Abstract
Gene targeting of the sperm nuclear proteins, the protamines, in mice leads to haploinsufficiency, abnormal chromatin compaction, sperm DNA damage, and male infertility. In order to investigate whether changes in amount or structure of the protamines could be a cause of human infertility, we sequenced the protamine genes of infertile men whose sperm appeared phenotypically similar to those of protamine deficient mice. We identified a heterozygous single nucleotide polymorphism (SNP) in the protamine (PRM1) gene in three infertile men (10% of the total infertile men analysed). This SNP disrupts one of the highly conserved arginine clusters needed for normal DNA binding. To rapidly screen for this SNP in infertile patients, we developed a simple PCR restriction fragment length polymorphism assay. This is the first report of a SNP in the PRM1 gene that appears associated with human male infertility.
Keywords: infertility, SNP, protamine, sperm and reproduction
Approximately 15% of couples are affected by infertility, with the man being responsible for nearly half of the cases.1 Despite advances in assisted reproduction technologies making paternity possible for many of these men, the genetic causes of the majority of male infertility remains unknown. Gene targeting in the mouse has provided valuable insights into the genetic aetiology of many human diseases and infertilities.2
During spermatogenesis, diploid spermatogonia replicate and differentiate into spermatocytes, which undergo meiosis, producing four spermatids. As the haploid spermatids develop into sperm, specialised structures such as the acrosome, tail, and a novel highly condensed nucleus are produced. In sperm nuclei, the DNA‐protamine complex compacts, stabilises, and protects the haploid genome. Protamines are small basic proteins widely conserved among species. All mammals have one protamine, PRM1, while some species including human and mouse have a second protamine, PRM2. Both proteins are rich in arginine and cysteine, amino acids needed for DNA binding and disulphide bridge formation.3,4 Disruption of either the Prm1 or Prm2 gene in mice leads to haploinsufficiency, abnormal chromatin compaction, sperm DNA damage, and male infertility.5,6 Because of the critical roles the protamines play in spermatid differentiation, aberrations in protamine expression or changes in protein structure could be causes of certain idiopathic human male infertilities.
To address this issue, we sequenced the genomic loci of the PRM1 gene in 30 infertile male patients whose sperm showed phenotypes similar to the sperm of the protamine deficient knockout mouse.
MATERIALS AND METHODS
Patients
All patients and donors participating in this study were recruited with full approval from and overseeing by the Baylor College of Medicine institutional review board for human subjects (Houston, TX, USA). DNA was extracted from blood samples of 30 unrelated infertile patients, and from 10 men of pregnancy proven fertility who served as controls. The sperm of the 30 infertile males were similar to the sperm derived from protamine deficient mice.5 The sperm counts were normal, but the sperm showed abnormal DNA fragmentation (>27% of the sperm fragmented) and/or abnormal morphology based upon Kruger's strict criteria (<4.4% normal forms).7 Known causes of infertility were excluded and the subjects' physical examinations, endocrine profiles, and semen parameters (except for strict morphology defects and/or DNA damage) were normal. Because their sperm count was normal, they were not analysed for Y chromosome micro deletions.
Genomic analysis of the PRM1 gene
To analyse the PRM1 gene locus, PCR directed sequencing was performed with DNA purified from blood samples from 10 normal fertile and 30 infertile men. Reverse transcription PCR was performed with a primer set designed to be homologous to the genomic regions for PRM1 on chromosome 168 (Ensembl gene code ENSG00000175646; NCBI locus code NM_002761) (Pr597 forward: 5′‐cataggcagcccctacactc‐3′; Pr087 reverse: 5′‐ccctctcaagaacaaggagagaa‐3′). Using the primer set, we were able to amplify fragments of 644 nucleotides inside the primer set (from nucleotides −160 to 9 bp after the end of transcription) of the PRM1 gene locus. The PCR conditions were as follows: one cycle of denaturation at 95°C for 5 minutes, followed by 35 cycles of denaturation at 95°C for 45 seconds, annealing at 59°C for 30 seconds, and extension at 72°C for 1 minute. The resultant PCR products were purified with a commerical kit (QIAquick PCR Purification kit; Qiagen Inc., Valencia, CA, USA) and directly sequenced in both orientations using the PCR primers as sequencing primers. When mutations were detected, the results were confirmed by the direct sequencing of two independently prepared PCR products to rule out PCR artefacts.
