Abstract
Aims
To determine the relative frequency of mutations in three different genes (low‐density lipoprotein receptor (LDLR), APOB, PCSK9), and to examine their effect in development of coronary heart disease (CHD) in patients with clinically defined definite familial hypercholesterolaemia in UK.
Patients and methods
409 patients with familial hypercholesterolaemia patients (158 with CHD) were studied. The LDLR was partially screened by single‐strand conformational polymorphism (SSCP) (exons 3, 4, 6–10 and 14) and by using a commercial kit for gross deletions or rearrangements. APOB (p.R3500Q) and PCSK9 (p.D374Y) were detected by specific assays. Coding exons of PCSK9 were screened by SSCP.
Results
Mutations were detected in 253 (61.9%) patients: 236 (57.7%) carried LDLR, 10 (2.4%) carried APOB p.Q3500 and 7 (1.7%) PCSK9 p.Y374. No additional mutations were identified in PCSK9. After adjusting for age, sex, smoking and systolic blood pressure, compared to those with no detectable mutation, the odds ratio of having CHD in those with an LDLR mutation was 1.84 (95% CI 1.10 to 3.06), for APOB 3.40 (0.71 to 16.36), and for PCSK9 19.96 (1.88 to 211.5; p = 0.001 overall). The high risk in patients carrying LDLR and PCSK9 p.Y374 was partly explained by their higher pretreatment cholesterol levels (LDLR, PCSK9 and no mutation, 10.29 (1.85), 13.12 and 9.85 (1.90) mmol/l, respectively, p = 0.001). The post‐statin treatment lipid profile in PCSK9 p.Y374 carriers was worse than in patients with no identified mutation (LDL‐C, 6.77 (1.82) mmol/l v 4.19 (1.26) mmol/l, p = 0.001, HDL‐C 1.09 (0.27) mmol/l v 1.36 (0.36) mmol/l, p = 0.03).
Conclusions
The higher CHD risk in patients carrying PCSK9 p.Y347 or a detected LDLR mutation supports the usefulness of DNA testing in the diagnosis and management of patients with familial hypercholesterolaemia. Mutations in PCSK9 appear uncommon in patients with familial hypercholesterolaemia in UK.
Familial hypercholesterolaemia is an autosomal dominant disorder associated with increased risk of coronary heart disease (CHD), with an estimated prevalence in the UK of 1 in 500 to 1 in 600.1 Roughly half of the men with familial hypercholesterolaemia, if untreated, will have developed clinically evident CHD by the age of 55 years. Affected women from the same families typically develop CHD about 9 years later than their affected male relatives, but again, often remarkably prematurely.2 The proportion of patients with familial hypercholesterolaemia identified and being treated in lipid clinics to date in the UK is, at best, 15% of the predicted number, with most of these being young people.1 Because lipid‐lowering drug treatment with statins substantially reduces coronary morbidity and mortality,3 identification of affected people by screening is crucially important. To this end, the Department of Health has recently funded five pilot sites in the UK to determine the efficiency of cascade testing in the current social structure and the framework of the National Health Service. Cascade testing is a cost‐effective method of finding additional patients with familial hypercholesterolaemia,4 and has been used extensively in other countries in Europe, most notably in Holland,5 for the past 5 years.
Key points
Patients with familial hypercholesterolaemia with a detectable LDLR mutation have a higher risk of early CHD than those in whom no mutation was detected.
Patients with the pD374Y mutation in PCSK9 have the highest pretreatment and post‐treatment levels of plasma cholesterol and the highest risk of early CHD.
Mutations in PCSK9 appear to be uncommon in patients with familial hypercholesterolaemia in UK.
The extent to which DNA testing for familial hypercholesterolaemia complements cholesterol measurement in cascade screening to identify affected patients is unclear, as is its role in determining the risk of CHD and response to treatment. In the current study, we carried out molecular genetic testing in patients recruited from the Simon Broome familial hypercholesterolaemia register6 in the UK as a cross‐sectional cohort study to identify risk factors for premature CHD in patients with familial hypercholesterolaemia.7 Primary results from this study indicated that the conventional cardiovascular risk factors of age, sex, smoking, pretreatment cholesterol levels and low levels of high‐density lipids (especially in women) were all associated with higher risk of CHD,7 confirming associations reported in other studies, for example.8 When this UK study was started, mutations at two loci causing familial hypercholesterolaemia had been identified, with mutations in the low‐density lipoprotein receptor gene (LDLR) accounting for most of the identified mutations,9 whereas one particular mutation in the gene encoding the ligand for the low density lipoprotein (LDL) receptor—namely apolipoprotein B (FDB)—occurs in about 5% of patients in the UK.10 This mutation, which alters a single amino acid (p.R3500Q), has been shown to reduce the affinity of the LDL cholesterol particle,10 where ApoB is the single‐protein component for the receptor. For the LDLR, currently >100 mutations have been reported in UK patients to date9,11 (see also www.ucl.ac.uk/FH). A commercially available kit for screening for deletions and rearrangements of the LDLR gene has become available, and it is known that up to 5% of patients with familial hypercholesterolaemia in patients in the UK may have such a deletion.12
Recently, defects in a third gene causing monogenic hypercholesterolaemia have been identified.13 The gene protein convertase subtilisin/kexin type 9 (PCSK9) codes for an enzyme that has also been called “neural apoptosis regulated convertase 1”, which has recently been proposed to be involved in degrading the LDLR protein in the lysosome of the cell and preventing it from recycling.14 Gain‐ of‐ function mutations in the PCSK9 gene could therefore cause increased degradation of LDLRs, reduced numbers of receptors on the surface of the cell, and monogenic hypercholesterolaemia. An alternative mechanism has also been proposed for the hypercholesterolaemic effect, whereby the gain of function causes increased secretion of apoB‐containing lipoproteins from the liver, with this being supported by in vivo turnover studies in patients carrying PCSK9 missense mutations15 and by in vitro studies in transiently transfected rat liver cells.16 One mutation in this gene, p.D374Y, has been reported in several independent families16,17,18 and we therefore also tested for this cause of familial hypercholesterolaemia in this group of patients, as well as using single‐strand conformational polymorphism (SSCP) analysis and direct sequencing to screen all coding exons of the gene.
