Abstract
Background
Brachydactyly type A2 (OMIM 112600) is characterised by hypoplasia/aplasia of the second middle phalanx of the index finger and sometimes the little finger. BDA2 was first described by Mohr and Wriedt in a large Danish/Norwegian kindred and mutations in BMPR1B were recently demonstrated in two affected families.
Methods
We found and reviewed Mohr and Wriedt's original unpublished annotations, updated the family pedigree, and examined 37 family members clinically, and radiologically by constructing the metacarpo‐phalangeal profile (MCPP) pattern in nine affected subjects. Molecular analyses included sequencing of BMPR1B, linkage analysis for STS markers flanking GDF5, sequencing of GDF5, confirmation of the mutation by a restriction enzyme assay, and localisation of the mutation inferred from the very recently reported GDF5 crystal structure, and by superimposing the GDF5 protein sequence onto the crystal structure of BMP2 bound to Bmpr1a.
Results
A short middle phalanx of the index finger was found in all affected individuals, but other fingers were occasionally involved. The fourth finger was characteristically spared. This distinguishes Mohr‐Wriedt type BDA2 from BDA2 caused by mutations in BMPR1B. An MCPP analysis most efficiently detected mutation carrier status. We identified a missense mutation, c.1322T>C, causing substitution of a leucine with a proline at amino acid residue 441 within the active signalling domain of GDF5. The mutation was predicted to reside in the binding site for BMP type 1 receptors.
Conclusion
GDF5 is a novel BDA2 causing gene. It is suggested that impaired activity of BMPR1B is the molecular mechanism responsible for the BDA2 phenotype.
Keywords: BDA2, brachydactyly, chondrogenesis, GDF5
Brachydactylies (BDs) are a group of inherited malformations characterised by shortening of the digits due to abnormal development of the phalanges and/or the metacarpals. They have been classified on an anatomic and genetic basis into five groups, A–E, including three subgroups (A1–A3) that usually manifest as autosomal dominant traits.1 Three types of brachydactyly, A1, A2, and C, mainly affect the middle phalanges.
BDA1 shows hypoplasia/aplasia of all middle phalanges, sometimes with joint fusion between the middle and the proximal phalanx, and is caused by mutations in Indian hedgehog (IHH),2 a signalling molecule with pivotal roles in the regulation of chondrocyte proliferation and differentiation. BDA2 patients display hypoplasia/aplasia of the middle phalanx of the second and, sometimes, the fifth fingers. It was described first by Mohr and Wriedt in a large Norwegian kindred of Danish descent.3 We recently showed that BDA2 is caused by mutations in bone morphogenetic receptor 1b (BMPR1B),4 a receptor with an important role in cell proliferation and differentiation, especially in limbs and the CNS. BDC is characterised by brachymesophalangia of the second, third, and fifth fingers, sometimes combined with hyperphalangia, usually of the second and third fingers. Heterozygous frameshift or nonsense mutations affecting growth/differentiation factor 5 (GDF5), a signal protein of the bone morphogenetic protein (BMP) family, have been identified in several individuals with BDC.5 Thus, these types of BD have certain overlapping features including hypoplasia/aplasia of the second middle phalanx and abnormal interdigital joint formation in the index finger, suggesting that the formation of joints and phalanges in this region is linked on a developmental and molecular basis.
We report on the updated kindred studied by Mohr and Wriedt, showing that this family constitutes a distinct subtype of BDA2 caused by a mutation in GDF5.
Methods
Patients
We studied 37 Norwegian members from a family with BDA2 segregating in nine generations (fig 1A) and one person from a Danish family with a similar phenotype. The study was approved by the Danish and Norwegian ethical committee systems and written, informed consent was obtained for all participants in accordance with the Helsinki II Declaration. The genealogy was unravelled, helped by a family book containing information on family members with “crooked fingers” and information from living family members. All subjects were examined clinically. Fourteen out of 23 molecularly confirmed mutation carriers were examined in more detail, and radiographs of the hands and feet were obtained in nine. Bone lengths were measured in 19 hand bones, and classified into normal or short according to population based references.6 We then performed metacarpal‐phalangeal profile (MCPP) analysis according to Poznanski,7 and evaluated the shape, structure, and position of the different bones.
