Abstract
Background
Joubert syndrome (JS) is an autosomal recessive disorder characterised by hypotonia, ataxia, mental retardation, altered respiratory pattern, abnormal eye movements, and a brain malformation known as the molar tooth sign (MTS) on cranial MRI. Four genetic loci have been mapped, with two genes identified (AHI1 and NPHP1).
Methods
We screened a cohort of 117 JS subjects for AHI1 mutations by a combination of haplotype analysis and sequencing of the gene, and for the homozygous NPHP1 deletion by sequencing and marker analysis.
Results
We identified a total of 15 novel AHI1 mutations in 13 families, including nonsense, missense, splice site, and insertion mutations, with some clustering in the WD40 domains. Eight families were consanguineous, but no single founder mutation was apparent. In addition to the MTS, retinal dystrophy was present in 11 of 12 informative families; however, no subjects exhibited variable features of JS such as polydactyly, encephalocele, colobomas, or liver fibrosis. In contrast to previous reports, we identified two families with affected siblings who developed renal disease consistent with nephronophthisis (NPH) in their 20s. In addition, two individuals with classic NPH were found to have homozygous NPHP1 deletions.
Conclusions
Overall, 11% of subjects had AHI1 mutations, while ∼2% had the NPHP1 deletion, representing a total of less than 15% in a large JS cohort. Some preliminary genotype‐phenotype correlations are possible, notably the association of renal impairment, specifically NPH, in those with NPHP1 deletions. Subjects with AHI1 mutations may be at risk of developing both retinal dystrophy and progressive kidney disease.
Keywords: AHI1 , cerebellar vermis hypoplasia, Joubert syndrome, nephronophthisis, NPHP1
Joubert syndrome (JS; MIM 213300) is an autosomal recessive brain malformation disorder characterised by cerebellar vermis hypoplasia with brainstem anomalies comprising the molar tooth sign (MTS).1 Clinical features include hypotonia, mental retardation, abnormal eye movements characterised by oculomotor apraxia or nystagmus, and abnormal breathing patterns consisting of alternating tachypnea and apnea, particularly in the newborn period.2,3,4 JS and related disorders (JSRD) represents a spectrum of conditions with the core features listed above plus more variable features including polydactyly, retinal dystrophy, ocular colobomas, renal disease (cysts or juvenile nephronophthisis), hepatic fibrosis, occipital encephalocele, and tongue papules or oral frenulae.5,6 Although estimated to occur in approximately 1 in 100 000 individuals, the actual incidence of JSRD may be greater, based on improved diagnosis and recognition of the MTS on axial MRI.
At least four loci have been mapped for JS, with two genes identified. One causative gene is AHI1 (Abelson helper integration site 1).7 The AHI1 protein contains a putative Src‐homology 3 (SH3) domain and six WD40 repeats proposed to mediate formation of large multiprotein complexes,8 and is expressed during murine embryonic development in the hindbrain and spinal cord.9 Five different point mutations including one missense mutation have been identified in the AHI1 gene in six consanguineous JS families.9,10 A second causative gene is the nephronophthisis 1 (NPHP1) gene, associated with a form of progressive renal disease characterised by corticomedullary cysts and known as juvenile nephronophthisis (MIM 256100). We previously identified a homozygous deletion of NPHP1 in subjects with JS and classic nephronophthisis (NPH) that was identical by mapping analysis to the deletion in subjects with NPH alone11; in addition, retinal dystrophy has been described in at least one JS subject with the same deletion.12 Previously, two JS loci at 9q3413 and pericentromeric chromosome 1114,15 were mapped, although the causal genes remain unknown. In this report, we determine the relative contribution of AHI1 and NPHP1 gene mutations within a large cohort of JS subjects and explore genotype‐phenotype correlations in JSRD.
Methods
Subjects
We ascertained a total of 117 families with JSRD based on previously described clinical criteria.2,3,4 Genomic DNA was isolated from peripheral blood by standard methods and lymphoblastoid cell lines were established from probands, their parents, and unaffected siblings when available. Informed consent for genetic studies from each patient or legal guardian was obtained under protocols approved by the Institutional Review Boards at the University of Washington, the University of Chicago, and Hacettepe University, Turkey.