Although patients with no identified mutation may have a monogenic cause of the disorder in an as yet undiscovered gene, it is also possible that some may have polygenic hypercholesterolaemia and have been misclassified using current clinical diagnostic criteria. These patients would be expected to have a milder degree of hyperlipidaemia, possibly not present from birth but only developing in later life, and would therefore be predicted to have a lower risk of CHD. The hypothesis we set out to test was that patients with identified mutations in the LDLR, PCSK9 or APOB genes would be at greater risk of CHD than patients with no identified mutation.
Materials and methods
Patient selection criteria
All patients were participating in the Simon Broome British Heart Foundation study, which is a cross‐sectional comparison of Caucasian patients aged ⩾18 years with treated heterozygous familial hypercholesterolaemia, with and without clinically documented CHD.7 Recruitment methods, and inclusion, exclusion and diagnostic criteria were as defined previously.6,7 Clinical CAD was defined as a definite myocardial infarction (new Q waves or ST elevation or new T wave inversion persisting in more than two leads with creatine kinase >400 IU/l or other equivalent enzyme changes), or having undergone a coronary artery bypass grafting, percutaneous transluminal coronary angioplasty, having angina with an ischaemic resting echocardiogram or a reported angiogram showing clinically important stenosis (percentage stenosis was not used to define this as some records were incomplete for patients who had undergone angiography up to 25 years previously). Cases were thus categorised as only those with an unequivocal diagnosis, and excluded patients with equivocal disease—that is, positive exercise echocardiogram, but either no angiogram had been carried out or the result was reported as normal. Of the 710 patients eligible, 410 met all inclusion criteria (see Results section7 for full details). Participants remained on their usual drug treatment and attended the clinic after an overnight fast of at least 12 h duration before measurement of blood pressure, height and weight. Pretreatment cholesterol levels were obtained from patient records. Currently prescribed drug treatment, alcohol consumption and smoking habit were documented (ever smoking was defined as having smoked at least one cigarette a day for at least 1 year), and a venous blood sample was collected into EDTA, fluoride and citrate vacutainers. Standard assays for plasma lipids were carried out as described.7 Low‐density lipoprotein concentrations were estimated using the Friedewald formula as described.7
Molecular genetic analysis
Genomic DNA was isolated from whole blood samples using standard methods.11 DNA was not available from one of the original 410 people.7LDLR (NM 000527.2) was screened by SSCP analysis11 only on exons that had previously been shown to contain a high prevalence of mutations—namely, exons 3, 4 (carried out in three overlapping fragments), 6–10 and 14. Oligonucleotide primers were designed to cover intron–exon junctions and 10–30‐bp regions of the introns.11 Polymerase chain reaction (PCR) products that showed SSCP band shifts potentially causing familial hypercholesterolaemia were subsequently sequenced using MegaBACE 1000, 96‐capillaries DNA sequencing system (Amersham Biosciences UK, Bucks, UK). The APOB (NM 174936) p.R3500Q mutation was screened by direct PCR assay.11 Gross deletions/insertions of the LDLR were analysed using a kit from MRC Holland (cat no SALSAP062), using the 96‐capillary MegaBACE machine (full details to be presented elsewhere). Detected variants were designated as not affecting function or as pathogenic, using criteria as described previously.11 Mutation nucleotide numbers were designated using the LDLR sequence reported (www.ucl.ac.uk/fh), with cDNA numbering beginning with A of ATG = 1. PCSK9 (NM 000384.1) coding exons were screened by SSCP using primers and conditions shown in Supplementary table A (available online at http://jmg.bmjjournals.com/supplemental). Genotyping for the PCSK9, exon 7 c.1120 G→T (p.D374Y) mutation was carried out using primers and conditions as follows: forward, 5′‐CTTAGGAGGGGACATTTGAGTGG‐3′; reverse, 5′‐TCTAATACAGCCCTGACCTCGTGT‐3′. The 20‐μl PCR reaction contained 16 mM (NH4)2SO4; 67 mM Tris‐HCl, Ph 8.3; 0.01% Tween 20; 0.2 mM dATP, dGTP, dTTP, dCTP; 1.5 mM MgCl2; 8 pM of each primer and 0.2 U of Taq polymerase (Bioline, UK), in addition to 15 ng genomic DNA. Samples were overlaid with mineral oil. The PCR protocol consisted of 95°C for 5 min; 32 cycles of 95°C for 30 s, 61°C for 30 s, and 72°C for 30 s; and 72°C for 5 min. Restriction digest was carried out with AluI (New England Biolabs) for 4 h at 37°C. GG (wild‐type allele); homozygotes produce band sizes of 58 bp (invariant site) and 460 bp, whereas GT (mutant allele carriers) heterozygotes produce band sizes of 58, 199, 261 and 460 bp. Products were separated on 7.5% polyacrylamide gel and visualised by staining with ethidium bromide using the microarray diagonal gel electrophoresis system. For both APOB and PCSK9, all gels included a positive heterozygous control sample (confirmed by direct sequencing), and genotypes were read by two independent observers blind to case–control status, and any discrepancies were confirmed by repeating PCR and digestion. Mutations were designated according to the Human Genome Variation Society guidelines (http://www.hgvs.org/mutnomen/).