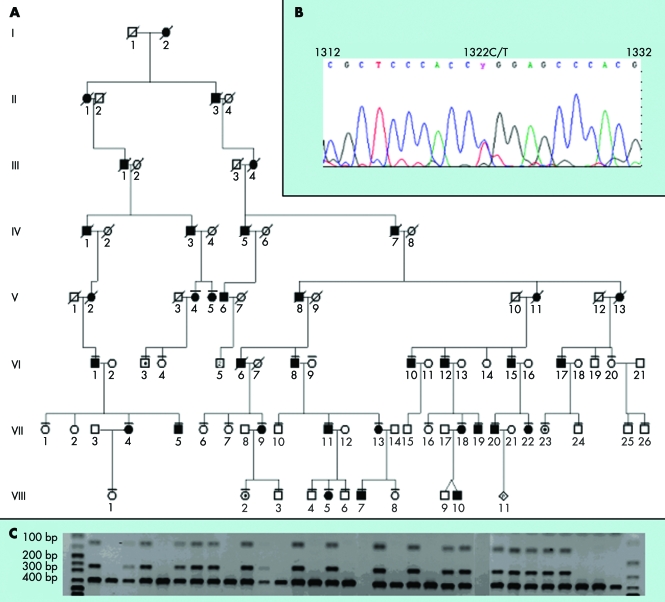
Figure 1 (A) Pedigree showing the clinically examined members of the family. The full pedigree is not shown. (B) Chromatogram showing the c.1322T>C mutation. (C) Digestion with HpaII results in a 379 bp fragment in unaffected subjects, whereas heterozygous mutation carriers in addition have two additional fragments of 124 and 255 bp.
Molecular testing
Mutation analysis of BMPR1B
DNA was extracted by a standard salt‐out method from a peripheral venous blood sample in adults, and from a filter blot or a buccal swap in children. The BMPR1B gene was tested for mutations as previously described.4
Linkage analysis
Linkage to the chromosomal region harbouring the GDF5 gene was tested with STS markers D20S847 and D20S106, located 827 Kb 5′ and 513 Kb 3′ from the gene, respectively. Primers for STS markers were radioactively labelled using c‐[33P]‐dATP (Hartmann Analytic, Braunschweig, Germany) and T4‐DNA polynucleotide kinase (Fermentas, Denmark) according to the manufacturer's protocols. Polymerase chain reaction (PCR) was carried out using 50 ng DNA template and Taq‐DNA polymerase (New England Biolabs, Beverly, MA, USA) under standard conditions according to the manufacturer's instructions, followed by separation by 5% acrylamide, 7 M urea, 1×TBE gel electrophoresis and overnight exposure to x ray films. A two point linkage analysis on affected individuals was performed using the computer program LIPED,8 and LOD scores were calculated using the MLINK and LINKMAP routines of FASTLINK software version 2.2,9 assuming an autosomal dominant mode of inheritance with a disease allele frequency of 0.0001 and equal female and male recombination rates.
Mutation analysis of GDF5
Both exons and exon‐intron border regions in the GDF5 gene were sequenced on both strands; primers were designed in the intron regions and amplified by PCR using Platinum Taq (Invitrogen, Carlsbad, CA, USA) or NEB Taq‐DNA polymerase (New England Biolabs). PCR conditions included an initial denaturation for 5 min at 94°C, followed by 40 cycles of 30 s at 94°C, 30 s at the individual annealing temperature determined for each primer set (table 1), 45 s at 72°C, and a final extension for 8 min at 72°C. PCR products were separated by 2% agarose, 1×TBE gel electrophoresis. Primers were removed by treatment with 1 U shrimp alkaline phosphatase (USB, Cleveland, OH, USA) and 10 U exonuclease I (New England Biolabs) followed by sequencing using the BigDye Terminator Kit (Applied Biosystems, Foster City, CA, USA) and analysed on an ABI 377 sequencer (Applied Biosystems).