AHI1 analysis
We determined the genotypes at polymorphic markers surrounding the AHI1 gene on chromosome 6q23 (D6S287, D6S262, D6S1656, D6S292, D6S1569) in 10 consanguineous and 18 multiplex (defined as having at least two affected children) JSRD families. We examined the haplotypes for regions of homozygosity in consanguineous pedigrees and for compatibility with linkage by segregation analysis in multiplex pedigrees.16
We sequenced the 28 exons encoding mRNA (GenBank mRNA accession number AJ4560824) and three alternatively spliced exons (GenBank accession numbers AJ459825, AK024085, and AI733147) of the AHI1 gene in DNA from a proband from each of 99 families, including those families not excluded by haplotype analysis for linkage to 6q23. Only a portion of the terminal 3′ untranslated region was sequenced. The amplicons for each exon included a minimum of 20 flanking nucleotides at each intron‐exon boundary of the gene. Products were amplified by two sequential rounds of nested polymerase chain reaction (PCR) using standard reaction conditions and sequenced bidirectionally (BigDye Terminator, Applied Biosystems, Foster City, CA; primers and PCR conditions are available upon request). Sequences were analysed using the DNAStar software package (SeqMan II, version 5.05; DNAStar, Madison, WI). We sequenced DNA from family members to evaluate for mutations identified in probands. To determine the prevalence of missense mutations, a minimum of 50 additional chromosomes was analysed from control subjects derived from an ethnically diverse population of mixed European origin.
NPHP1 analysis
We previously screened 25 probands with retinal and/or renal involvement for mutations in each of the coding exons of the NPHP1 gene; additional polymorphic markers were evaluated to exclude the possibility of a heterozygous deletion of the NPHP1 gene in conjunction with a point mutation.11 In this study, an additional 92 subjects were tested for a homozygous deletion in the NPHP1 gene at 2q13 by evaluating the presence or loss of STS marker (9657T) 3′ to the NPHP1 gene and within the common deleted region.17,18 The total number of subjects evaluated was 117.
Results
AHI1 analysis
Eighteen of the 28 multiplex and/or consanguineous families were excluded from linkage to 6q23 by haplotype analysis. Six of the families were compatible with linkage to 6q23, and four were indeterminate; all of these families were tested for AHI1 mutations by sequencing DNA from an affected individual. Four of the six potentially linked families were consanguineous, exhibited identity by descent, and harboured AHI1 mutations (table 1). The remaining families did not have detectable AHI1 mutations. Given the lack of consanguinity and the small size of these families, the haplotype analysis likely indicates cosegregation of shared haplotypes and disease by chance.16
Table 1 Summary of clinical features for JS subjects with AHI1 and NPHP1 mutations*.
| Pedi‐ gree | Ethnicity | Sequence change† | Protein change‡/ effect | Con‐ sang | Age§ | Sex | CNS¶ | Eye move‐ ments | Ret‐ ina | Respir‐ atory | Renal | Other** | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| New AHI1 mutations | |||||||||||||
| K8062 | Caucasian | Ex 6: 517 A>T | K246X | No | 18 | F | MTS | OMA, | RD | – | Left | DM | |
| Ex 18: 1995 T>G | L832X | nys | MCDK | ||||||||||
| K8090 | Caucasian | Ex 6: 662 C>G | S221X | No | 4 | M | MTS | – | RD | Tachy | – | – | |
| Ex 13: 1898 ins GG | fsX648 | ||||||||||||
| K8067 | Caucasian | IVS8 (−2 A>G) | Presumed | No | 6 | F | MTS | OMA, | RD | Apnea | – | – | |
| splice error | nys | ||||||||||||
| Ex 9: 1260 G>A | W420X | ||||||||||||
| K8019†† | Caucasian | Ex 9: 1267 C>T | Q423X | Yes | 5 | F | MTS‡‡ | NA | “Poor | Tachy | – | – | |
| homozygous | vision” | ||||||||||||
| K8052†† | Armenian | Ex 9: 1267 C>T | Q423X | Yes | 21 | M | MTS | Nys | RD | Tachy | – | – | |
| homozygous | |||||||||||||
| K8075 | Caucasian | Ex 9: 1267 C>T | Q423X | No | 5 | M | MTS | OMA, nys | RD | Tachy | – | – | |
| Ex 15: 2212 C>T | R738X | ||||||||||||
| K8107 | Turkish | IVS11 (+5 ins | Presumed | Yes | 26/9 | M/F | NA/MTS | NA/NA | RD/− | +/NA | Cysts/− | −/− | |
| TTAC) homozygous | splice error | ||||||||||||
| K8012†† | Icelandic | IVS14 (+1 G>T) | Presumed | Yes | 30/26 | F/M | MTS/MTS | Nys/nys | RD/RD | Tachy/ | NPH/ | −/− | |
| homozygous | splice error | tachy | NPH, | ||||||||||
| cysts | |||||||||||||
