Abstract
Background
The molecular basis of autosomal recessive non‐syndromic mental retardation (NSMR) is poorly understood, mostly owing to heterogeneity and absence of clinical criteria for grouping families for linkage analysis. Only two autosomal genes, the PRSS12 gene on chromosome 4q26 and the CRBN on chromosome 3p26, have been shown to cause autosomal recessive NSMR, each gene in only one family.
Objective
To identify the gene causing autosomal recessive NSMR on chromosome 19p13.12.
Results
The candidate region established by homozygosity mapping was narrowed down from 2.4 Mb to 0.9 Mb on chromosome 19p13.12. A protein truncating mutation was identified in the gene CC2D1A in nine consanguineous families with severe autosomal recessive NSMR. The absence of the wild type protein in the lymphoblastoid cells of the patients was confirmed. CC2D1A is a member of a previously uncharacterised gene family that carries two conserved motifs, a C2 domain and a DM14 domain. The C2 domain is found in proteins which function in calcium dependent phospholipid binding; the DM14 domain is unique to the CC2D1A protein family and its role is unknown. CC2D1A is a putative signal transducer participating in positive regulation of I‐κB kinase/NFκB cascade. Expression of CC2D1A mRNA was shown in the embryonic ventricular zone and developing cortical plate in staged mouse embryos, persisting into adulthood, with highest expression in the cerebral cortex and hippocampus.
Conclusions
A previously unknown signal transduction pathway is important in human cognitive development.
Keywords: CC2D1A, non‐syndromic mental retardation
Mental retardation affects approximately 1% to 3% of the general population.1,2 Defining features of mental retardation include intellectual functioning level (IQ) below 70, significant limitations in two or more adaptive skill areas, and the condition present from childhood (defined as age 18 years or less).3 Genetic aetiologies are found in approximately two thirds of cases of mental retardation.4 An autosomal recessive mode of inheritance may account for nearly a quarter of all individuals with non‐syndromic mental retardation (NSMR).5,6,7 NSMR is the diagnosis of exclusion in mentally retarded individuals who do not have major physical abnormalities, dysmorphism, or neurological abnormalities. Biological processes involved in neuronal differentiation and synaptic plasticity, synaptic vesicle cycling, and gene expression regulation are considered to be important in the causation of mental retardation.8
While various X linked genes associated with NSMR have been identified, there is a large gap in our knowledge about the genetic basis of autosomal mental retardation, because of heterogeneity and the absence of clinical criteria for grouping the NSMR families for linkage analysis. Only two autosomal genes—the PRSS12 gene on chromosome 4q26 (MIM 606709) and the CRBN gene on chromosome 3p26 (MIM 607417)—have been shown to cause autosomal recessive NSMR, each gene in only one family.9,10 No mutations in these genes have been described in additional families with mental retardation.
We have previously identified and clinically evaluated four consanguineous families with severe autosomal recessive NSMR, comprising 10 affected and 24 unaffected individuals (MRT3 (MIM 608443)). We mapped the disease locus on chromosome 19p13.12–p13.2 to an interval of 2.4 Mb, between the loci D19S547 proximally and D19S1165 distally.11 In the current study, we have further reduced the candidate region to ∼900 kb between the markers D19S564 and D19S547. We have identified the disease causing mutation in the gene CC2D1A in all the families.
Methods
Patients
The study was reviewed and approved by the human subjects committee of the Rabin Medical Centre. The initial clinical presentation in all affected family members was psychomotor developmental delay during early childhood. All affected individuals had no or only single words and were severely mentally retarded, whereas their general physical examination was normal. There were no autistic features or seizures. The general appearance was normal, with no dysmorphic features; height, weight, and head circumference were normal in all except one subject who had progressive microcephaly. Neurological examination of the patients was normal.
Formal psychological evaluation was carried out on one patient at the age of eight years. He was assessed by the Leiter International Performance Scale12 and was found to function in the severely mentally retarded range.
Microsatellite marker analysis
DNA was isolated from the blood samples by standard methods.13 Four hundred microsatellite markers, spaced at 10 cM intervals, from the ABI PRISM linkage mapping set, version 2.5 (Applied Biosystems, Foster City, California, USA) were amplified by multiplex polymerase chain reaction (PCR), using standard protocols. Amplified markers were electrophoresed on an ABI 3700 DNA capillary sequencer and were analysed with Genescan and Genotyper software (Applied Biosystems).