Table 1 Primers used for sequencing of GDF5.
| Exon | Forward | Reverse | DNA Polymerase | Annealing temp (°C) | Product length (bp) |
|---|---|---|---|---|---|
| 1 | CAGCATTACGCCATTCTTCCT | CCGGTGCCTCCCTTTG | Platinum Taq | 63 | 613 |
| 1 | GGGACCAGGCCAGGATTG | GGGCCAGATGCACACAGTG | Platinum Taq | 63 | 637 |
| 2 | GACTGGCTCCCTTGGT | GGAATGCGGTTAGAGGTGAGC | Platinum Taq | 63 | 1688 |
| 2 | ATGAGATTAAGGCCCGCTCTG | TGTCTCCCTGGACCTGTGC | Platinum Taq | 60 | 692 |
| 2 | GGAGGCGGGCGGGCTGCCCA | GCAGAGTCAATGAAGAGGAT | NEB Taq | 55 | 680 |
| 2 | GGCCCCCTTTTATCCACAAGT* | GTCGAACACCTCCCAGCCAG* |
*Sequencing primer.
Confirmation of the mutation
The mutation was confirmed by HpaII (NEB) restriction enzyme digestion of the PCR product generated by the primers 5′‐ATGAGATTAAGGCCCGCTCTG‐3′ and 5′‐GCAGAGTCAATGAAGAGGAT‐3′. Fifty normal individuals (100 alleles) served as a control group. Digestion of the PCR product from an unaffected individual resulted in a 379 bp product, whereas heterozygous mutation carriers had an additional two products of 124 and 255 bp. The digested PCR products were separated on a 20% acrylamide 1×TBE gel.
Results
Mutation analysis of BMPR1B
Initially, sequence analysis of the known BDA2 causing gene, BMPR1B, in the first identified affected family member, VI:1, did not identify any mutations.
Linkage analysis
Hands and feet were clinically examined in 37 family members, 19 of whom had a short second middle finger as compared to the fourth middle finger. In most affected cases this was obvious, even though the trait was not always recognised by the person themselves. Affected individuals were then included in the linkage analysis.
As GDF5 is the primary ligand for BMPR1B, we subsequently focused on this gene. Linkage analysis with STS markers D20S847 and D20S106 spanning a ∼1.35 Mb region containing GDF5 showed a shared haplotype in all affected individuals, and a two point LOD score of 3.98 was calculated in affected subjects only.
Molecular analysis of GDF5
Bidirectional sequencing of GDF5 demonstrated heterozygosity for a single base pair substitution, c.1322T>C (fig 1B), causing a change in amino acid residue 441 from a leucine to a proline. The mutation induced a novel cleavage site for the restriction enzyme HpaII, and digestion confirmed the mutation in 22 subjects. Three of these (VI:3, VIII:2, VII:23; fig 2C) were clinically unaffected (fig 1C). Furthermore, the same mutation was identified in the Danish subject. The mutation was not observed in a panel of 100 Danish control alleles.
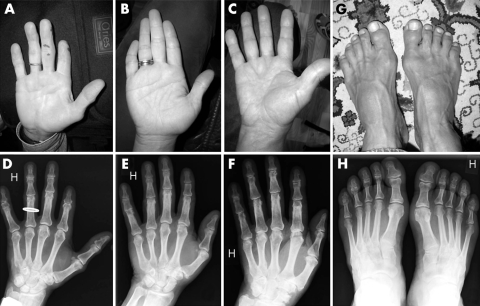
Figure 2 (A−F) Different degrees of BDA2 observed in the family (A, D: VI:8; B, E: VI:10; C, F: VI:15). (G, H). Foot phenotype with shortened or absence of middle phalanges. (Photographs are reproduced with consent) (A colour version of this figure can be found as supplemental data at http://jmedgenet.com/supplemental.)