| K8103†† | Saudi | Ex 15: 2156 A>G | D719G | Yes | 4/3 | M/M | MTS/MTS | Nys/nys | RD/NA | Tachy/− | Left hydro‐ | −/− | |
| Arabian | homozygous | neph/− | |||||||||||
| K8018 | Caucasian | Ex 15: 2452 T>C | W725R | No | 10 | F | MTS | OMA, nys | RD | – | – | – | |
| Unknown | Unknown | ||||||||||||
| K8127 | Turkish | Ex 14: 1917 T>A | Y639X | Yes | 4 | F | MTS | Nys | RD | Tachy | – | – | |
| homozygous | |||||||||||||
| K8131 | Turkish | Ex 14: 2012 C>T | T671I | Yes | 8 | F | MTS | Nys | RD | NA | – | – | |
| homozygous | |||||||||||||
| K8134 | Turkish | Ex 19: 2687 A>G | H896R | Yes | 9/2 | F/M | MTS/MTS | Nys/nys | −/− | −/− | −/− | −/− | |
| homozygous | |||||||||||||
| Totals | 8/13 | 2–30 years | 9F/8M | 12/13 | 9/10 | 11/12 | 8/12 | 3/13 | 0/13 | ||||
| Published AHI1 mutations | |||||||||||||
| MTI‐01010 | Palestinian | Ex 7: 787 ins C | fsX270 | Yes | 1 aff child | M | MTS | OMA | NA | + | – | – | |
| homozygous | |||||||||||||
| MTI‐11510 | Kuwaiti | Ex 9: 1188‐9 del TG | fsX408 | Yes | 2 aff | M/F | MTS, PMG, | OMA | RD/RD | – | – | – | |
| homozygous | thin CC | ||||||||||||
| MTI‐14410 | Turkish | Ex 9: 1328 T>A | V443D | Yes | 1 aff | F | MTS, PMG, | OMA | NA | + | NA | ASD | |
| homozygous | thin CC | ||||||||||||
| Pedigree 39 | Saudi | Ex 9: 1328 T>A | V443D | Yes | 1 aff | M | MTS | NA | NA | NA | NA | – | |
| Arabian | homozygous | child | |||||||||||
| Pedigree 19 | Saudi | Ex 8: 1051 C>T | R351X | Yes | 3 aff | 3M | MTS | OMA, | NA | NA | NA | Mirror | |
| Arabian | homozygous | children | nys | mvts | |||||||||
| Pedigree 29 | Saudi | Ex 9: 1303 C>T | R435X | Yes | 2 | M/M | MTS | OMA | NA | NA | NA | Mirror | |
| Arabian | homozygous | children | or nys | mvts | |||||||||
| Family 129 | Turkish | Not identified | Not | Yes | 5 aff | 3F/2M | MTS | Nys in 1 | RD in 1 | NA | – | GR, | |
| identified | children | scoli‐ | |||||||||||
| (17–28) | osis, sz | ||||||||||||
| Family 229 | Swiss | Not identified | Not | Yes | 2 aff | F/F | MTS | Nys in 1 | RD in 1 | + | – | – | |
| identified | children | ||||||||||||
| (2, 23) | |||||||||||||
| Published NPHP1 mutations | |||||||||||||
| K807611 | Caucasian | Homozygous | No protein | No | 12/8 | F/F | MTS§§/NA | OMA/ | −/− | −/− | NPH/− | −/− | |
| deletion | made | OMA | |||||||||||
| K808411 | Caucasian | Homozygous | No protein | No | 17 | M | MTS§§ | – | – | – | NPH | – | |
| deletion | made | ||||||||||||
| Patient EC12 | Italian | Homozygous | No protein | No | 3 | F | MTS§§ | OMA | RD | – | NPH | – | |
| deletion | made | ||||||||||||
*+, present; −, absent; ASD, atrial septal defect; CC, corpus callosum; CNS, central nervous system involvement; Consang, consanguinity; DM, diabetes mellitus; Ex, exon; F, female; GR, growth retardation; hydroneph, hydronephrosis; IVS, intron; M, male; MCDK, multicystic dysplastic kidney; Mirror mvts, mirror movements; MTS, molar tooth sign on MRI; OMA, oculomotor apraxia; NA, information not available; NPH, nephronophthisis; Nys, nystagmus; PMG, polymicrogyria; RD, retinal dystrophy; sz, seizures; Tachy, tachypnea
†DNA mRNA sequences are numbered starting from the A of the ATG initiator codon (nucleotide +1); exons (Ex) are numbered by convention of the Ensembl assembly for GenBank accession number AJ459824.
‡Proteins are numbered starting from the initiator methionine codon 1.
§Age in years when last ascertained; aff, affected.
¶Note that all subjects had hypotonia, ataxia, and mental retardation or developmental delay, which was milder in those with NPHP1 deletions.
**Other includes polydactyly, hepatic fibrosis, occipital encephaloceles, and ocular coloboma.
††Family compatible with linkage to 6q23 by haplotype analysis.
‡‡Unable to confirm MTS but early studies showed cerebellar vermis hypoplasia prior to description of MTS.
§§Molar tooth has distinctive appearance with elongated but not thickened superior cerebellar peduncles.
We identified 15 different AHI1 mutations distributed in exons 6–19 in 13 families with JS (table 1; fig 1). None of the mutations had been described previously. The segregation pattern of all mutations within families is consistent with autosomal recessive inheritance. All eight consanguineous families (one Icelandic, one Saudi Arabian, four Turkish) had homozygous mutations, although none of the families from Turkey shared the same mutation. Only two consanguineous families had an identical mutation (Q423X).