Statistical analysis
All statistical analyses were carried out using STATA (Intercooled Stata 8.2, Stata Corporation, TX, USA). Pretreatment cholesterol levels were not available for 110 patients, and this is reflected in differences in the numbers shown in the tables. Concentrations of serum cholesterol, LDL‐C, HDL‐C, ApoA1, ApoB and triglyceride were not normally distributed, and are presented as geometric means with an approximate standard deviation. Differences in baseline characteristics between patients with or without CHD were assessed using t tests with log transformation where distribution was non‐gaussian for continuous variables and χ2 tests for binary variables. Differences in the proportions of each mutation type were tested by Fisher's exact test. Logistic regression models were fitted to obtain odds ratios after adjustment for confounders. Pairwise comparisons were made without adjustment for multiple comparisons because, although the type I error is reduced, such adjustment increases the likelihood of type II error and fewer misinterpretations are made when no adjustment is carried out.19 A p value of <0.05 was taken as significant.
Results
The characteristics of the patients recruited with CHD‐positive and CHD‐negative definite familial hypercholesterolaemia have been presented elsewhere7 (table 1). A higher proportion of the CHD‐positive patients were male, and were on average 11 years older than the CHD‐negative group, with a higher body mass index, A greater proportion of CHD‐positive patients had a history of smoking and their systolic blood pressure and pretreatment serum total cholesterol levels were considerably higher. Their lower mean LDL‐cholesterol at recruitment reflects the more aggressive lipid‐lowering treatment being used in these patients.
Table 1 Baseline characteristics of patients with definite familial hypercholesterolaemia, with and without documented coronary heart disease.
| CHD negative n = 251 | CHD positive n = 158 | p Value | |
|---|---|---|---|
| % Male (n) | 42.6 (107) | 65.2 (103) | <0.001 |
| Age (years)* | 44.5 (13.6) | 56.2 (10.3) | <0.001 |
| BMI (kg/m2)* | 23.7 (4.1) | 25.0 (3.5) | 0.004 |
| % ever smokers (n) | 37.8 (95) | 60.8 (96) | <0.001 |
| % current smokers (n) | 15.1 (38) | 11.4 (18) | 0.28 |
| SBP (mm Hg)* | 124.3 (16.0) | 130.7 (19.9) | <0.001 |
| DBP (mm Hg)* | 76.8 (9.8) | 78.6 (12.4) | 0.12 |
| Pretreatment cholesterol (mmol/l)* | 9.89 (1.73) (n = 185) | 10.55 (2.11) (n = 114) | 0.004 |
| Recruitment LDL‐chol (mmol/l)* | 4.77 (1.26) | 4.21 (1.23) | <0.001 |
| Recruitment HDL‐chol (mmol/l)* | 1.39 (0.35) | 1.28 (0.36) | 0.004 |
BMI, body mass index; CHD, coronary heart disease; DBP, diastolic blood pressure; HDL‐chol, high‐density lipoprotein‐cholesterol; LDL‐chol, low‐density lipoprotein‐cholesterol; N, number of subjects; SBP, systolic blood pressure.
LDL‐chol and HDL‐chol values at recruitment are available for all patients.
*For traits marked (values in parentheses = standard deviations)
Number of patients with pretreatment cholesterol data available is shown.
Overall, 236 of the patients had a detectable mutation in LDLR, 10 carried the p.Q3500 mutation in the APOB gene and 7 the p.Y374 mutation in PCSK9. Of the LDLR mutations, 32 (7.8%) were gross deletions or rearrangements (manuscript in preparation), with 11 patients showing deletion of exons 2–6, and the remainder being small insertions, deletions or single‐base changes that were detected by the SSCP analysis. A full list of mutations identified is presented in supplementary table B (available online at http://jmg.bmjjournals.com/supplemental). Table 2 shows a list of the 21 most common mutations. Testing for the nine most common LDLR mutations and the deletion of exons 2–6, and the APOB and PCSK9 mutations, would identify the genetic cause of the disorder in 54.2% of those with an identified mutation and in 33.5% of the whole patient group.
Table 2 Distribution and relative frequency of common mutations (occurring in >2 patients) in 253 UK patients with familial hypercholesterolaemia with detected mutations.
| Mutation | Mutation(old name) | n (%)* |
|---|---|---|
| i3 splice donor c.313+1 G→A | 313+1 G→A | 27 (11.4) |
| p.E101K | p.E80K | 21 (8.9) |
| p.P685L | p.P664L | 16 (6.8) |
| p.G218del | p.G197del | 12 (5.1) |
| Del exon 2–6 | 11 (4.7) | |
| APOB–p.R3500Q | APOB–p.R3500Q | 10 |
| p.R350X | p.R329X | 9 (3.8) |
| PCSK9 – p.D374Y | PCSK9–p.D374Y | 7 |
| p.E228X | p.E207X | 7 (3.0) |
| p.Q384X/p.D386E | p.Q363X/p.D365E | 7 (3.0) |
| p.D221G | p.D200G | 5 (2.1) |
| p.D482H | p.D461H | 5 (2.1) |
| p.C89Y | p.C68Y | 4 (1.7) |
| p.D227fs | p.D206fs | 4 (1.7) |
| Del exon 5 | 4 (1.7) | |
| p.D221N | p.D200N | 3 (1.3) |
| p.C313X | p.C292X | 3 (1.3) |
| p.C358Y | p.C337Y | 3 (1.3) |
| p.G374fs | p.G353fs | 3 (1.3) |
| p.L479P | p.L458P | 3 (1.3) |
| p.C677R | p.C656R | 3 (1.3) |
| p.I687fs | p.I666fs | 3 (1.3) |
| Del exon 7 | 3 (1.3) | |
| ⩽2 | 80 |
*Percentage of all detected LDLR mutations.
Mutations were designated according to http://www.hgvs.org/mutnomen/.
Screening was carried out on all coding exons of PCSK9 in those 156 patients in whom no LDLR or APOB mutation had been identified. Exon 1 is highly polymorphic,20 and this exon was examined by direct sequencing. Apart from previously identified18,20,21 common variants in exons 1, 4, 5, 7, 9 and 12, no pathogenic sequence changes were identified (table 3).
Table 3 Nucleotide changes identified by SSCP or direct sequencing (for exon 1) in PCSK9 in 156 patients with familial hypercholesterolaemia with no detected LDLR or APOB mutation.
| Fragment | Position | Sequence variant | Amino acid | Allele frequency* | rs number | |
|---|---|---|---|---|---|---|
| Ex1 | Exon 1 5′UTR | c.‐64C→T | – | 0.110 | – | |
| Exon 1 | c.42_43insCTG | p.15_16insL | 0.120 | – | ||
| Exon 1 | c.137G→T | – | 0.004 | |||
| Exon 1 | c.141C→T | p.S47S | 0.004 | – | ||
| Exon 1 | c.158C→T | p.A53V | 0.110 | rs11583680 | ||
| Intron 1 | c.207+15G→A | – | 0.050 | rs2495482 | ||
| Ex2 | None found | |||||
| Ex3 | None Found | |||||
| Ex4 | Intron 3 | c.524–11G→A | – | 0.040 | – | |
| Intron 4 | c.657+9G→A | – | 0.040 | – | ||
| Ex5 | Intron 4 | c.658–7C→T | – | 0.320 | rs2483205 | |
| Intron 5 | c.799+3A→G | – | 0.320 | rs2495477 | ||
| Ex6 | None found | |||||
| Ex75′ | Exon 7 | c.1035G→A | p.P345P | 0.003 | – | |
| Ex73′ | Exon 7 | c.1120G→T | p.D374Y | 0.020 | – | |
| Ex8 | None found | |||||
| Ex9 | Exon 9 | c.1380G→A | p.V460V | 0.090 | rs540796 | |
| Exon 9 | c.1420A→G | p.I474V | 0.090 | rs562556 | ||
| Ex10 | None found | |||||
| Ex11 | None found | |||||
| Ex12 | Exon 12 | c.2009A→G | p.E670G | 0.055 | rs505151 | |
Table 4 shows the prevalence of CHD in patients with the three different genetic causes of familial hypercholesterolaemia. Overall, there was significant evidence for an association between gene‐mutation group and CHD status (p = 0.03). After adjusting for age, sex, smoking (never versus ex plus current) and systolic blood pressure, compared with the patients with no identified mutation, those with any LDLR mutation had an odds ratio (confidence interval (CI)) of 1.84 (1.10 to 3.06; p = 0.02) of being CHD positive and those with the PCKS9 mutation of 19.96 (1.88 to 211.6; p = 0.01). Although carriers of the APOB mutation appeared to have an increased risk of CHD, this was not significant. As shown in table 4, adjustment for recruitment levels of LDL and HDL, or for other measured risk factors such as recruitment levels of fibrinogen and homocysteine,7 did not considerably alter these risk estimates. Untreated total cholesterol levels were not available for 110 people whose age, body mass index, prevalence of current smoking, systolic and diastolic blood pressure and treated lipid levels were not significantly different from those with complete data (not shown). When missing data were replaced with the mean untreated total cholesterol level for all patients in that mutation group and compared with those with no detected mutation, the odds ratio (CI) for CHD (1.70 after this adjustment, for those with any LDLR mutation, remained significant (table 4; 1.70 (1.01 to 2.86), p = 0.05).
Table 4 Odds ratio for coronary heart disease by mutation type in men and women combined, unadjusted and adjusted for risk factors.
| CHD+ve/CHD−ve (% CHD+ve) | OR* (95% CI) | OR† (95% CI) | OR‡ (95% CI) | OR§ (95% CI) | |
|---|---|---|---|---|---|
| None | 55/101 (35.2) | 1.00 | 1.00 | 1.00 | 1.00 |
| LDLR (any) | 91/145 (38.6) | 1.84 (1.10 to 3.06) p = 0.02 | 1.81 (1.08 to 3.01) p = 0.02 | 2.23 (1.30 to 3.83) p = 0.004 | 1.70 (1.01 to 2.86) p = 0.05 |
| APOB (R3500Q) | 6/4 (60) | 3.40 (0.71 to 16.36) p = 0.13 | 3.44 (0.71 to 16.8) p = 0.13 | 4.06 (0.84 to19.68) p = 0.08 | 3.76 (0.76 to 18.76) p = 0.11 |
| PCSK9 (D374Y) | 6/1 (85.7) | 19.96 (1.88 to 211.55) p = 0.01 | 16.22 (1.56 to 168.3) p = 0.02 | 47.73 (3.79 to 601) p = 0.003 | 14.74 (1.35 to 161) p = 0.03 |
| p Value | p = 0.03¶ | p = 0.003 | p = 0.006 | p = 0.001 | p = 0.01 |
*Model 1 adjusted for age, sex, smoking (never v ex and current) and systolic blood pressure at recruitment.