Phenotypic spectrum in mutation carriers
Hands were more commonly affected than the feet (table 2). Relative shortening of the second mesophalanx was the predominant trait in all affected individuals. In two of nine radiographically evaluated cases this bone was absent, and in five other individuals it was shortened by more than 2 SD. In two cases (VI:15, VII:23) the second mesophalanx was of normal length. Clinodactyly occurred in four cases. Occasionally, other bones were also affected. A short third, fourth, or fifth mesophalanx, or first proximal phalanx was observed in three, two, two, and two cases, respectively. Metacarpal 1 and distal phalanx 1 and 2 were each short in one of the nine cases. Three mutation carriers appeared clinically normal. Radiographs were only available for one of these (VII:23), and confirmed normal bone length.
Table 2 Phenotype in 14 patients, determined by clinical examination, and an additional radiological examination in nine of the 14.
| ID | Hands | Feet | ||
|---|---|---|---|---|
| Right | Left | Right | Left | |
| V:4 | Clinically described in the original paper by Mohr and Wriedt | |||
| V:5 | Clinically described in the original paper by Mohr and Wriedt | |||
| VI:8 | Absent 2nd mesophalanx, milder shortening of 3rd, 4th, | Absent 2nd mesophalanx, 3rd meso‐ | Hypoplastic middle | Similar to |
| and 5th mesophalanges with slightly camptodactyly in the | phalanx short with camptodactyly in | phalanges 2–5 | right foot | |
| 4th DIP joint. Radiographs showed short 1st and 2nd | the MP and DIP joints. Flexion was | |||
| proximal phalanges bilaterally | limited in the 4th DIP joint | |||
| VI:10 | Short 2nd mesophalanx and clinodactyly of the 5th DIP | Short 2nd mesophalanx and | Broad foot, hallux | Normal |
| joint. Radiographs showed short 1st proximal phalanx | clinodactyly of the 5th DIP joint | overriding 2nd toe | ||
| bilaterally | ||||
| VI:12 | The 2nd mesophalanx slightly short compared to the 4th | Similar to right hand | Normal | Normal |
| mesophalanx. Radiographs showed short 1st metacarpals | ||||
| VI:15 | The 2nd mesophalanx slightly short compared to the 4th | Normal | Normal | Normal |
| mesophalanx | ||||
| VI:17 | Short 2nd mesophalanx, limited flexion of the 4th DIP joint | Short 2nd mesophalanx | Short 2nd toe | Normal |
| VII:11 | Absent 2nd mesophalanx and ulnar deviation in the 2nd DIP | Similar to the right hand | Hypoplastic middle | Similar to |
| joint. The 3rd, 4th and 5th mesophalanges were short with | phalanges 2, 4, | right foot | ||
| clinodactyly and limited flexion in the 5th DIP joint. Radio‐ | and 5 | |||
| graphs showed long 2nd distal phalanges | ||||
| VII:18 | Short 2nd and 5th mesophalanges with clinodactyly of the | Similar to the right hand | Normal | Normal |
| 5th DIP joint | ||||
| VII:19 | The 2nd mesophalanx slightly short compared to the 4th | Similar to the right hand | Normal | Normal |
| mesophalanx. Radiographs in addition showed a short 3rd | ||||
| mesophalanx and short 1st distal phalanx bilaterally | ||||
| VII:20 | The 2nd mesophalanx slightly short compared to the 4th | Similar to the right hand | Normal | Normal |
| mesophalanx | ||||
| VII:22 | Short 2nd mesophalanx and clinodactyly of the | Short 2nd mesophalanx | Broad | Broad |
| 5th DIP joint | ||||
| VIII:5 | Short 2nd mesophalanx | Similar to the right hand | Normal | 2nd |
| mesophalanx | ||||
| hypoplastic | ||||
DIP, distal interphalangeal; MP, metacarpal phalangeal.