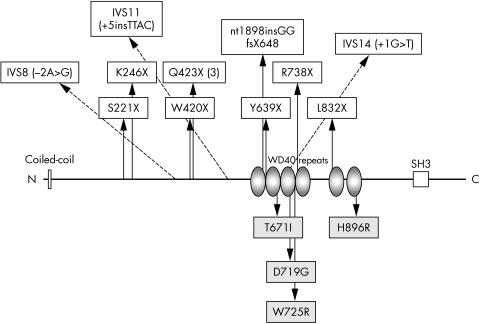
Figure 1 AHI1 protein with mutations identified in this study. Protein motifs include an amino‐terminal coiled‐coil domain (“Coiled‐coil”), six WD40 domains (“WD40 repeats”), and a carboxy‐terminal Src‐homology 3 (“SH3”) domain indicated by a white box. Nonsense mutations or those predicted to result in transcription termination are indicated above the horizontal bar at positions relative to the total peptide length of 1196 amino acids. Splice site mutations are indicated by dotted arrows. Missense mutations are indicated as shaded boxes below the protein.
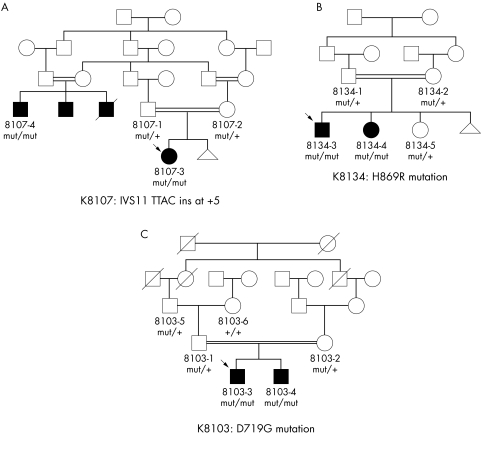
We found eight different nonsense mutations; one (Q423X) is shared by three families of distinct ethnic origins. In family K8090, a 2 bp insertion results in a frameshift with the prediction of 18 aberrant amino acids prior to a stop codon at position 648. Two intronic mutations alter a nucleotide in the invariant splice acceptor and donor sequences at positions −2 and +1, respectively, and thus are likely disease causing. In consanguineous family K8107, a homozygous 4 bp intronic insertion at the +5 position in intron 11 changes a highly conserved G residue to a T and cosegregates with the phenotype in two affected individuals from distinct branches of the family (fig 2A). This G is conserved in 82% of splice donor sites.19 We evaluated the wild type and mutated intron 11 donor site using several splice site prediction algorithms, and the mutated site scored markedly lower than the wild type.19,20,21,22 These calculations, combined with the cosegregation of the mutation with the disease in two distinct branches of the family, strongly suggest that this mutation is pathogenic.
Figure 2 Pedigrees for three homozygous AHI1 mutations showing familial segregation patterns. The presence of the mutation specified in the panel legend is indicated by “mut/+” in the heterozygous state and by “mut/mut” in the homozygous state. Wild type mutations sequences are represented by “+/+”. Arrows indicate the proband. and in (A), two of the black boxes indicate males presumed to have JS, including one male who died of renal failure, although full clinical details are not available.
We identified four different missense mutations in exons 14, 15, and 19. In two consanguineous pedigrees, these homozygous missense mutations segregated with the JS phenotype (fig 2B,C) and were not identified in control samples. All of the missense mutations fall within highly conserved residues that are flanked by less conserved residues based on comparison with the orthologous genes in four other vertebrate species (see supplemental figure available at http://www.jmedgenet.com/supplemental). These missense mutations are found within three of the six WD40 repeat domains of the AHI1 protein (fig 1); these motifs of approximately 40 amino acids usually terminate in a tryptophan‐aspartic acid dipeptide (“WD”) and may coordinate assembly of multi‐protein complexes.8,23 One proband (K8108) carried a W‐to‐R change at the conserved tryptophan of the third WD40 repeat, but we did not identify a mutation on the second allele in this patient despite sequencing all the coding exons and flanking intronic sequences. There was no evidence of either a homozygous or heterozygous deletion of NPHP1 in this subject based on assessment of polymorphic markers within the common NPHP1 deleted region.11
We identified two changes (exon 12: 1643 G>A (R548H) and exon 17: 2488 C>T (R830W)) that do not segregate with JS. These presumed polymorphisms were present in multiple JSRD families and in control samples.
NPHP1 analysis
Of the total of 117 probands with JS screened, no additional subjects were identified with a homozygous deletion aside from the family previously described with two affected daughters and one subject with a very mild MTS and mild learning disability.11 None of the subjects with AHI1 mutations was identified as having the NPHP1 deletion.