Restriction fragment length polymorphism assay to detects the G197T SNP
In the analyses of the PRM1 gene, a heterozygous G→T single nucleotide polymorphism (SNP) at nucleotide 197 was identified in three men. The G197T mutation disrupts the recognition site of the restriction enzyme BseRI (changing GAGGAG to GAGTAG). To develop a restriction fragment length polymorphism (RFLP) assay for this SNP, the PCR products were digested with BseRI for 1 h at 37°C and separated on a 1.5% agarose gel. The DNA fragments were visualised by ethidium bromide staining.
RESULTS
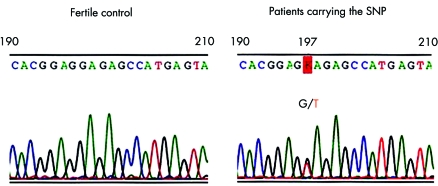
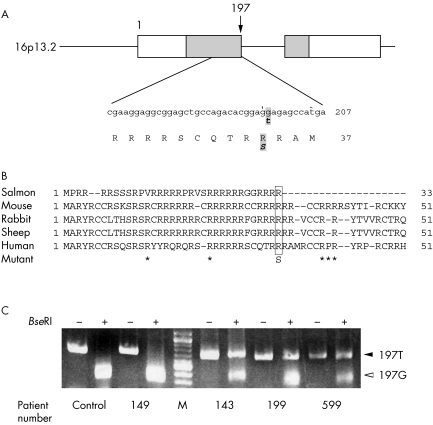
One heterozygous single nucleotide polymorphism (SNP) at nucleotide 197 was identified in the DNA of three of the 30 infertile men examined (10%). To rule out PCR artefacts, the SNP was confirmed by the direct sequencing of at least two independently prepared PCR products. A single wave of G was seen at nucleotide 197 in samples from fertile control men, while two overlapping patterns, one common type (G) and one variant (T), were seen in the three unrelated infertile men indicating a polymorphism of one PRM1 allele (fig 1). This SNP causes an amino acid change of arginine to serine at codon 34 in a highly conserved arginine cluster (fig 2A, B). No identical SNP was observed in a total of 779 men (the 37 men tested in the current study: 10 pregnancy proven fertile control and 27 infertile), in a total of 522 men from previous studies (270 fertile, 226 sterile, and 26 unselected) or in 220 individuals from the Ensembl human GeneSNPView and NCBI SNP databases (Ensembl gene code ENSG00000175).8,9 Although two other SNPs in the coding region of PRM1, at codons 45 (SNP ID; rs11544792) and 47 (SNP ID; rs737008), have been reported (Ensembl gene code ENSG00000175), and three other SNPs at codons 14, 23, and 46 were reported in a previous study,8 all five SNPs are synonymous, causing no amino acid change.
Figure 1 Sequence profiles of PRM1 (190 to 210 nucleotides) from a normal fertile man and the three infertile patients carrying the SNP.
Figure 2 (A) Schematic representation of the PRM1 gene and the site of the nucleotide and predicted amino acid sequences adjacent to the SNP. The transcription start site is defined as +1. White boxes indicate the 5′ and 3′ untranslated regions. The solid bar indicates the genomic region of PRM1 and contains the two exons and an intron of the gene. The grey boxes indicate the coding region of PRM1 with the SNP position marked by an arrow. The amino acid sequence is shown in capital letters under the nucleotide sequence. The numbers in the right margin indicate the positions of nucleotides and amino acids. The altered sequences are shown in bold italicised letters. (B) Comparison of protamine 1 protein sequences among species. The amino acid substitution from the G197T SNP disrupts one of the conserved arginine clusters. Asterisks indicate the amino acid positions, previously reported as synonymous SNPs. (C) PCR restriction fragment length polymorphism analysis of genomic DNA from a fertile man (control) and four infertile patients. Patient 149 is infertile but lacks a G197T SNP, while patients 143, 199, and 599 have the G197T SNP. M indicates DNA size markers. Samples were incubated with (+) or without (−) the restriction enzyme, BseRI.
By sequence analysis, we found the SNP disrupts the cutting site of the restriction enzyme BseRI (GAGGAG). PCR products were digested with BseRI and separated on agarose gels. One rapidly migrating band, major type 197G, containing two fragments, 369 and 315 bp in size, was detected with DNA from a fertile control and an infertile patient with two alleles carrying the G in the PRM1 gene. Analysis of DNA from three patients carrying a G197T SNP (patients 143, 199, and 599) revealed one slower migrating DNA band of 684 bp (fig 2C). Thus, this RFLP assay provides a simple and rapid screening method to detect this polymorphism.
DISCUSSION
We identified a novel SNP, G197T, in the PRM1 gene in three of our selected 30 infertile men. This SNP causes an amino acid change from arginine to serine (R34S) in a highly conserved arginine cluster. Based upon the absence of this SNP in more than 770 individuals (220 from the Ensembl human GeneSNP database; 522 from previous reports, and 10 proven fertile controls and 27 infertile men from this study), it appears to be a uncommon SNP (Ensembl gene code ENSG00000175).8,9 In addition to disrupting an arginine core essential for DNA binding, the SNP creates a new RS sequence, a putative phosphorylation site for the enzyme, SR protein specific kinase 1, known to phosphorylate serines in the RS motifs of PRM1 (fig 1B).10,11 Such phosphorylations are associated with the deposition of protamines on sperm chromatin and subsequent chromatin condensation.10,11 Thus, improper phosphorylation could substantially alter both DNA binding and protamine to protamine interactions in the sperm nucleus. Furthermore, Robson's secondary structure prediction program (GENETYX; Genetyx Corporation, Tokyo, Japan) suggests the R34S amino acid substitution disrupts a beta sheet structure in PRM1 (data not shown), an event likely to change protein conformation. The high incidence of DNA fragmentation in the sperm from the infertile patients carrying the SNP supports our hypothesis of altered protamine‐DNA interactions. Taken together, the mutant protein caused by the G197T SNP may affect its binding to DNA and affect chromatin compaction. However, we also cannot exclude the possibility that this SNP may not directly cause the infertility.
PRM2 defects have been reported to be associated with human male infertility (0.4% of 226 infertile men).8 On the other hand, the SNP that we identified may be the first case to link an amino acid substitution in PRM1 with human infertility. The fact that the sperm of all mammals investigated to date contain PRM1, but only a subset contain PRM2, may in part explain the small number of SNPs reported for the PRM1 gene and is additional evidence for the critical functions of PRM1 in male fertility. As seen with protamine deficient mice,5 the SNP has a deleterious effect as a heterozygous mutation, perhaps because the protamines are first expressed post‐meiotically12 and the germ cells are interconnected in a clonal syncytium by intercellular bridges.13,14,15
To develop a rapid assay to detect this SNP in infertile patients, we have taken advantage of the fact that the G197T SNP disrupts the cutting site of the restriction enzyme, BseRI. A PCR fragment of 197T was not digested by BseRI, while the fragment of major type 197G was. Because murine embryonic development is unsuccessful following intracytoplasmic sperm injection (ICSI) with DNA damaged sperm resulting from a protamine insufficiency,6 we speculate that an arrest of embryo development may contribute to the infertility of men carrying the DNA damaged sperm of the G197T SNP. Protamine deficiencies and aberrant protamine ratios have been linked to reduced fertilisation rates,16,17,18 and PRM1 has been proposed to be a potentially critical factor in post‐ICSI human embryonic development.19 Our PCR‐RFLP assay provides an easy means to screen and detect G197T SNPs in idiopathic infertile males.
ACKNOWLEDGEMENTS
We thank L Murthy and W Chuang for preparation of DNA samples. This work was supported by grant P01HD36289 from the National Institutes of Health to Professor D J Lamb.
Abbreviations
ICSI - intracytoplasmic sperm injection
RFLP - restriction fragment length polymorphism
SNP - single nucleotide polymorphism
Footnotes
Competing interests: there are no competing interests.
References
- 1.Sigman M, Lipshultz L I, Howards S S. Evaluation of the subfertile male. In: Lipshultz LI, Howards SS, eds. Infertility in the male. St. Louis: Mosby, 1997173–193.
- 2.Matzuk M M, Lamb D J. Genetic dissection of mammalian fertility pathways. Nat Cell Biol 20024(suppl)41–49. [DOI] [PubMed] [Google Scholar]
- 3.Hecht N B. Molecular biology of structural proteins of the mammalian testis. In: Adolph KW, ed. Molecular biology of chromosome function. New York, NY: Springer, 1989396–420.
- 4.Oliva R, Dixon G H. Vertebrate protamine genes and the histone‐to‐protamine replacement reaction. Prog Nucleic Acid Res Mol Biol 19914025–94. [DOI] [PubMed] [Google Scholar]
- 5.Cho C, Willis W D, Goulding E H, Jung‐Ha H, Choi Y C, Hecht N B, Eddy E M. Haploinsufficiency of protamine‐1 or ‐2 causes infertility in mice. Nat Genet 20012882–86. [DOI] [PubMed] [Google Scholar]
- 6.Cho C, Jung‐Ha H, Willis W D, Goulding E H, Stein P, Xu Z, Schultz R M, Hecht N B, Eddy E M. Protamine 2 deficiency leads to sperm DNA damage and embryo death in mice. Biol Reprod 200369211–217. [DOI] [PubMed] [Google Scholar]
- 7.Kruger T F, Acosta A A, Simmons K F, Swanson R J, Matta J F, Oehninger S. Predictive value of abnormal sperm morphology in in vitro fertilization. Fertil Steril 198849112–117. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka H, Miyagawa Y, Tsujimura A, Matsumiya K, Okuyama A, Nishimune Y. Single nucleotide polymorphisms in the protamine‐1 and ‐2 genes of fertile and infertile human male populations. Mol Hum Reprod 2003969–73. [DOI] [PubMed] [Google Scholar]
- 9.Wyckoff G J, Wang W, Wu C I. Rapid evolution of male reproductive genes in the descent of man. Nature 2000403304–309. [DOI] [PubMed] [Google Scholar]
- 10.Papoutsopoulou S, Nikolakaki E, Chalepakis G, Kruft V, Chevaillier P, Giannakouros T. SR protein‐specific kinase 1 is highly expressed in testis and phosphorylates protamine 1. Nucleic Acids Res 1999272972–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drosou V, Brancorsini S, Nikolakaki E, Sassone‐Corsi P, Giannakouros T. Temporal association of protamine 1 with the inner nuclear membrane protein lamin B receptor during spermiogenesis. J Biol Chem 200427911626–11631. [DOI] [PubMed] [Google Scholar]
- 12.Wykes S M, Nelson J E, Visscher D W, Djakiew D, Krawetz S A. Coordinate expression of the PRM1, PRM2, and TNP2 multigene locus in human testis. DNA Cell Biol 199514155–161. [DOI] [PubMed] [Google Scholar]
- 13.Dym M, Fawcett D W. Further observations on the numbers of spermatogonia, spermatocytes, and spermatids connected by intercellular bridges in the mammalian testis. Biol Reprod 19714195–215. [DOI] [PubMed] [Google Scholar]
- 14.Braun R E, Behringer R R, Peschon J J, Brinster R L, Palmiter R D. Genetically haploid spermatids are phenotypically diploid. Nature 1989337373–376. [DOI] [PubMed] [Google Scholar]
- 15.Caldwell K A, Handel M A. Protamine transcript sharing among postmeiotic spermatids. Proc Natl Acad Sci USA 1991882407–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balhorn R, Reed S, Tanphaichitr N. Aberrant protamine 1/protamine 2 ratios in sperm of infertile human males. Experientia 19884452–55. [DOI] [PubMed] [Google Scholar]
- 17.Mengual L, Ballesca J L, Ascaso C, Oliva R. Marked differences in protamine content and P1/P2 ratios in sperm cells from Percoll fractions between patients and controls. J Androl 200324438–447. [DOI] [PubMed] [Google Scholar]
- 18.Aoki V W, Liu L, Carrell D T. Identification and evaluation of a novel sperm protamine abnormality in a population of infertile males. Hum Reprod 2005201298–1306. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell V, Steger K, Marchetti C, Herbaut J C, Devos P, Rigot J M. Cellular expression of protamine 1 and 2 transcripts in testicular spermatids from azoospermic men submitted to TESE‐ICSI. Mol Hum Reprod 200511373–379. [DOI] [PubMed] [Google Scholar]