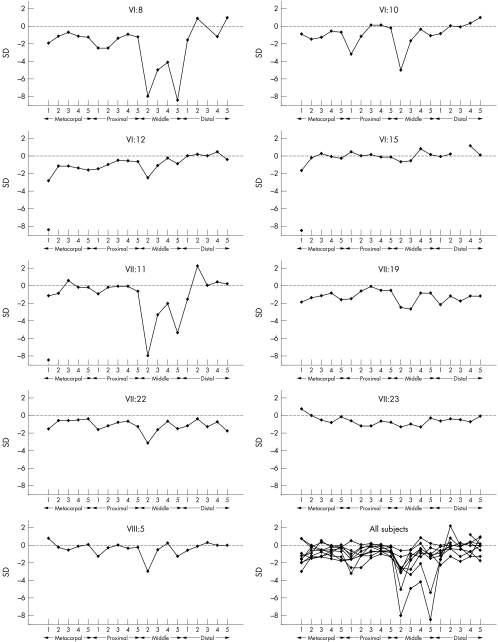
The MCPP demonstrated that the second mesophalanx was constantly (eight out of nine subjects) relatively shorter than mesophalanges 3, 4, and 5, and the fourth was relatively longer than the others (table 3, fig 3). The third mesophalanx was shorter than the fifth mesophalanx in half the cases. Another common finding was a relatively short first proximal phalanx. The characteristic MCPP was evident in VI:15 who otherwise had bones of normal length, suggesting that this analysis is the most certain method for determining carrier status. The other phenotypically normal mutation carrier (VII:23) had an uncharacteristic profile pattern (fig 3), demonstrating reduced penetrance.
Table 3 Measurements of hand bone length in nine mutation carriers, given by standard deviations (SD) from normal6 in 19 hand bones.
| ID | Sex | Age | Mc1 | Mc2 | Mc3 | Mc4 | Mc5 | Pp1 | Pp2 | Pp3 | Pp4 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| VI:8 | M | 76 | −1.93 | −1.24 | −0.79 | −1.17 | −1.33 | −2.63* | −2.59* | −1.35 | −1.09 |
| VI:10 | M | 76 | −0.90 | −1.5 | −1.32 | −0.60 | −0.67 | −3.16* | −1.23 | 0.19 | 0.22 |
| VI:12 | M | 67 | −2.97* | −1.24 | −1.32 | −1.46 | −1.67 | −1.58 | −1.23 | −0.58 | −0.65 |
| VI:15 | M | 65 | −1.59 | −0.18 | 0.26 | −0.03 | −0.33 | 0.53 | 0.14 | 0.19 | −0.22 |
| VII:11 | M | 57 | −1.24 | −0.97 | 0.53 | −0.31 | −0.33 | −1.05 | −0.32 | −0.19 | −0.22 |
| VII:19 | M | 30 | −1.93 | −1.5 | −1.32 | −0.89 | −1.67 | −1.58 | −0.77 | −0.19 | −0.65 |
| VII:22 | F | 35 | −1.62 | −0.67 | −0.65 | −0.57 | −0.53 | −1.7 | −1.30 | −0.87 | −0.75 |
| VII:23 | F | 35 | 0.69 | 0.02 | −0.65 | −0.86 | −0.25 | −0.7 | −1.30 | −1.30 | −0.75 |
| VIII:5 | F | 12 | 0.8 | −0.27 | −0.50 | −0.22 | 0.10 | −1.33 | −0.38 | 0.01 | −0.31 |
| Pp5 | Mp2 | Mp3 | Mp4 | Mp5 | Dp1 | Dp2 | Dp3 | Dp4 | Dp5 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| VI:8 | −1.15 | Miss* | −5.06* | −4.125* | −8.5* | −1.57 | 0.86 | nm | −1.25 | 1.00 | |
| VI:10 | −0.15 | −5.06* | −1.72 | −0.38 | −1.00 | −0.86 | 0.14 | −0.08 | 0.42 | 1.00 | |