Genotype‐phenotype correlations
Table 1 summarises the clinical data for the families with AHI1 and NPHP1 mutations. All the affected subjects with AHI1 mutations had hypotonia, ataxia, and developmental delay with cognitive impairment. Breathing abnormalities were identified in 8/12 families, typically consisting of neonatal onset tachypnea with or without apneic pauses. Only four subjects had documented oculomotor apraxia, although nystagmus was common. The MTS was clearly documented in subjects from 12 families with AHI1 mutations, but imaging was not available for the remaining subject. While frontal polymicrogyria and other brain anomalies including corpus callosal thinning have been reported in patients with AHI1 mutations,10 none of our subjects had evidence of either of these brain malformations based on detailed evaluation of five full MRI scans and review of selected images from seven other MRI scans. At least one subject had retinal dystrophy in 11 of 12 families with AHI1 mutations. One family was discordant for retinal dystrophy; the 26 year old affected male in family K8107 had retinal disease, while his 9 year old cousin did not. Ocular colobomas, hepatic fibrosis, polydactyly, occipital encephaloceles, and tongue tumours or oral frenulae were not observed in any of these subjects.
We identified two consanguineous families with AHI1 mutations and progressive renal disease. In K8012, two siblings with a homozygous splice site mutation developed salt‐losing renal insufficiency with polydipsia and polyuria in their 20s. Renal ultrasound revealed increased cortical echogenicity and one to three macroscopic cysts. The male is 26 years old and is receiving haemodialysis. Biopsy of primarily cortical renal parenchyma revealed global glomerular sclerosis, periglomerular fibrosis, and tubular atrophy with interstitial fibrosis, compatible with a diagnosis of NPH.24 While less severely affected, his 30 year old sister has anaemia and progressive renal insufficiency and is awaiting transplant. In family K8107, two affected individuals share the homozygous splice site mutation, IVS11 +5 ins TTAC (fig 2A). A 26 year old affected male has renal cysts. No DNA is available from his two adult brothers with renal cysts, one of whom died of renal insufficiency at 25 years of age. Their first cousin once removed is a 9 year old girl with JS who thus far does not exhibit renal disease. One other subject with two different AHI1 nonsense mutations (K8062) has a unilateral multicystic dysplastic kidney with stable renal function at 18 years of age; however, this may represent an unrelated developmental renal malformation.
Discussion
In this large and diverse cohort of JS patients, we identified 15 novel AHI1 mutations causative for JS, including the first splice site mutations for this gene. Surprisingly, even consanguineous families with a shared ethnic background (that is, four Turkish families) had different AHI1 mutations, demonstrating the lack of a founder effect for this gene. Although AHI1 is expressed in the developing mouse brain, the role of the protein is not known. The previously identified missense mutation in AHI1 is located significantly 5′ to the WD40 domains,9,10 and thus identification of four novel missense mutations in the WD40 domains suggests the importance of these domains for AHI1 function. In fact, one of the AHI1 missense mutations (W725R) alters the conserved tryptophan of the “WD” dipeptide in the third repeat. Three of the four mutated residues in the AHI1 WD40 domains align with conserved amino acids in the WD40 domains of PEX7 that are mutated in patients with autosomal recessive chondrodysplasia punctata.25,26 Missense mutations in the WD40 domains of LIS1 and CKN1 have been shown to cause classic lissencephaly27 and Cockayne syndrome,28 respectively. Although this ancient domain arose during early eukaryotic development and has a well conserved propeller‐like structure, the functions of the WD40 containing proteins are highly variable, ranging from signal transduction to RNA processing and vesicular trafficking.23 Thus, identifying the role of the WD40 domains and the AHI1 protein in cerebellar, retinal, and renal development remains a future challenge.
In this study, 11 of 12 informative families (92%) with AHI1 mutations manifested retinal dystrophy, significantly more than the three of eight families reported in other series.9,10,29 It is possible that this represents ascertainment differences between the studies. Other pertinent JSRD features such as ocular colobomas, hepatic fibrosis, polydactyly, encephaloceles, and tongue tumours or oral frenulae described in some JS subtypes5,6 were notably absent from our cohort and not reported by others.9,10,29 The absence of other cortical anomalies such as polymicrogyria in our cohort does not necessarily negate an association between AHI1 mutations and polymicrogyria, as this is a rare observation in JSRD; additional studies will be necessary to clarify this question.
Our observation of renal impairment in conjunction with retinal dystrophy in patients with AHI1 mutations supports the hypothesis of a distinctive retinal‐renal subtype in JS, also termed Joubert syndrome type B.4,30 We establish for the first time the presence of renal disease in two pedigrees with AHI1 mutations. The renal involvement is consistent with NPH resulting in progressive renal failure with an apparent later age of onset than classic juvenile NPH (typically, between 10 and 20 years of age).31
The identification of renal disease in older patents with AHI1 mutations has significant implications for long term care and management of children with JS due to mutations in AHI1. It is possible that the AHI1 protein plays a role in the function of the primary cilia as do the other proteins that have been implicated in the development of NPH.32,33,34,35 Indeed, the AHI1 protein is highly expressed in the brain and kidney in human fetuses,9 and this may indicate an important role for AHI1 in renal development. Thus, regular monitoring of renal function in children with AHI1 mutations is recommended until the long term risk of developing renal impairment can be elucidated.