†Model 2 plus HDL at recruitment.
‡Model 3 plus LDL at recruitment.
δModel 1 plus recorded pretreatment. Total cholesterol or group average value if data not recorded.
¶Fisher exact test.
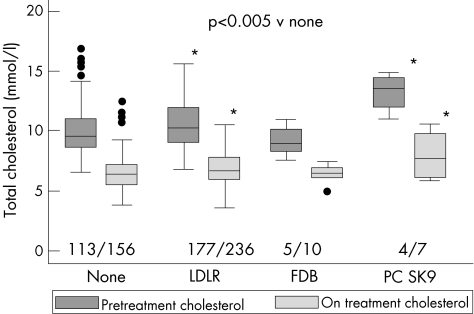
As would be predicted from the higher risk in the patients with LDLR and PCSK9 genes, the pretreatment total cholesterol levels in these groups of patients were considerably higher than in those with no identified mutation (fig 1 and supplementary table C; available online at http://jmg.bmjjournals.com/supplemental). At recruitment, the “statin‐treated” total cholesterol levels had fallen considerably in all groups of patients, but the LDLR and PCSK9 groups still had considerably higher levels than those with no identified mutation. However, as shown in fig 1, the percentage change in all groups was between 30.5% and 40.5% and was not significantly different between groups. This suggests that, although the LDLR, and particularly the PCSK9, patients are more refractory to treatment, all patients are capable of showing a reasonably good lipid‐lowering response. With regard to other recruitment‐level plasma lipid traits compared with those with no detected mutation, patients carrying the APOB mutation had among the lowest plasma triglycerides (1.03 (0.27) v 1.42 (0.64) mmol/l, respectively; p = 0.03) and patients with PCSK9 had among the lowest HDL (1.09 (0.27) v 1.36 (0.36) mmol/l, respectively; p = 0.03) and ApoAI levels (1.15 (0.24) v 1.37 (0.32) g/l, respectively; p = 0.04). These effects were maintained after adjustment for age, sex and ever smoking status (supplementary table C; available online at http://jmg.bmjjournals.com/supplemental).
Figure 1 Median (inter quartile range) untreated total cholesterol levels in Simon Broome patients by types of mutation. Boxes show median and interquartile range, with 95% range shown by bars. Outliers (more than 1.5 times the interquartile range from the edge of the box) are shown as dots. p Values shown from two‐tailed unpaired Student's t tests. None (n = 113; 9.5 (8.6, 11.0)); LDLR (any; n = 177, 10.2 (9.0, 11.9)); FDB APOB (p.R3500Q; n = 5; 9.0 (8.3, 10.1)); PCSK9 (p.D374Y; n = 4, 13.5 (12.0, 14.4)) and total cholesterol levels at recruitment, None (n = 156; 6.4 (5.6, 7.25)); LDLR (any; n = 236, 6.6 (6.0, 7.7)) FDB APOB (p.R3500Q; n = 10; 6.6 (6.1, 7.2)); PCSK9 (p.D374Y; n = 7, 9.3 (6.4, 10.4)). Absolute change (SD) in mmol/l, No mutation −3.53 (2.16), LDLR (any) −3.43 (2.34), APOB −2.82 (0.88), PCSK9 −5.27 (1.31).
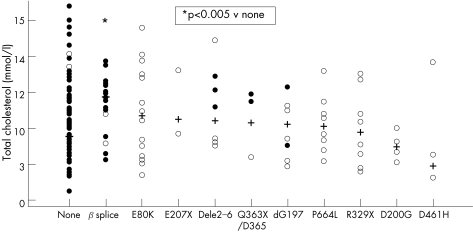
To examine whether particular LDLR mutations were associated with higher or lower than average lipid levels or CHD risk, results were analysed in patients grouped according to their detected mutation. Figure 2 shows the considerable range of values seen in all mutation groups, but the median pretreatment cholesterol levels in the intron 3 G→A carriers (c.313+1 G→A) was considerably higher than in the no‐mutation group (mean (SE) 11.19 (0.37) v 9.85 (0.18) mmol/l; p = 0.007), with recruitment (treated) LDL‐C also being higher in this group (4.81 (0.36) v 4.19 (0.12) mmol/l; p = 0.03). This group had a non‐significantly higher risk of having CHD than the no‐mutation group (1.52 (0.58–4.01); p = 0.39). No other mutation was associated with any significant difference in any lipid trait or CHD risk than the no‐mutation group.
Figure 2 Scatter plot of untreated total cholesterol levels in patients with familial hypercholesterolaemia with different LDLR mutations (old nomenclature), showing the median and individual data points.
Discussion
The major novel findings of this study are the 84% higher risk of CHD in those with an identified LDLR mutation compared with those with no detected mutation, and the relatively high frequency and extremely high risk of CHD in carriers of the PCSK9 p.D374Y mutation, confirming recent reports.21 The mechanism through which the LDLR and PCSK9 mutations influence the risk of CHD is probably through causing raised plasma lipid levels and thus promoting early development of clinically important atherosclerosis. However, the measured pre‐treatment total‐cholesterol levels did not explain the high risk in these compared with patients in the no‐mutation group, as the risk estimates were not materially affected by adjusting for pretreatment cholesterol data. This is possibly because a single measure of total cholesterol pretreatment may considerably underestimate an individual's usual levels, and measures of the more clinically relevant LDL‐cholesterol might be a better predictor of risk. The non‐significant threefold higher CHD risk in carriers of the APOB p.3500Q mutation may be a result of the strict inclusion criteria of the study, as the reported risk associated with this mutation is lower in other studies.10 The use of the no‐mutation patients as the reference group for risk comparison is likely to underestimate the true size of the effects seen, as at least a proportion of these patients are likely to have LDLR mutations that were not detected. This comparison also grossly underestimates the true risk associated with carriage of an identified mutation, as the reference group of patients has itself a high prevalence of CHD.3,6 The results confirm the genetic heterogeneity of familial hypercholesterolaemia in the UK, with 70 different LDLR mutations and 14 different duplications/deletions being identified. However, a subset of mutations are relatively common, of which the intron3 c.313+1 G→A is the most common (6.6% of all patients) and the most severe in terms of untreated plasma cholesterol levels and risk of CHD.
Several studies have shown that particular “severe” LDLR mutations have higher lipid levels and CHD risk.22,23 In genetically heterogeneous populations such as the UK, such comparisons are difficult, owing to the relatively low frequency of any single mutation. In this UK cohort, the most common mutation was in intron3 (c.313+1G→A), which has been shown to disrupt correct splicing and result in a functionally “null” allele, as the predicted mRNA is in frame with exon 3 deleted.24 In agreement with previous data,24 carriers of this mutation in the current cohort had mean untreated cholesterol levels 1.35 mmol/l higher, and LDL‐C levels 0.62 mmol/l higher than those with no mutation, and a 51% higher risk of having CHD (after taking into account age, sex and smoking history). For such an effect to be significant, it would require a sample 2–3 times larger, and confirmation of this estimate is required.
As reported previously,10,11 the frequency of the APOB p.Q3500 mutation was 5%, and 7% of patients had a deletion or duplication in the LDLR gene (reported 5%12). Overall, in this group of patients with definite familial hypercholesterolaemia, a clear molecular cause of familial hypercholesterolaemia was identified in 61.4% of patients, which compares favourably with previous reports of 40–79% of patients with clinically definite familial hypercholesterolaemia who had detectable mutations.11,22,25 Care was taken to assign “causality” using strictly approved criteria,11 and several other LDLR sequence changes potentially causing familial hypercholesterolaemia were detected, including those predicting synonymous codon changes and those within the screened intron sequences, but outside canonical splice junction sequences. There is precedent, however, for such changes to affect correct splicing,26 and splice assays are being developed to investigate the potential functionality of those changes that occurred in only one or two patients and that were absent in 100 healthy people.
There are several limitations to the study. The SSCP method used here is reported to be 85–90% sensitive compared with direct sequencing,27 so an additional 10–15% of LDLR (or PCSK9) mutations may have been detected in these patients using such an approach. In addition, in the current study, several exons in which a low mutation detection rate has been reported were not examined. The detection rate would clearly have increased if all exons had been included, and based on published data,9,11 we estimate that this would possibly have increased the detection rate by a further 10–15%. For both LDLR and PCSK9, only 10–30 bp of the introns were examined by the primers used here, and mutations in regions not covered may exist, although few have been reported.9 Although any additional mutations would be of clinical interest, we do not believe they would alter the main interpretations of the data. Where mutations have been missed because of technical reasons, this would mean that the no‐mutation detected group will contain a small number of false‐negative mutation carriers, and this would result in the group having higher mean lipid levels than true no‐mutation patients. The difference in plasma lipid levels seen here between the no‐mutation and the LDLR groups would thus be an underestimate of the true difference.
A second limitation is in the sample size available. The number of patients with CHD is relatively small, and only effects of relatively large size could be detected with statistical certainty. Thus, the failure of the higher risk of CHD in carriers of the APOB p.3500Q mutation to reach statistical significance is likely to be due to a type II error, and it is also possible that the higher risk in LDLR and PCSK9 mutation carriers is a false positive finding. Finally, although the accuracy of the diagnosis of CHD would be enhanced by angiographically determined information on these patients, dependence on the clinical documentation of CHD means that some apparently unaffected people (with undiagnosed disease) will have been misclassified, which again will result in an underestimate of effect size.
The finding that 2% of the patients with familial hypercholesterolaemia are carrying a single mutation (p.D374Y) in the PCSK9 gene is of interest. It raises the possibility that other mutations in this gene may occur in UK patients with familial hypercholesterolaemia, but an SSCP/sequencing screen of all coding exons of the gene in the 156 patients where no other mutation had been detected failed to identify any pathogenic sequence changes. To date, 11 likely causative mutations have been reported in the PCSK9 gene in patients with autosomal dominant hypercholesterolaemia.13,16,28,29 None of these mutations were detected in the Simon Broome cohort, although we cannot be completely certain that the SSCP method would have detected them, as no positive controls were available, but other (non‐functional) variants were detected in these exons. As discussed above, the SSCP method is not 100% sensitive, but this suggests that mutations in the PCSK9 gene are not a common cause of familial hypercholesterolaemia in the UK, confirming other reports.26,30
Although it may be possible that a proportion of the patients have a mutation in an as yet unidentified gene, some of the patients in whom no mutation can yet be identified probably do not have true monogenic autosomal dominant hypercholesterolaemia. The clinical diagnostic criteria for familial hypercholesterolaemia are not completely specific, and no family studies have been carried out in any of the patients with no mutations detected to determine this.
The particularly high untreated and treated cholesterol levels in carriers of the PCSK9 p.D374Y mutation have been noted previously.16,17,18,21 Partly at least, the higher mean level of LDL‐cholesterol after statin treatment reflects the higher baseline values, as the percentage fall in these patients was comparable with that seen in the other groups, although it should be noted that this is an observational study and was not designed to address response to treatment. The possible molecular mechanism by which this mutation has these effects has been previously discussed in detail.21 The higher CHD risk in carriers of the p.D374Y mutation, reported previously,16,21 is confirmed here. The 3–4‐fold risk of CHD in the patients with APOB p.3500Q compared with those with no detectable mutation is not significant in the small sample, but is in line with previously published reports.10
Although the clinical utility of DNA testing in the diagnosis of patients with familial hypercholesterolaemia has been reported by many groups,5,25 its utility in patient management and its cost effectiveness have yet to be determined unequivocally. More information will need to be collected regarding the negative and positive predictive values of DNA testing and a detailed cost–benefit analysis, to support its widespread introduction to complement diagnosis of familial hypercholesterolaemia by lipid levels alone.
Acknowledgements
We thank the research nurses at each of the centres: Jane Crawley, Lorna Day, Jane Donnarumma, Sue Neil, Clare Neuwirth and Janet Morgan, (and the late Sue McCarthy), and all the patients for participating in the study.
Abbreviations
CHD - coronary heart disease
LDL - low‐density lipoprotein
LDLR - low‐density lipoprotein receptor
PCR - polymerase chain reaction
SSCP - single‐strand conformational polymorphism
Footnotes
Funding: The study was supported by a grant from the British Heart Foundation (grant RG93008). SEH acknowledges BHF support (RG 2005/014) and a grant from the Department of Health to the London IDEAS Genetics Knowledge Park.
Competing interests: None declared.
References
- 1.Neil H A, Hammond T, Huxley R, Matthews D R, Humphries S E. Extent of underdiagnosis of familial hypercholesterolaemia in routine practice: prospective registry study. BMJ 2000321148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slack J. Risks of ischaemic heart‐disease in familial hyperlipoproteinaemic states. Lancet 196921380–1382. [DOI] [PubMed] [Google Scholar]
- 3.Betteridge D J, Broome K, Durrington P N, Hawkins M M, Humphries S E, Mann J I, Miller J P, Neil H A W, Thompson G R, Thorogood M, Scientific Steering Committee on behalf of the Simon Broome Register Group Mortality in treated heterozygous familial hypercholesterolaemia: implications for clinical management. Atherosclerosis 1999142105–112. [PubMed] [Google Scholar]
- 4.Marks D, Wonderling D, Thorogood M, Lambert H, Humphries S E, Neil H A. Cost effectiveness analysis of different approaches of screening for familial hypercholesterolaemia. BMJ 20023241303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Umans‐Eckenhausen M A, Defesche J C, Sijbrands E J, Scheerder R L, Kastelein J J. Review of first 5 years of screening for familial hypercholesterolaemia in the Netherlands. Lancet 2001357165–168. [DOI] [PubMed] [Google Scholar]
- 6.Scientific Steering Committee on behalf of the Simon Broome Register Group Risk of fatal coronary heart disease in familial hypercholesterolaemia. BMJ 1991303893–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neil H A, Seagroatt V, Betteridge D J, Cooper M P, Durrington P N, Miller J P, Seed M, Naoumova R P, Thompson G R, Huxley R, Humphries S E. Established and emerging coronary risk factors in patients with heterozygous familial hypercholesterolaemia. Heart 2004901431–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sauvage Nolting P R, Defesche J C, Buirma R J, Hutten B A, Lansberg P J, Kastelein J J. Prevalence and significance of cardiovascular risk factors in a large cohort of patients with familial hypercholesterolaemia. J Intern Med 2003253161–168. [DOI] [PubMed] [Google Scholar]
- 9.Villeger L, Abifadel M, Allard D, Rabes J P, Thiart R, Kotze M J, Beroud C, Junien C, Boileau C, Varret M. The UMD‐LDLR database: additions to the software and 490 new entries to the database. Hum Mutat 20022081–87. [DOI] [PubMed] [Google Scholar]
- 10.Myant N B. Familial defective apolipoprotein B‐100: a review, including some comparisons with familial hypercholesterolaemia. Atherosclerosis 19931041–18. [DOI] [PubMed] [Google Scholar]
- 11.Heath K E, Humphries S E, Middleton‐Price H, Boxer M. A molecular genetic service for diagnosing individuals with familial hypercholesterolaemia (FH) in the United Kingdom. Eur J Hum Genet 20019244–252. [DOI] [PubMed] [Google Scholar]
- 12.Sun X M, Webb J C, Gudnason V, Humphries S, Seed M, Thompson G R, Knight B L, Soutar A K. Characterization of deletions in the LDL receptor gene in patients with familial hypercholesterolemia in the United Kingdom. Arterioscler Thromb 199212762–770. [DOI] [PubMed] [Google Scholar]
- 13.Abifadel M, Varret M, Rabes J P, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, Derre A, Villeger L, Farnier M, Beucler I, Bruckert E, Chambaz J, Chanu B, Lecerf J M, Luc G, Moulin P, Weissenbach J, Prat A, Krempf M, Junien C, Seidah N G, Boileau C. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet 200334154–156. [DOI] [PubMed] [Google Scholar]
- 14.Maxwell K N, Fisher E A, Breslow J L. Overexpression of PCSK9 accelerates the degradation of the LDLR in a post‐endoplasmic reticulum compartment. Proc Natl Acad Sci USA 20051022069–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouguerram K, Chetiveaux M, Zair Y, Costet P, Abifadel M, Varret M, Boileau C, Magot T, Krempf M. Apolipoprotein B100 metabolism in autosomal‐dominant hypercholesterolemia related to mutations in PCSK9. Arterioscler Thromb Vasc Biol 2004241448–1453. [DOI] [PubMed] [Google Scholar]
- 16.Sun X M, Eden E R, Tosi I, Neuwirth C K, Wile D, Naoumova R P, Soutar A K. Evidence for effect of mutant PCSK9 on apolipoprotein B secretion as the cause of unusually severe dominant hypercholesterolaemia. Hum Mol Genet 2005141161–1169. [DOI] [PubMed] [Google Scholar]
- 17.Timms K M, Wagner S, Samuels M E, Forbey K, Goldfine H, Jammulapati S, Skolnick M H, Hopkins P N, Hunt S C, Shattuck D M. A mutation in PCSK9 causing autosomal‐dominant hypercholesterolemia in a Utah pedigree. Hum Genet 2004114349–353. [DOI] [PubMed] [Google Scholar]
- 18.Leren T P. Mutations in the PCSK9 gene in Norwegian subjects with autosomal dominant hypercholesterolemia. Clin Genet 200465419–422. [DOI] [PubMed] [Google Scholar]
- 19.Rothman K J. No adjustments are needed for multiple comparisons. Epidemiology 1990143–46. [PubMed] [Google Scholar]
- 20.Shioji K, Mannami T, Kokubo Y, Inamoto N, Takagi S, Goto Y, Nonogi H, Iwai N. Genetic variants in PCSK9 affect the cholesterol level in Japanese. J Hum Genet 200449109–114. [DOI] [PubMed] [Google Scholar]
- 21.Naoumova R P, Tosi I, Patel D, Neuwirth C, Horswell S D, Marais A D, van Heyningen C, Soutar A K. Severe hypercholesterolemia in four British families with the D374Y mutation in the PCSK9 gene: long‐term follow‐up and treatment response. Arterioscler Thromb Vasc Biol 2005252654–2660. [DOI] [PubMed] [Google Scholar]
- 22.Graham C A, McClean E, Ward A J, Beattie E D, Martin S, O'Kane M, Young I S, Nicholls D P. Mutation screening and genotype:phenotype correlation in familial hypercholesterolaemia. Atherosclerosis 1999147309–316. [DOI] [PubMed] [Google Scholar]
- 23.Kotze M J, De Villiers W J, Steyn K, Kriek J A, Marais A D, Langenhoven E, Herbert J S, Graadt Van Roggen J F, Van der Westhuyzen D R, Coetzee G A. Phenotypic variation among familial hypercholesterolemics heterozygous for either one of two Afrikaner founder LDL receptor mutations. Arterioscler Thromb 1993131460–1468. [DOI] [PubMed] [Google Scholar]
- 24.Sun X M, Patel D D, Bhatnagar D, Knight B L, Soutar A K. Characterization of a splice‐site mutation in the gene for the LDL receptor associated with an unpredictably severe clinical phenotype in English patients with heterozygous FH. Arterioscler Thromb Vasc Biol 199515219–227. [DOI] [PubMed] [Google Scholar]
- 25.Damgaard D, Larsen M L, Nissen P H, Jensen J M, Jensen H K, Soerensen V R, Jensen L G, Faergeman O. The relationship of molecular genetic to clinical diagnosis of familial hypercholesterolemia in a Danish population. Atherosclerosis 2005180155–160. [DOI] [PubMed] [Google Scholar]
- 26.Fujimaru M, Tanaka A, Choeh K, Wakamatsu N, Sakuraba H, Isshiki G. Two mutations remote from an exon/intron junction in the beta‐hexosaminidase beta‐subunit gene affect 3′‐splice site selection and cause Sandhoff disease. Hum Genet 1998103462–469. [DOI] [PubMed] [Google Scholar]
- 27.Jensen H K, Jensen L G, Hansen P S, Faergeman O, Gregersen N. High sensitivity of the single‐strand conformation polymorphism method for detecting sequence variations in the low‐density lipoprotein receptor gene validated by DNA sequencing. Clin Chem 1996421140–1146. [PubMed] [Google Scholar]
- 28.Allard D, Amsellem S, Abifadel M, Trillard M, Devillers M, Luc G, Krempf M, Reznik Y, Girardet J P, Fredenrich A, Junien C, Varret M, Boileau C, Benlian P, Rabes J P. Novel mutations of the PCSK9 gene cause variable phenotype of autosomal dominant hypercholesterolemia. Hum Mutat 200526497. [DOI] [PubMed] [Google Scholar]
- 29.Pisciotta L, Oliva C P, Cefalu A B, Noto D, Bellocchio A, Fresa R, Cantafora A, Patel D, Averna M, Tarugi P, Calandra S, Bertolini S. Additive effect of mutations in LDLR and PCSK9 genes on the phenotype of familial hypercholesterolemia. Atherosclerosis 2006186433–440. [DOI] [PubMed] [Google Scholar]
- 30.Damgaard D, Jensen J M, Larsen M L, Soerensen V R, Jensen H K, Gregersen N, Jensen L G, Faergeman O. No genetic linkage or molecular evidence for involvement of the PCSK9, ARH or CYP7A1 genes in the familial hypercholesterolemia phenotype in a sample of Danish families without pathogenic mutations in the LDL receptor and apoB genes. Atherosclerosis 2004177415–422. [DOI] [PubMed] [Google Scholar]