| VI:12 | −0.65 | −2.56* | −1.17 | −0.38 | −1.00 | −0.14 | 0.14 | −0.08 | 0.42 | −0.54 | |
| VI:15 | −0.15 | −0.69 | −0.61 | 0.88 | 0.25 | −0.14 | 0.20 | nm | 1.25 | 0.23 | |
| VII:11 | −0.65 | Miss* | −3.39* | −2.25* | −5.38* | −1.57 | 2.29* | −0.08 | 0.42 | 0.23 | |
| VII:19 | −0.65 | −2.56* | −2.83* | −1.00 | −1.00 | −2.29* | −1.29 | −1.75 | −1.25 | −1.31 | |
| VII:22 | −1.32 | −3.25* | −1.71 | −0.82 | −1.59 | −1.31 | −0.46 | −1.31 | −0.77 | −1.83 | |
| VII:23 | −0.79 | −1.38 | −1.12 | −1.41 | −0.41 | −0.69 | −0.46 | −0.54 | −0.77 | −0.17 | |
| VIII:5 | −0.23 | −3.05* | −0.63 | 0.17 | −1.29 | −0.53 | −0.13 | 0.29 | −0.07 | 0.00 |
Dp, distal phalanx; Mc, Metacarpal; Mp, mesophalanx; Pp, proximal phalanx; nm, not measurable.
*Left hands, >2 SD.
Figure 3 Metacarpal‐phalangeal profile analysis in nine mutation carriers.
Structure of GDF5 and localisation of the mutation
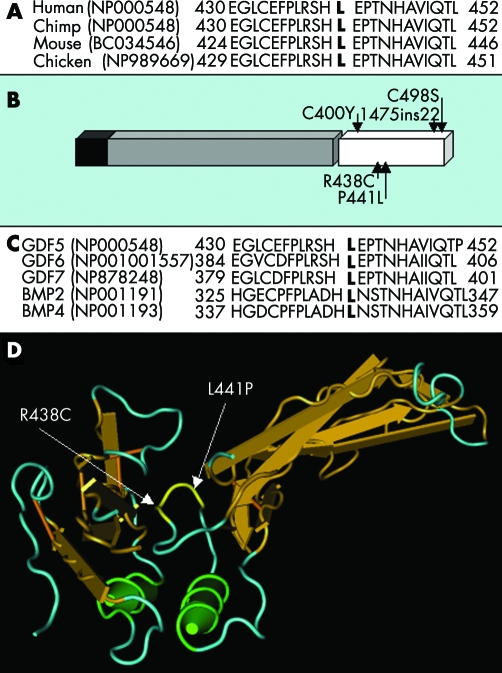
The amino acid L441 in GDF5 is conserved in human, chimp, mouse, and chicken (fig 4A) and resides in the active signalling domain of GDF5 (fig 4B). Furthermore, a leucine corresponding to amino acid residue 441 is conserved in the paralogous proteins GDF−5/−6/−7 and BMP−2/−4 (fig 4C), which suggests a crucial function for this residue. The corresponding leucine in Bmp2 is located in the receptor binding site as revealed by the crystal structure of Bmp2 bound to the Bmpr1a receptor (fig 4D). This suggests that the L441 residue in GDF5 is also located in the receptor binding site and may therefore interfere with the normal binding of BMP type 1 receptors. This was supported by the very recently reported crystal structure of GDF5.10
Figure 4 (A) Conservation of L441 in human, chimp, mouse, and chicken. (B) GDF5 monomer contains a signal peptide (black), a prodomain (grey), and an active signalling domain (white). The mutations reported in the active signalling domain are shown. (C) Conservation of the mutated proline residue in human GDF−5/−6/−7 and BMP−2/−4. (D) The mutations R438C and L441P superimposed on the crystal structure of Bmp2 bound to its receptor. Both mutations are located in the receptor binding site.