In contrast to the renal disease observed in patients with AHI1 mutations, three out of four individuals with JS and a homozygous NPHP1 deletion have developed classic juvenile NPH. Two subjects have required renal transplantation at 9 and 12 years of age (K8076 and K8084). Retinal involvement appears to be variable, with one of four subjects showing retinal pigmentary changes.12 For those with NPHP1 mutations, the most consistent clinical findings are development of NPH with or without retinal dystrophy, milder developmental delay, and a distinctive MTS on radiologic imaging.11,32
Among our cohort, only 13 families out of 117 tested harboured pathogenic AHI1 mutations, representing 11% of the total. The analysis of NPHP1 deletions reveals that this gene is likely to be a minor contributor as well, on the order of 2%, a result confirmed by the finding of a homozygous NPHP1 deletion in only 1/40 probands tested in an Italian cohort.12 Overall, these results suggest that mutations in AHI1 and NPHP1 probably represent less than 15% of the total genetic causes of JSRD. These findings confirm substantial genetic heterogeneity for this group of disorders and indicate that major genetic loci for JS remain to be identified. Thus far, neither gene is associated with ocular colobomas, hepatic fibrosis, encephaloceles, or polydactyly. It is possible that mutations in AHI1 or NPHP1 do not cause any of these findings. Discovery of the causative genes at the loci on chromosomes 9 and 11, as well as identification of novel JSRD loci, will facilitate further genotype‐phenotypic correlations, generate prognostic information, and guide the medical monitoring of children with JSRD.
Electronic‐database information
URLs for data presented are as follows: Ensembl Genome Browser, http://www.ensembl.org/; GenBank sequence database, http://www.ncbi.nlm. nih.gov/entrez/query.fcgi?db = Nucleotide; GeneReviews at GeneTests, http://www.geneclinics. org or http://www.genetests.org; Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm. nih.gov/Omim/; UCSC human genome browser, http://genome.ucsc.edu/cgi‐bin/hgGateway?org = human; Single nucleotide polymorphisms (SNPs) are reported in dbSNP, http://www.ncbi.nlm.nih.gov/SNP/; Joubert Syndrome Foundation and Related Cerebellar Disorders, http://www.joubertsyndrome.org
Acknowledgements
We thank the patients and their families, the Joubert Syndrome Foundation & Related Cerebellar Disorders, and Bernard L Maria, MD. We acknowledge Karen Barnett, MS and Dana Knutzen, MS for coordinating the genetic studies and Eugene Lee and Nell Niewiadomski for technical assistance.
Abbreviations
JS - Joubert syndrome
JSRD - Joubert syndrome and related disorders
MTS - molar tooth sign
NPH - nephronophthisis
PCR - polymerase chain reaction
SH3 - Src‐homology 3
Footnotes
This work was supported by National Institutes of Health grants P30‐HD02274, K23‐NS45832 (MAP), and K24‐HD46712 (IAG); the March of Dimes Endowment for Healthier Babies at Children's Hospital in Seattle; and the Center for Neurogenetics and Neurotherapeutics, University of Washington
Competing interests: none declared
Ethics approval was provided under a protocol approved by the University of Washington Human Subject Division (#97‐6328‐B 07) and at Hacettepe University, Turkey. Approval to participate in these research studies was provided under a protocol of informed consent as outlined in the Methods section
URLs for data presented are as follows: Ensembl Genome Browser, http://www.ensembl.org/; GenBank sequence database, http://www.ncbi.nlm. nih.gov/entrez/query.fcgi?db = Nucleotide; GeneReviews at GeneTests, http://www.geneclinics. org or http://www.genetests.org; Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm. nih.gov/Omim/; UCSC human genome browser, http://genome.ucsc.edu/cgi‐bin/hgGateway?org = human; Single nucleotide polymorphisms (SNPs) are reported in dbSNP, http://www.ncbi.nlm.nih.gov/SNP/; Joubert Syndrome Foundation and Related Cerebellar Disorders, http://www.joubertsyndrome.org
References
- 1.Maria B L, Hoang K B, Tusa R J, Mancuso A A, Hamed L M, Quisling R G, Hove M T, Fennell E B, Booth‐Jones M, Ringdahl D M, Yachnis A T, Creel G, Frerking B. “Joubert syndrome” revisited: key ocular motor signs with magnetic resonance imaging correlation. J Child Neurol 199712(7)423–430. [DOI] [PubMed] [Google Scholar]
- 2.Maria B L, Boltshauser E, Palmer S C, Tran T X. Clinical features and revised diagnostic criteria in Joubert syndrome. J Child Neurol 199414(9)583–90 discussion 5901. [DOI] [PubMed] [Google Scholar]
- 3.Parisi M A, Glass I A. Joubert syndrome. In: GeneReviews at GeneTests: medical genetics information resource [online database], Seattle: University of Washington, 1997–2005. Available at http://www.geneclinics.org
- 4.Saraiva J M, Baraitser M. Joubert syndrome: a review. Am J Med Genet 199243(4)726–731. [DOI] [PubMed] [Google Scholar]
- 5.Gleeson J G, Keeler L C, Parisi M A, Marsh S E, Chance P F, Glass I A, Graham J M, Jr, Maria B L, Barkovich A J, Dobyns W B. Molar tooth sign of the midbrain‐hindbrain junction: occurrence in multiple distinct syndromes. Am J Med Genet 2004125A(2)125–34 discussion 117. [DOI] [PubMed] [Google Scholar]
- 6.Satran D, Pierpont M E, Dobyns W B. Cerebello‐oculo‐renal syndromes including Arima, Senior‐Loken and COACH syndromes: more than just variants of Joubert syndrome. Am J Med Genet 199986(5)459–469. [PubMed] [Google Scholar]
- 7.Jiang X, Hanna Z, Kaouass M, Girard L, Jolicoeur P. Ahi‐1, a novel gene encoding a modular protein with WD40‐repeat and SH3 domains, is targeted by the Ahi‐1 and Mis‐2 provirus integrations. J Virol 200276(18)9046–9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Close J, Game L, Clark B, Bergounioux J, Gerovassili A, Thein S L. Genome annotation of a 1.5 Mb region of human chromosome 6q23 encompassing a quantitative trait locus for fetal hemoglobin expression in adults. BMC Genomics 20045(1)33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferland R J, Eyaid W, Collura R V, Tully L D, Hill R S, Al‐Nouri D, Al‐Rumayyan A, Topcu M, Gascon G, Bodell A, Shugart Y Y, Ruvolo M, Walsh C A. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat Genet 200436(9)1008–1013. [DOI] [PubMed] [Google Scholar]
- 10.Dixon‐Salazar T, Silhavy J L, Marsh S E, Louie C M, Scott L C, Gururaj A, Al‐Gazali L, Al‐Tawari A A, Kayserili H, Sztriha L, Gleeson J G. Mutations in the AHI1 gene, encoding jouberin, cause Joubert syndrome with cortical polymicrogyria. Am J Hum Genet 200475(6)979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parisi M A, Bennett C L, Eckert M L, Dobyns W B, Gleeson J G, Shaw D W, McDonald R, Eddy A, Chance P F, Glass I A. The NPHP1 gene deletion associated with juvenile nephronophthisis is present in a subset of individuals with Joubert syndrome. Am J Hum Genet 200475(1)82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castori M, Valente E M, Donati M A, Salvi S, Fazzi E, Procopio E, Galluccio T, Emma F, Dallapiccola B, Bertini E. NPHP1 gene deletion is a rare cause of Joubert syndrome related disorders. J Med Genet 200542(2)e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saar K, Al‐Gazali L, Sztriha L, Rueschendorf F, Nur E, Kamal M, Reis A, Bayoumi R. Homozygosity mapping in families with Joubert syndrome identifies a locus on chromosome 9q34.3 and evidence for genetic heterogeneity. Am J Hum Genet 199965(6)1666–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keeler L C, Marsh S E, Leeflang E P, Woods C G, Sztriha L, Al‐Gazali L, Gururaj A, Gleeson J G. Linkage analysis in families with Joubert syndrome plus oculo‐renal involvement identifies the CORS2 locus on chromosome 11p12–q13.3. Am J Hum Genet 200373(3)656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valente E M, Salpietro D C, Brancati F, Bertini E, Galluccio T, Tortorella G, Briuglia S, Dallapiccola B. Description, nomenclature, and mapping of a novel cerebello‐renal syndrome with the molar tooth malformation. Am J Hum Genet 200373(3)663–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett C L, Parisi M A, Eckert M L, Huynh H M, Chance P F, Glass I A. Joubert syndrome: a haplotype segregation strategy and exclusion of the zinc finger protein of cerebellum 1 (ZIC1) gene. Am J Med Genet 2004125A(2)117–24 discussion 117. [DOI] [PubMed] [Google Scholar]
- 17.Heninger E, Otto E, Imm A, Caridi G, Hildebrandt F. Improved strategy for molecular genetic diagnostics in juvenile nephronopthisis. Am J Kidney Dis 200137(6)1131–1139. [DOI] [PubMed] [Google Scholar]
- 18.Hildebrandt F, Nothwang H G, Vossmerbaumer U, Springer C, Strahm B, Hoppe B, Keuth B, Fuchshuber A, Querfeld U, Neuhaus T J, Brandis M. Lack of large, homozygous deletions of the nephronophthisis 1 region in Joubert syndrome type B. Pediatr Nephrol 199812(1)16–19. [DOI] [PubMed] [Google Scholar]
- 19.Shapiro M B, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 198715(17)7155–7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunak S, Engelbrecht J, Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol 1991220(1)49–65. [DOI] [PubMed] [Google Scholar]
- 21.Yeo G, Burge C. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. In: Miller W, Vingron M, Istrail S, et al, eds. Proceedings of the 7th Annual International Conference on Computational Molecular Biology (RECOMBO3). New York: ACM Press, 2003322–331.