Sequencing of candidate genes
We identified the genes in the candidate locus by means of available databases (NCBI and UCSC genome browser). Sequencing of the candidate genes was undertaken with primer sets designed using standard software Primer 3. All exons, including exon–intron junctions, were amplified from genomic DNA with primers designed from the genomic sequences available from NCBI and UCSC genome browser. Both strands of the PCR products were sequenced with BigDye Terminators (Applied Biosystems) on an ABI 3100 sequencer. Sequence chromatograms were analysed using SeqScape software, version 1.1 (Applied Biosystems). We initially tested one affected individual, one heterozygous parent, and a non‐carrier sibling (as defined by haplotype analysis).
Molecular analysis of the CC2D1A gene
On the basis of the predicted genomic sequence, the 21 primer pairs were used for PCR amplification of exons and splicing junctions (primer sequences available on request).
CC2D1A deletion mutation screening was undertaken on genomic DNA by PCR using the following primers: for normal alleles: primer A (forward): GTGGATGTCGCTGAATTGC; primer B (reverse): CAAGCGATCCTCCCATCTT; for mutant alleles: primer A (forward): GTGGATGTCGCTGAATTGC; primer C (reverse): TACAGTCCTCCGCCAACTTT. Primer A was derived from exon 13 (upstream of the deletion), primer B from intron 14 (within the deletion), and primer C from exon 19 (downstream of the deletion).
RNA isolation
We obtained RNA from the blood of the affected individual by use of Trizol reagent and carried out reverse transcriptase reactions with both random hexamers and oligo dT primers in order to obtain cDNA.
Western blot
Whole cell extracts were separated on 7.5% SDS‐PAGE and analysed by immunoblotting with CC2D1A specific antibody. The cell proteins were prepared. Briefly, cells were washed twice with phosphate buffered saline (PBS) and lysed at 4°C with lysis buffer (0.6% NP40 and protease inhibitors). After 15 minutes of incubation, cell lysates were centrifuged for five minutes at 16 000 xg and supernatants were collected. Protein concentrations were determined using the BCA protein assay reagent kit (Pierce Biotechnology, Rockford, Illinois, USA). Whole cell proteins (approximately 20 μg) were subjected to 7.5% SDS‐PAGE and transferred to nitrocellulose membranes. The membranes were blocked with 5% skimmed milk in Tris buffer (10 mM Tris, 150 mM NaCl, and 0.1% Tween20) for 30 minutes at room temperature and subsequently incubated with anti‐CC2D1A rabbit polyclonal antibody (Bethyl Laboratories, Montgomery, Texas, USA) diluted 1:20 000, or rabbit polyclonal anti‐emerin diluted 1:5000, or mouse monoclonal antibodies anti‐NFκB p50 diluted 1:1000/p65 diluted 1:5000 (Santa Cruz Biotechnology, Santa Cruz, California, USA). After washing and subsequent incubation for one hour with horseradish peroxidase coupled to anti‐rabbit IgG or mouse IgG, respectively (Jackson ImmunoResearch Laboratories, West Grove, Pennsylvania, USA), immunoreactive proteins were visualised by an ECL (enhanced chemiluminescence super signal) detection kit (Pierce Biotechnology). Antibody to emerin protein (Santa Cruz Biotechnology) was used to demonstrate the loading level.
Immunofluorescence
Human bone osteosarcoma line U2OS cells were grown on cover slips overnight. The cells were fixed with ice cold methanol (five minutes) and ice cold acetone (five minutes), washed with tris buffered saline (TBS; 100 mM Tris‐HCl pH 7.5, 150 mM NaCl) and blocked using 5% skimmed milk in TBS containing 0.1% Tween 20 (TBS‐T) for 30 minutes. Incubations with primary (rabbit polyclonal anti‐CCD21A (Bethyl Laboratories; 1:100 dilution), and secondary (Cy3‐conjugated donkey anti‐rabbit (Jackson ImmunoResearch Laboratories; 1:500 dilution) antibodies were carried out in blocking solution for one hour each. Between and after the incubation with the antibodies, cells were washed three times with TBS‐Tween. The cover slips were mounted using DAPI containing Fluoromount‐G (SouthernBiotech, birmingham, Alabama, USA) and the cells photographed after fluorescent microscopy analysis.
Immunoprecipitation
A sample of approximately 200 μg total protein from the lymphoblasts of a patient and a control subject were incubated with rabbit anti‐CC2D1A antibody (diluted 1:100) overnight, with rotation at 4°C. Subsequently 50 μl anti‐rabbit IgG (whole molecule) agarose antibody (Sigma Chemical Company, St Louis, Missouri, USA) was added and the mixture was incubated for one hour at room temperature. The agarose beads were washed once with a lysis buffer and three additional times with PBS. Absorbed proteins were solubilised with SDS‐PAGE sample buffer and subjected to electrophoresis on 12% polyacrylamide gels and immunoblotted as described in the western blot method.
In situ hybridisation
Non‐radioactive in situ hybridisation was carried out as previously described in frozen mouse brains, using digoxigenin (DIG)‐labelled cRNA probes.14 Briefly, the murine CC2D1A probes were generated by PCR from IMAGE clone 5363547 (BC027028). Probes were generated at the following nucleotide (nt) sequence positions in the CC2D1A cDNA: probes CC2D1A‐1 (nt 38–506; 469 bp) and CC2D1A‐2 (nt 2657–3021; 365 bp; 3′‐UTR). All CC2D1A nucleotide probe sequences were from nucleotides downstream of the ATG start site. The probes were made from PCR amplified sequences using primers constructed to amplify the sequences described above (primer sequences are available upon request). All forward primers contained T3 promoter sequences on their 5′ ends in order to drive the transcription of the cRNA sense probes, and all reverse primers contained T7 promoter sequences on their 5′ ends in order to drive the transcription of the cRNA antisense probes.
Frozen brains were sectioned (10–20 μm) in a cryostat and placed on Superfrost plus microscope slides. Brain sections were fixed, acetylated, and hybridised at 68°C (approximate concentration of the probe 100 ng/ml). Hybridised probes were visualised using alkaline phosphatase conjugated anti‐DIG Fab fragments (Roche, Indianapolis, Indiana, USA) and 5‐bromo‐4‐chloro‐3‐indolyl‐phosphate/nitro blue tetrazolium (BCIP/NBT) substrate (Kierkegard and Perry Laboratories (KPL), Gaithersburg, Maryland, USA). Sections were washed and cover slipped with glycerol gelatin (Sigma). Both CC2D1A antisense probes yielded comparable expression patterns.
Phylogenetic analysis
The phylogenetic analysis included multiple alignment using ClustalX,15 and dendogram creation with MEGA216 using neighbour joining.17 Accession numbers for the family members are: XP542027 (Dog1), XP237884 (Rat1), NP666082 (Mouse1), XP_614352 (Cow1), CAG06037 (Tetraodon1), XP_513407 (Chimpanzee2), XP588670 (Cow2), XP233342 (Rat2), NP796019 (Mouse2), CAG077113 (Tetraodon2), and MGC68887 (Xenopus laevis). The protein Chimpanzee1 was defined from the chimpanzee genome by aligning the human mRNA using Sim418 and translation of the prediction. Tetraodon2 is a result of Genescan prediction19 the region chr15_random: 1575804‐1583914.
Results
Refinement of the candidate region
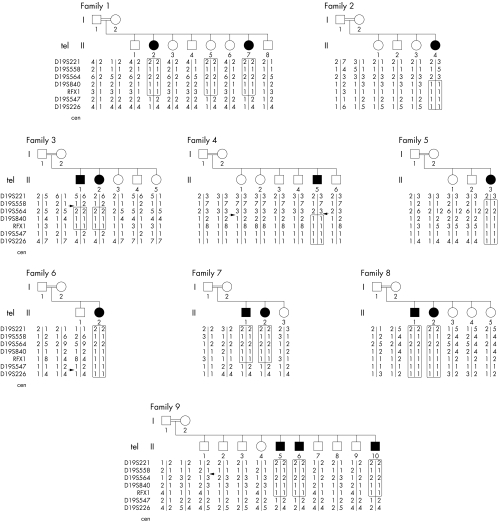
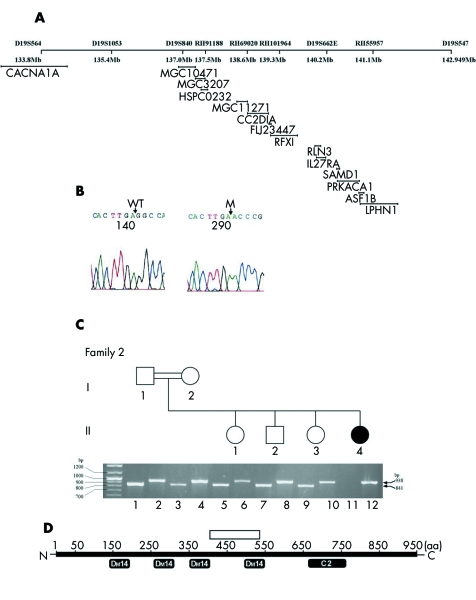
A common homozygous disease bearing haplotype for the polymorphic markers RFX1 and D19S840 defined a critical region of 0.9 Mb between the markers D19S564 and D19S547 in the four original and five additional families, with a total of 16 affected and 48 unaffected individuals, all living in the same village and bearing the same family name (fig 1). The presence of the common disease haplotype suggests that the disease is caused by a single mutation derived from a single ancestral founder in all the families. A minimal candidate region contains 14 genes (fig 2A).
Figure 1 Fine mapping of the candidate region on 19p13.12. The genotypes of the pedigrees with autosomal recessive NSMR. Black symbols in the pedigree represent affected individuals. The boxed regions indicate the homozygous regions in each affected individual. The polymorphic markers are shown on the left. Arrows indicate informative recombinations.
Figure 2 Mutational analysis of CC2D1A. (A) Minimum candidate interval on chromosome 19, showing the relative positions of the genes in this region. (B) Sequence chromatogram from a wild‐type (WT) and disease (M) genotype on cDNA from control and affected individuals, showing the deletion mutation. (C) Segregation of the deletion mutation in CC2D1A as assayed on the genomic DNA. Two separate polymerase chain reaction (PCR) with primers A+B and A+C were done (see methods). Amplification with primers A+B (lanes 1, 3, 5, 7, and 9) produces the 841 base pair (bp) PCR product only in normal alleles, while amplification with primers A+C (lanes 2, 4, 6, 8, 10, and 12) produces the 938 bp PCR product only in alleles carrying the deletion. (D) Schematic diagram of the structure of the CC2D1A gene showing the main domains of the protein; the white bar depicts the relative position of the deletion mutation.
Mutation analysis
All the genes in the candidate region were analysed. Sequencing of exons 14, 15 and 16 of the CC2D1A gene showed failure of amplification in all the affected family members as opposed to successful amplification in heterozygous and homozygous normal individuals. Sequencing of the cDNA with the primers derived from exons 13 and 17 confirmed complete deletion of exons 14, 15, and 16; sequencing of the genomic DNA with the primers flanking the exon–intron junctions for exons 13 and 17 revealed the deletion of 3589 nucleotides beginning in intron 13 and ending in intron 16 (IVS13_IVS16del). The deletion causes removal of aa 408 to 547 and introduces a frameshift of the encoded protein immediately after the deletion, creating a nonsense peptide of 30 aa and a stop codon at position 438 of the mutant protein (G408fsX437). In each pedigree analysed, the CC2D1A mutation segregated with the disease; affected patients displayed homozygous CC2D1A mutations, whereas parents who were clinically normal displayed heterozygosity for a normal and a disease allele, consistent with recessive inheritance (fig 2B and 2C). The mutation abolishes DM14 and C2 domains of the gene (fig 2D). The mutation was not observed in 300 control chromosomes including 100 Jewish and 50 Israeli Arab individuals.
Protein analysis
We confirmed the absence of the wild type protein product by testing patient derived lymphoblastoid cells by western blot analysis with CC2D1A antibody, which detects the N‐terminus of the protein, and found that antibodies identified a 104 kDa band in normal individuals but not in the patients; the smaller protein, most probably representing the truncated protein, was observed in the patients (fig 3A).
Figure 3 CC2D1A protein analysis. (A) Western blot analysis of CC2D1A (arrow on the right) showing the absence of the normal protein in the lymphoblasts of affected individuals. The smaller protein of approximately 85 kDa (arrowhead on the left) was observed in the patients. (B) CCD21A protein is localised to the cytoplasm of U2OS. Cells were fixed, immunostained (orange) with rabbit anti‐CCD21A antibodies, and analysed by fluorescent microscopy. The nuclei are blue stained with DAPI.
To determine the subcellular localisation of CC2A1 protein, immunolabelling experiments were carried out by using polyclonal antibody U2OS cells (fig 3B). CC2D1A protein is distributed throughout the cytoplasm; no signal was detected in the nuclei. It is a soluble protein, as no transmembrane regions were detected. The absence of signal peptide in the protein sequence suggests that it is not secreted.
We examined possible direct interactions between CC2D1A protein and the p65 and p50 subunits of the NFκB complex by using co‐immunoprecipitation of the endogenous proteins in control lymphoblastoid cells. CC2DA1 protein does not interact directly with the NFκB p65 subunit or the p50 subunit in vivo (data not shown), as detected by immunoprecipitation.
Expression analysis
We carried out in situ hybridisation in staged mouse embryos to determine the spatial expression patterns of CC2D1A during brain development (fig 4). CC2D1A mRNA was observed at E12 throughout the ventricular zone and developing cortical plate and ganglionic eminences. By E16, CC2D1A mRNA was found throughout the mouse brain, but most strongly in the cortical plate. At postnatal (P) day 3, CC2D1A mRNA continued to be widely expressed with strong expression in the cerebral cortex and hippocampus (particularly in the hippocampal CA3 region). Continued expression was observed in the brain into adulthood.
Figure 4 In situ hybridisation of the CC2D1A mRNA. Expression of CC2D1A mRNA is seen in murine brain from E12 into adulthood. CC2D1A mRNA expression is found throughout the ventricular zone progenitor cells and developing cortical plate. Later in development at E16, CC2D1A mRNA is found throughout the cortical plate and developing hippocampus. During the postnatal period of brain development (P3), CC2D1A mRNA expression is widely expressed throughout the brain, but particularly in the cerebral cortex and hippocampus, especially CA3 of the hippocampus. In adult brain, the expression of CC2D1A mRNA is widespread but appears restricted mostly to neurones. The scale bar in the E12, E16, and P3 panels is 500 microns, and the one in the adult panel 1 mm. CBL, cerebellum; CC, corpus callosum; CP, cortical plate; CTX, cerebral cortex; GE, ganglionic eminences; HPC, hippocampus; OB, olfactory bulb; STR, striatum; TH, thalamus.
Gene structure and phylogenetic analysis
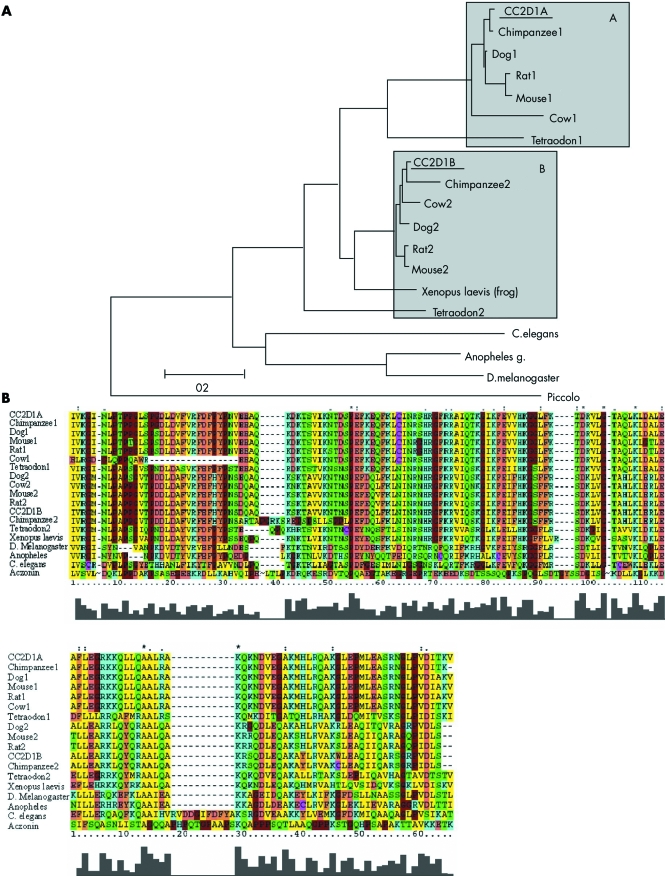
The CC2D1A gene (GenBank accession number NM_017721) covers 37 kb of genomic DNA on 19p13.12. It encodes 3715 bp long mRNA, containing 31 exons. At least 12 splice variants have been identified (UCSC genome browser). The coding sequence is 2853 bp long. The gene is widely expressed in various tissues. The CC2D1A protein contains 950 amino acids (National Center for Biotechnology Information (NCBI) Entrez Protein accession number NP_060191), although a shorter isoform of the protein exists, which contains only 388 amino acids. A search against the CDart database showed that the protein family carries two conserved motifs. The most conserved motif is a C2 domain (IPR008973, IPR006608, IPR000008). The other motif, DM14, is unique to the CC2D1A protein family and its role is unknown.
A phylogenetic analysis of the CC2D1A protein family is shown in fig 5A. All protein family members are predicted and their function is unknown. The two invertebrate branches were inferred, using the mouse protein aczonin (presynaptic cytomatrix protein) as an out group. Sequence similarity searches of the CC2D1A protein, using BLASTP against swiss‐prot and non‐redundant databases, revealed similarity to many potential orthologues in mammals, in other vertebrates (Xenopus lavis, tetraodon), as well as in invertebrates (Drosophila melanogaster and Caenorhabditis elegans).20 No candidate orthologue was found in the yeast genome. In vertebrates the gene was duplicated, as two paralogs are found in the tetraodon genome and the mammalian genome. Although only one copy was found in the frog genome, this is probably because the full genome sequence was yet not determined. A multiple alignment of the C2 domain and one of the DM14 domains is shown in fig 5B.
Figure 5 CC2D1A protein family. (A) A phylogenetic analysis of the CC2D1A protein family. The two invertebrate branches, A and B, are emphasised by the grey boxes. (B) A multiple alignment of the C2 domain (upper panel) and one of the DM14 domains (lower panel). Strongly conserved positions are marked on top, where ‘*' indicates positions that are occupied by a single amino acid, while ‘:' and ‘.' are for positions where a conserved substitution have occurred. The conservation graph is shown below. Note that some positions in the aczonin protein were truncated to allow a better alignment of the C2 domain. These are marked by the ‘∼' sign.
Discussion
In this study we have identified a protein truncating mutation in the novel CC2D1A gene with unknown function causing autosomal recessive NSMR. The protein has two main isoforms containing 950 amino acids and 388 amino acids. The shorter isoform of the protein does not contain exons 14 to 16, which are deleted in the patients. Thus only the long protein isoform seems to play a central role in disease pathogenesis in the patients. C2 motif, which is located at the C‐terminus of CC2D1A at positions 661–762, is found in proteins which function in calcium dependent phospholipid binding,21 where the C2 domain itself participates in the binding pocket of the Ca2+ cation. Proteins which have a C2 domain include phospolipases, protein kinases C, and also the synaptotagmin protein family. Synaptotagmins are synaptic vesicle membrane trafficking proteins. The role of the other motif, DM14, is unknown. It repeats four times in the human CC2D1A sequence but only three times in the Caenorhabditis elegans orthologue sequence. The DM14 motif also appears only in the long isoform of CC2D1A, while the C2 domain, which is present in both isoforms, probably plays a central role in its protein activity. The protein truncating mutation in our patients abolishes one of the four DM14 domains and the C2 domain. The absence of signal peptide in the protein sequence suggests that it is not secreted. Phylogenetic analysis shows similarity to many potential orthologues in mammals, in other vertebrates, and in invertebrates, but no candidate orthologue was found in the yeast genome.
Matsuda et al originally identified CC2D1A as a putative signal transducer participating in positive regulation of I‐κB kinase/NFκB cascade.22 Their large scale study was designed to identify human genes that activate the NFκB and MAPK signalling pathways, and they reported that the CC2D1A gene was found to induce by at least fourfold the transcription of a luciferase reporter gene driven by a promoter containing the consensus NFκB binding sites. CC2DA1 protein does not directly interact with the NFκB p65 subunit or the p50 subunit in vivo as detected by immunoprecipitation, suggesting that NFκB expression is regulated by the CC2D1A gene through other signalling proteins that belong to the IKK/IκB dependent or the IKK/IκB independent pathway. Functional NFκB complexes are present in all cell types in the nervous system, including neurones, microglia, oligodendrocytes, and astrocytes.23 Stimulation of glutamate receptors and membrane depolarisation stimulate NFκB activation in neurones, probably through a calcium mediated mechanism. NFκB is also activated in association with long term potentiation of synaptic transmission, a process believed to be related to learning and memory.24 Electrophysiological measurements of synaptic plasticity in hippocampal cells from mice lacking tumour necrosis α receptors suggest an important function of NFκB in the long term depression (LTD) of synaptic transmission.25 Along with the potential role of NFκB in neuronal function in the brain, there is an indication that NFκB might be important for neuronal development based on the pattern of NFκB activity in the brain during mouse development.26 Basal synaptic input in mature mouse hippocampal neurones activates the NFκB transcription factor and induces κB DNA binding activity, consisting of p50:p65 and p50:p50 dimers. Mice lacking p65 show a selective learning deficit in the spatial version of the radial arm maze. These observations suggest that long term changes to adult neuronal function caused by synaptic stimulation can be regulated by NFκB nuclear translocation and gene activation.27
The expression patterns of the CC2D1A mRNA appear to be restricted mostly to the ventricular zone progenitors and neurons. Given these expression patterns in the cerebral cortex and hippocampus and the phenotype in patients, CC2D1A appears to be important in cognitive development in humans.
Electronic database information
Accession numbers and URLs for data presented in this paper are as follows:
NCBI Map Viewer, http://www.ncbi.nlm.nih.gov/mapview/
UCSC Genome Browser, http://genome.ucsc.edu/ (May 2004 build)
Primer3, http://frodo.wi.mit.edu/cgi‐bin/primer3/primer3_www.cgi
NCBI Entrez Protein Database, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db = Protein (for CC2A1 protein [accession number NP_060191])
GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for CC2D1A mRNA [accession number NM_017721]
Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for MRT3)
Acknowledgements
We are grateful to the families who participated in this study and clinical nurses Hitam and Inshirah. We thank Dr D Gurevitch for establishing the lymphoblastoid cell line. We also thank Dr G Halpern for her help with editing the manuscript and Rafi Rainshtein for his help with preparing the figures. This study was funded by the Yeshaya Horovitz grant for Neurogenetics research and Israeli Science Foundation (grant No 690/04).
Abbreviations
NSMR - non‐syndromic mental retardation
Footnotes
Competing interests: none declared.
References
- 1.McLaren J, Bryson S E. Review of recent epidemiological studies of mental retardation: prevalence, associated disorders, and etiology. Am J Ment Retard 198792243–254. [PubMed] [Google Scholar]
- 2.Leonard H, Wen X. The epidemiology of mental retardation: challenges and opportunities in the new millennium. Ment Retard Dev Disabil Res Rev 20028117–134. [DOI] [PubMed] [Google Scholar]
- 3.American Pyschiatric Association Diagnostic and statistical manual of mental disorders (DSM‐IV). 4th ed. Washington, DC: American Psychiatric Association, 199439–46.
- 4.Curry C J, Stevenson R E, Aughton D, Byrne J, Carey J C, Cassidy S, Cunniff C, Graham J M, Jones M C, Kaback M M, Moeschler J, Schaefer G B, Schwartz S, Tarleton J, Opitz J. Evaluation of mental retardation: recommendations of a consensus conference: American College of Medical Genetics. Am J Med Genet 199772468–477. [DOI] [PubMed] [Google Scholar]
- 5.Wright S W, Tarjan G, Eyer L. Investigation of families with two or more mentally defective siblings: clinical observations. Am J Dis Child 195997445–456. [DOI] [PubMed] [Google Scholar]
- 6.Priest J H, Thuline H C, Laveck G D, Jarvis D B. An approach to genetic factors in mental retardation. Studies of families containing at least two siblings admitted to a state institution for the retarded. Am J Ment Defic 19616642–50. [PubMed] [Google Scholar]
- 7.Bartley J A, Hall B D. Mental retardation and multiple congenital anomalies of unknown etiology: frequency of occurrence in similarly affected sibs of the proband. Birth Defects Orig Artic Ser 197814127–137. [PubMed] [Google Scholar]
- 8.Renieri A, Pescucci C, Longo I, Ariani F, Mari F, Meloni I. Non‐syndromic X‐linked mental retardation: From a molecular to a clinical point of view. J Cell Physiol 20052048–20. [DOI] [PubMed] [Google Scholar]
- 9.Molinari F, Rio M, Meskenaite V, Encha‐Razavi F, Auge J, Bacq D, Briault S, Vekemans M, Munnich A, Attie‐Bitach T, Sonderegger P, Colleaux L. Truncating neurotrypsin mutation in autosomal recessive nonsyndromic mental retardation. Science 20022981779–1781. [DOI] [PubMed] [Google Scholar]
- 10.Higgins J J, Pucilowska J, Lombardi R Q, Rooney J P. A mutation in a novel ATP‐dependent Lon protease gene in a kindred with mild mental retardation. Neurology 2004631927–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basel‐Vanagaite L, Alkelai A, Straussberg R, Magal N, Inbar D, Mahajna M, Shohat M. Mapping of a new locus for autosomal recessive non‐syndromic mental retardation in the chromosomal region 19p13.12–p13.2: further genetic heterogeneity. J Med Genet 200340729–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leiter R G.Leiter International Performance Scale: instruction manual. Chicago: Stoelting Company, 1980
- 13.Sambrook J, Fritsch E, Maniatis T. Isolation of high‐molecular‐weight DNA from mammalian cells. In: Nolan C, ed. Molecular cloning: a laboratory manual. 2nd ed. New York: Cold Spring Harbor Laboratory Press, 19899.14–9.19.
- 14.Ferland R J, Eyaid W, Collura R V, Tully L D, Hill R S, Al‐Nouri D, Al‐Rumayyan A, Topcu M, Gascon G, Bodell A, Shugart Y Y, Ruvolo M, Walsh C A. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat Genet 2004361008–1013. [DOI] [PubMed] [Google Scholar]
- 15.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997244876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput Appl Biosci 199410189–191. [DOI] [PubMed] [Google Scholar]
- 17.Saitou N, Nei M. The neighbor‐joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 19874406–425. [DOI] [PubMed] [Google Scholar]
- 18.Pidoux A L, Richardson W, Allshire R C. Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J Cell Biol 2003161295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol 199726878–94. [DOI] [PubMed] [Google Scholar]
- 20.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol 1990215403–410. [DOI] [PubMed] [Google Scholar]
- 21.Davletov B A, Suedhof T C. A single C2 domain from synaptotagmin I is sufficient for high affinity Ca2+/phospholipid binding. J Biol Chem 199326826386–26390. [PubMed] [Google Scholar]
- 22.Matsuda A, Suzuki Y, Honda G, Muramatsu S, Matsuzaki O, Nagano Y, Doi T, Shimotohno K, Harada T, Nishida E, Hayashi H, Sugano S. Large‐scale identification and characterization of human genes that activate NF‐kappaB and MAPK signaling pathways. Oncogene 2003223307–3318. [DOI] [PubMed] [Google Scholar]
- 23.O'Neill L A, Kaltschmidt C. NF‐kappa B: a crucial transcription factor for glial and neuronal cell function. Trends Neurosci 199720252–258. [DOI] [PubMed] [Google Scholar]
- 24.Meberg P J, Kinney W R, Valcourt E G, Routtenberg A. Gene expression of the transcription factor NF‐κB in hippocampus: regulation by synaptic activity. Brain Res Mol Brain Res 199638179–190. [DOI] [PubMed] [Google Scholar]
- 25.Albensi B C, Mattson M P. Evidence for the involvement of TNF and NF‐kappaB in hippocampal synaptic plasticity. Synapse 200035151–159. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt‐Ullrich R, Memet S, Lilienbaum A, Feuillard J, Raphael M, Israel A. NF‐kappaB activity in transgenic mice: developmental regulation and tissue specificity. Development 19961222117–2128. [DOI] [PubMed] [Google Scholar]
- 27.Meffert M K, Chang J M, Wiltgen B J, Fanselow M S, Baltimore D. NF‐kappa B functions in synaptic signaling and behavior. Nat Neurosci 200361072–1078. [DOI] [PubMed] [Google Scholar]