Discussion
The reported family is the original Mohr‐Wriedt family
Brachydactyly type A2 was first described by two pioneers in human genetics, OL Mohr and C Wriedt, in a five‐generation Norwegian family of Danish descent.3 We searched for their original material in the archives of Ullevaal Hospital, Oslo, and found an original letter sent by Mohr to Wriedt during their field study. A copy of a family book containing important phenotypic and genealogical notes was also found. We were then able to update the pedigree and find the living family members (fig 1A). Of the finally included individuals, V:4 and V:5 had been examined by Mohr and Wriedt when they were children.
The identification of the same mutation in the Norwegian and Danish families suggests a founder effect. This is supported by the fact that the father of I:2 was a salesman from the city of Aarhus in Denmark. The single Danish affected person was living in this area, and his family had been living there for generations. His father was reported to have short index fingers and to have in possession a book with the pedigree of the family. However, he refused to participate in the study.
GDF5 is the second BDA2 causing gene and a third locus exists
We recently reported mutations in BMPR1B in two German BDA2 families.4 However, a third family did not map to this locus suggesting genetic heterogeneity. The Danish/Norwegian family presented here did not have any mutation in BMPR1B either. Instead, our data showed linkage to the region containing the GDF5 gene, and subsequent sequencing demonstrated heterozygosity for a mutation c.1322T>C causing the substitution of leucine 441 with proline. The third family from our previous study was also tested but did not show linkage to the GDF5 region, suggesting that a third BDA2 locus exists.
The family represents a specific Mohr‐Wriedt subtype of BDA2
We found that the second mesophalanx was the only constantly shortened bone, confirming the BDA2 phenotype. However, several findings distinguish the phenotype of the reported family from that of the families with BMPR1B mutations. The length of the second mesophalanx was apparently normal in three of 22 mutation carriers, and radiographs showed that in two of nine cases the length was within normal range. Furthermore, medial deviation of the index finger was only observed once. In the two previously reported BDA2 families caused by BMPR1B mutations, all affected individuals had medially deviated or shortened index fingers.4 Furthermore, we observed shortened mesophalanges in fingers 3, 4, and 5 in a limited number of patients, and the MCPP demonstrated a relatively short mesophalanx 2, a relatively long mesophalanx 4, and a relatively short first proximal phalanx. These findings suggest that the original Mohr‐Wriedt family represents a distinct subtype of BDA2.
Mohr‐Wriedt type BDA2 is different from BDC
The relatively “spared” fourth mesophalanx also occurs in another condition caused by GDF5 mutations, brachydactyly type C (BDC). BDC mainly affects the middle phalanges of the second, third, and fifth fingers and the first metacarpal bone. However, the constantly more severe involvement of the second mesophalanx is a distinct feature of BDA2, whereas no specific mesophalanx could be identified as most severely affected in BDC.11 The difference is further demonstrated by the limited number of cases with affected third or fifth mesophalanges, short first metacarpal, or ulnar deviation of the index finger, and the absence of hypersegmentation or other defects commonly seen in BDC.
Three heterozygous BDC causing missense mutations have been reported in the mature domain of GDF5 (C400Y, R438C, C498S). They all removed or introduced a cysteine residue important for the normal folding of the protein, resulting in early degradation, which suggests that the molecular cause of BDC is haploinsufficiency.5,12 The phenotypic difference between BDC and BDA2 makes it highly unlikely that the BDA2 phenotype described here is caused by simple haploinsufficiency for GDF5.
The L441P mutation resides in the BMP receptor binding domain of GDF5 and likely impairs normal activation of BMPR1B
GDF5 belongs to the TGFbeta superfamily and is most closely related to GDF6/7 and BMP2/4, which can all bind BMP type 1 receptors. The conservation of L441 in all these molecules suggests it has an important biological function. The crystal structure of Bmp2 bound to Bmpr1a is known: the BMP2 residue corresponding to L441 of GDF5 (fig 4C) resides in the domain forming a pocket, which allows proper contact with BMP type 1 receptors (fig 4D). The very recently reported crystal structure of GDF5 confirmed that the homologue residue in GDF5 is also located in the binding site of BMP type 1 receptors. Binding affinity for the Bmpr1a receptor can be altered by targeted mutagenesis in the receptor binding site of its ligands.13 In contrast to the severe mutations reported previously, the substitution of a leucine with a proline within this site is expected to have a mild conformational effect as both are non‐polar amino acids. Gdf5 binds with high affinity to Bmpr1b (but not Bmpr1a),14,15 and dominant negative mutations in BMPR1B were previously shown to cause BDA2. Thus, although unproved, it is very likely that the L441P mutation impairs the normal activation pattern of BMPR1B.
Recessive conditions caused by GDF5 mutations
Two recessive conditions, duPan syndrome and the Hunter‐Thompson type of chondrodysplasia (HTC), were reported to be caused by mutations in the mature domain of GDF5. Homozygosity for the L441P mutation which we found in our BDA2 family, caused duPan syndrome with aplasia of fibula and severe acromesomelic limb shortening with small, non‐functional toes,16,17 and a homozygous frameshift mutation (1475ins22) caused HTC.18 Heterozygous L441P carriers in the duPan syndrome families were described as normal, but our study suggests that they may have BDA2, and heterozygous 1475ins22 carriers have not been described. Interestingly, duPan syndrome represents a milder phenotype than HTC, which is in accordance with the prediction that the L441P mutation only alters the protein to a very limited extent, whereas the 1475ins22 mutation is suspected to dramatically alter the normal properties of the mature protein.
In conclusion, we have identified GDF5 as a novel BDA2 causing gene in the original BDA2 family reported by Mohr and Wriedt, and we have found evidence for the existence of a third BDA2 locus. The constantly shortened index finger and occasional involvement of other fingers, and the relatively spared fourth finger define a specific Mohr‐Wriedt subtype of BDA2. The mutation is suggested to affect the binding affinity of GDF5 to BMP type 1 receptors, and thereby possibly impair the normal activity of the BMP receptor 1B.
Acknowledgements
The Wilhelm Johannsen Centre for Functional Genome Research was established by the Danish National Research Foundation.
Abbreviations
BD - brachydactyly
BMP - bone morphogenetic protein
HTC - Hunter‐Thompson type of chondrodysplasia
MCPP - metacarpo‐phalangeal profile
PCR - polymerase chain reaction
Footnotes
Klaus W Kjaer was supported by a grant from the IMK Almene Fond
Competing interests: none declared
Consent has been given for the publication of patient details
References
- 1.Bell J. In: Penrose LS, ed. Treasury of human inheritance. Vol 5. London: Cambridge University Press, 19511–31.
- 2.Gao B, Guo J, She C, Shu A, Yang M, Tan Z, Yang X, Guo S, Feng G, He L. Mutations in IHH, encoding Indian hedgehog, cause brachydactyly type A‐1. Nat Genet 200128386–388. [DOI] [PubMed] [Google Scholar]
- 3.Mohr O L, Wriedt C.A new type of hereditary brachyphalangy in man. Washington, DC: Institution of Washington, 1919
- 4.Lehmann K, Seemann P, Stricker S, Sammar M, Meyer B, Suring K, Majewski F, Tinschert S, Grzeschik K H, Muller D, Knaus P, Nurnberg P, Mundlos S. Mutations in bone morphogenetic protein receptor 1B cause brachydactyly type A2. Proc Natl Acad Sci U S A 200310012277–12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polinkovsky A, Robin N H, Thomas J T, Irons M, Lynn A, Goodman F R, Reardon W, Kant S G, Brunner H G, van der Burgt I, Chitayat D, McGaughran J, Donnai D, Luyten F P, Warman M L. Mutations in CDMP1 cause autosomal dominant brachydactyly type C. Nat Genet 19971718–19. [DOI] [PubMed] [Google Scholar]
- 6.Garn S M, Hertzog K P, