3 becomes available. In particular, one patient was diagnosed with HLH when he was already 18 years old. For FLH3, as we have previously reported for patients with FHL2,20 clinicians must be aware that, although most patients will be diagnosed when infants or young children,2 the diagnosis of FHL—at least for these two genetic subtypes—must also be considered for young adult patients with fever, cytopenia, splenomegaly and hypercytokinemia of unknown origin.32 Thus, such information should also be widely shared with adult haematologists who may be expected care for such patients.
The mutation 1847A→G appears to be linked with the age of the disease onset, as all four patients in whom it was found (two homozygous and two with compound heterozygosity), developed the disease when 7.9 years or older. Our study shows that this mutation disrupts splicing and gives rise to a number of different splice forms, some of which may encode functional protein.
CNS involvement is a frequent finding in HLH.2,3 In a recent analysis of the HLH‐2004 trial,33 122 of 193 patients enrolled (63%) had neurological symptoms or abnormal CSF at onset. In our small series, CNS involvement appears to be a major feature of patients with FHL3, and may reflect the important role played by Munc family proteins in the CNS.34
Patients with FHL3 are characterised by a profound impairment of natural killer cell activity, a finding that is confirmed in all of our patients. Therefore assaying natural killer cell activity provides a reliable tool for identification of HLH patients who are candidates for Munc13–4 mutation.
In conclusion, mutations of the Munc13–4 gene are found in half of the patients with HLH in whom PRF1 mutations had been previously ruled out. If one takes into consideration the impact of Munc13–4 mutations among patients with all subsets of HLH, this subset accounts for about 30% of all HLH patients in Italy (data not shown). This rate appears comparable with that reported in Japan,35,36 but not with other countries, such as Germany.2 The disease may present at any age including young adulthood, often with major CNS involvement, and is usually associated with a marked defect of natural killer cell activity. At present, other tools (immunological, flow‐cytometric) for rapid screening of Munc13–4‐defective patients are not available. Thus, mutation analysis remains the gold standard to confirm the diagnosis of FHL3. Two mutations appear to be of special interest in our experience: 753+1G→T, which has been reported by the French26 and German22 groups and occurred in three of our families, and the 1847A→G (E616G), a novel mutation identified in four Italian families. Both mutations are responsible for impaired splicing of the gene product. Although we did not find any hot spot or mutation clustering, it is remarkable that all mutations with defined pathogenic role fall within the functional domains of the Munc protein. Despite the high number of coding exons, we consider that mutation analysis of all the coding exons of Munc13–4 a necessary, although expensive and time consuming, tool to confirm the diagnosis, to refine the therapeutic choice including indication for haematopoietic stem‐cell transplantation, and to identify the carriers and thus select familial donors. Information on the familial genetic markers may allow also prenatal diagnosis. Identification of pre‐symptomatic affected siblings may represent another challenge for the medical team, which will provide a tailored clinical and functional monitoring to determine if and when treatment may be indicated.
Abbreviations
BSA - bovine serum albumin
CTL - cytotoxic T lymphocyte
FHL - familial haemophagocytic lymphohistiocytosis
HLH - haemophagocytic lymphohistiocytosis
HRP - horseradish peroxidase
MIM - mendelian inheritance in man
PBMC - peripheral blood mononuclear cells
PBS - phosphate‐buffered saline
PCR - polymerase chain reaction
PHA - phyto haemagglutinin
PRF - prolactin‐releasing factor
Footnotes
Funding: This work was funded by grants from the MIUR‐FIRB(LDN), AIRC (MA, AS, DP, LM), Associazione Ricerca Sindromi Emofagocitiche ‐ARSE (MA, GB), Ricerca finalizzata 2004 (MA, AS, DP, FD, LDN), Fondazione Compagnia di San Paolo (DP, LM) and the Wellcome Trust, UK (GMG, GB, JCS, FG).
Competing interests: None declared.
References
- 1.Farquhar J, Claireaux A. Familial haemophagocytic reticulosis. Arch Dis Child 195227519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aricò M, Janka G, Fischer A, Henter J I, Blanche S, Elinder G, Martinetti M, Rusca M P. Hemophagocytic lymphohistiocytosis. Report of 122 children from the International Registry. FHL Study Group of the Histiocyte Society. Leukemia 199610197–203. [PubMed] [Google Scholar]
- 3.Janka G E. Familial hemophagocytic lymphohistiocytosis. Eur J Pediatr 1983140221–230. [DOI] [PubMed] [Google Scholar]
- 4.Henter J I, Samuelsson‐Horne A, Aricò M, Egeler R M, Elinder G, Filipovich A H, Gadner H, Imashuku S, Komp D, Ladisch S, Webb D, Janka G, Histocyte Society Treatment of hemophagocytic lymphohistiocytosis with HLH‐94 immunochemotherapy and bone marrow transplantation. Blood 20021002367–2373. [DOI] [PubMed] [Google Scholar]
- 5.Janka G E, Schneider E M. Modern management of children with haemophagocytic lymphohistiocytosis. Br J Haematol 20041244–14. [DOI] [PubMed] [Google Scholar]
- 6.Stephan J L, Donadieu J, Ledeist F, Blanche S, Griscelli C, Fischer A. Treatment of familial hemophagocytic lymphohistiocytosis with antithymocyte globulins, steroids, and cyclosporin A. Blood 1993822319–2323. [PubMed] [Google Scholar]
- 7.Jabado N, de Graeff‐Meeder E R, Cavazzana‐Calvo M, Haddad E, Le Deist F, Benkerrou M, Dufourcq R, Caillat S, Blanche S, Fischer A. Treatment of familial hemophagocytic lymphohistiocytosis with bone marrow transplantation from HLA genetically nonidentical donors. Blood 1997904743–4748. [PubMed] [Google Scholar]
- 8.Durken M, Horstmann M, Bieling P, Erttmann R, Kabisch H, Loliger C, Schneider E M, Hellwege H H, Kruger W, Kroger N, Zander A R, Janka G E. Improved outcome in haemophagocytic lymphohistiocytosis after bone marrow transplantation from related and unrelated donors: a single‐centre experience of 12 patients. Br J Haematol 19991061052–1058. [DOI] [PubMed] [Google Scholar]
- 9.Horne A, Janka G, Egeler M R, Gadner H, Imashuku S, Ladisch S, Locatelli F, Montgomery S M, Webb D, Winiarski J, Filipovich A H, Henter J I, Histiocyte Society Haematopoietic stem cell transplantation in haemophagocytic lymphohistiocytosis. Br J Haematol 2005129622–630. [DOI] [PubMed] [Google Scholar]
- 10.Aricò M, Allen M, Brusa S, Clementi R, Pende D, Maccario R, Moretta L, Danesino C. Haemophagocytic lymphohistiocytosis: proposal of a diagnostic algorithm based on perforin expression. Br J Haematol 2002119180–188. [DOI] [PubMed] [Google Scholar]
- 11.Henter J I, Elinder G, Ost A. Diagnostic guidelines for hemophagocytic lymphohistiocytosis. The FHL Study Group of the Histiocyte Society. Semin Oncol 19911829–33. [PubMed] [Google Scholar]
- 12.Perez N, Virelizier J L, Arenzana‐Seisdedos F, Fischer A, Griscelli C. Impaired natural killer activity in lymphohistiocytosis syndrome. J Pediatr 1984104569–573. [DOI] [PubMed] [Google Scholar]
- 13.Arico M, Nespoli L, Maccario R, Montagna D, Bonetti F, Caselli D, Burgio G R. Natural cytotoxicity impairment in familial haemophagocytic lymphohistiocytosis. Arch Dis Child 198863292–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider E M, Lorenz I, Muller‐Rosenberger M, Steinbach G, Kron M, Janka‐Schaub G E. Hemophagocytic lymphohistiocytosis is associated with deficiencies of cellular cytolysis but normal expression of transcripts relevant to killer‐cell‐induced apoptosis. Blood 20021002891–2898. [DOI] [PubMed] [Google Scholar]
- 15.Ohadi M, Lalloz M R, Sham P.et al Localization of a gene for familial hemophagocytic lymphohistiocytosis at chromosome 9q21.3–22 by homozygosity mapping. Am J Hum Genet 199964165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dufurcq‐Lagelouse R, Jabado N, Le Deist F.et al Linkage of familial hemophagocytic lymphohistiocytosis to 10q21–22 and evidence for heterogeneity. Am J Hum Genet 199964172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stepp S E, Dufourcq‐Lagelouse R, Le Deist F.et al Perforin gene defects in familial hemophagocytic lymphohistiocytosis. Science 19992861957–1959. [PubMed] [Google Scholar]
- 18.Arico M, Danesino C, Pende D, Moretta L. Pathogenesis of haemophagocytic lymphohistiocytosis. Br J Haematol 2001114761–769. [DOI] [PubMed] [Google Scholar]
- 19.Stinchcombe J, Bossi G, Griffiths G M. Linking albinism and immunity: the secrets of secretory lysosomes. Science 200430555–59. [DOI] [PubMed] [Google Scholar]
- 20.Clementi R, zur Stadt U, Savoldi G, Varotto S, Conter V, De Fusco C, Notarangelo L D, Schneider M, Klersy C, Janka G, Danesino C, Arico M. Six novel mutations in the PRF1 gene in children with haemophagocytic lymphohistiocytosis. J Med Genet 200138643–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goransdotter Ericson K, Fadeel B, Nilsson‐Ardnor S, Soderhall C, Samuelsson A, Janka G, Schneider M, Gurgey A, Yalman N, Revesz T, Egeler R, Jahnukainen K, Storm‐Mathiesen I, Haraldsson A, Poole J, de Saint Basile G, Nordenskjold M, Henter J. Spectrum of perforin gene mutations in familial haemophagocytic lymphohistiocytosis. Am J Hum Genet 200168590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zur Stadt U, Beutel K, Kolberg S, Schneppenheim R, Kabisch H, Janka G, Hennies H C. Mutation spectrum in children with primary hemophagocytic lymphohistiocytosis: molecular and functional analyses of PRF1, UNC13D, STX11, and RAB27A. Hum Mutat 20062762–68. [DOI] [PubMed] [Google Scholar]
- 23.Molleran Lee S, Villanueva J, Sumegi J, Zhang K, Kogawa K, Davis J, Filipovich A H. Characterisation of diverse PRF1 mutations leading to decreased natural killer cell activity in North American families with haemophagocytic lymphohistiocytosis. J Med Genet 200441137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueda I, Morimoto A, Inaba T, Yagi T, Hibi S, Sugimoto T, Sako M, Yanai F, Fukushima T, Nakayama M, Ishii E, Imashuku S. Characteristic perforin gene mutations of haemophagocytic lymphohistiocytosis patients in Japan. Br J Haematol 2003121503–510. [DOI] [PubMed] [Google Scholar]
- 25.Risma K A, Frayer R W, Filipovich A H, Sumegi J. Aberrant maturation of mutant perforin underlies the clinical diversity of hemophagocytic lymphohistiocytosis. J Clin Invest 2006116182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feldmann J, Callebaut I, Raposo G, Certain S, Bacq D, Dumont C, Lambert N, Ouachee‐Chardin M, Chedeville G, Tamary H, Minard‐Colin V, Vilmer E, Blanche S, Le Deist F, Fischer A, de Saint Basile G. Munc13–4 is essential for cytolytic granules fusion and is mutated in a form of familial hemophagocytic lymphohistiocytosis (FHL3). Cell 2003115461–473. [DOI] [PubMed] [Google Scholar]
- 27.zur Stadt U, Schmidt S, Kasper B, Beutel K, Diler A S, Henter J I, Kabisch H, Schneppenheim R, Nurnberg P, Janka G, Hennies H C. Linkage of familial hemophagocytic lymphohistiocytosis (FHL) type‐4 to chromosome 6q24 and identification of mutations in syntaxin 11. Hum Mol Genet 200514827–834. [DOI] [PubMed] [Google Scholar]
- 28.Shirakawa R, Higashi T, Tabuchi A, Yoshioka A, Nishioka H, Fukuda M, Kita T, Horiuchi H. Munc13–4 is a GTP‐Rab27‐binding protein regulating dense core granule secretion in platelets. J Biol Chem 200427910730–10737. [DOI] [PubMed] [Google Scholar]
- 29.Stinchcombe J C, Page L J, Griffiths G M. Secretory lysosome biogenesis in cytotoxic T lymphocytes from normal and Chediak Higashi syndrome patients. Traffic 20001435–444. [DOI] [PubMed] [Google Scholar]
- 30.Pagani F, Raponi M, Baralle F E. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution Proc Natl Acad Sci USA 20051026368–6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishii E, Ueda I, Shirakawa R, Yamamoto K, Horiuchi H, Ohga S, Furuno K, Morimoto A, Imayoshi M, Ogata Y, Zaitsu M, Sako M, Koike K, Sakata A, Takada H, Hara T, Imashuku S, Sasazuki T, Yasukawa M. Genetic subtypes of familial hemophagocytic lymphohistiocytosis: correlations with clinical features and cytotoxic T lymphocyte/natural killer cell functions. Blood 20051053442–3448. [DOI] [PubMed] [Google Scholar]
- 32.Allen M, De Fusco C, Legrand F, Clementi R, Conter V, Danesino C, Janka G, Arico M. Familial hemophagocytic lymphohistiocytosis: how late can the onset be? Haematologica 200186499–503. [PubMed] [Google Scholar]
- 33.Horne A C, Trottestam H, Aricò M, Egeler M, Filipovich A, Gadner H, Imashuku S, Ladisch S, Webb D, Janka G, Henter J ‐ I. Frequency and spectrum of CNS involvement in 193 children with hemophagocytic lymphohistiocytosis. Histiocyte Society Meeting 2005 [DOI] [PubMed]
- 34.Betz A, Telemenakis I, Hofmann K, Brose N. Mammalian Unc–13 homologues as possible regulators of neurotransmitter release. Biochem Soc Trans 199624661–666. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto K, Ishii E, Sako M, Ohga S, Furuno K, Suzuki N, Ueda I, Imayoshi M, Yamamoto S, Morimoto A, Takada H, Hara T, Imashuku S, Sasazuki T, Yasukawa M. Identification of novel MUNC13–4 mutations in familial haemophagocytic lymphohistiocytosis and functional analysis of MUNC13–4‐deficient cytotoxic T lymphocytes. J Med Genet 200441763–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueda I, Ishii E, Morimoto A, Ohga S, Sako M, Imashuku S. Correlation between phenotypic heterogeneity and gene mutational characteristics in familial hemophagocytic lymphohistiocytosis (FHL). Pediatr Blood Cancer 200646482–488. [DOI] [PubMed] [Google Scholar]