- 22.Reese M G, Eeckman F H, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol 19974(3)311–323. [DOI] [PubMed] [Google Scholar]
- 23.Smith T F, Gaitatzes C, Saxena K, Neer E J. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci 199924(5)181–185. [DOI] [PubMed] [Google Scholar]
- 24.Kumada S, Hayashi M, Arima K, Nakayama H, Sugai K, Sasaki M, Kurata K, Nagata M. Renal disease in Arima syndrome is nephronophthisis as in other Joubert‐related cerebello‐oculo‐renal syndromes. Am J Med Genet A 2004131(1)71–76. [DOI] [PubMed] [Google Scholar]
- 25.Braverman N, Chen L, Lin P, Obie C, Steel G, Douglas P, Chakraborty P K, Clarke J T, Boneh A, Moser A, Moser H, Valle D. Mutation analysis of PEX7 in 60 probands with rhizomelic chondrodysplasia punctata and functional correlations of genotype with phenotype. Hum Mutat 200220(4)284–297. [DOI] [PubMed] [Google Scholar]
- 26.Braverman N, Steel G, Obie C, Moser A, Moser H, Gould S J, Valle D. Human PEX7 encodes the peroxisomal PTS2 receptor and is responsible for rhizomelic chondrodysplasia punctata. Nat Genet 199715(4)369–376. [DOI] [PubMed] [Google Scholar]
- 27.Cardoso C, Leventer R J, Dowling J J, Ward H L, Chung J, Petras K S, Roseberry J A, Weiss A M, Das S, Martin C L, Pilz D T, Dobyns W B, Ledbetter D H. Clinical and molecular basis of classical lissencephaly: mutations in the LIS1 gene (PAFAH1B1). Hum Mutat 200219(1)4–15. [DOI] [PubMed] [Google Scholar]
- 28.Ren Y, Saijo M, Nakatsu Y, Nakai H, Yamaizumi M, Tanaka K. Three novel mutations responsible for Cockayne syndrome group A. Genes Genet Syst 200378(1)93–102. [DOI] [PubMed] [Google Scholar]
- 29.Lagier‐Tourenne C, Boltshauser E, Breivik N, Gribaa M, Betard C, Barbot C, Koenig M. Homozygosity mapping of a third Joubert syndrome locus to 6q23. J Med Genet 200441(4)273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King M D, Dudgeon J, Stephenson J B. Joubert's syndrome with retinal dysplasia: neonatal tachypnoea as the clue to a genetic brain‐eye malformation. Arch Dis Child 198459(8)709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antignac C, Kleinknecht C, Habib R. Nephronophthisis. In: Cameron D, Davison AM, Cameron JS, et al, eds. Clinical nephrology. Oxford: Oxford University Press, 19982417–2426.
- 32.Valente E M, Marsh S E, Castori M, Dixon‐Salazar T, Bertini E, Al‐Gazali L, Messer J, Barbot C, Woods C G, Boltshauser E, Al‐Tawari A A, Salpietro C D, Kayserili H, Sztriha L, Gribaa M, Koenig M, Dallapiccola B, Gleeson J G. Distinguishing the four genetic causes of Jouberts syndrome‐related disorders. Ann Neurol 200557(4)513–519. [DOI] [PubMed] [Google Scholar]
- 33.Watnick T, Germino G. From cilia to cyst. Nat Genet 200334(4)355–356. [DOI] [PubMed] [Google Scholar]
- 34.Otto E A, Schermer B, Obara T, O'Toole J F, Hiller K S, Mueller A M, Ruf R G, Hoefele J, Beekmann F, Landau D, Foreman J W, Goodship J A, Strachan T, Kispert A, Wolf M T, Gagnadoux M F, Nivet H, Antignac C, Walz G, Drummond I A, Benzing T, Hildebrandt F. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left‐right axis determination. Nat Genet 200334(4)413–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olbrich H, Fliegauf M, Hoefele J, Kispert A, Otto E, Volz A, Wolf M T, Sasmaz G, Trauer U, Reinhardt R, Sudbrak R, Antignac C, Gretz N, Walz G, Schermer B, Benzing T, Hildebrandt F, Omran H. Mutations in a novel gene, NPHP3, cause adolescent nephronophthisis, tapeto‐retinal degeneration and hepatic fibrosis. Nat Genet 200334(4)455–459. [DOI] [PubMed] [Google Scholar]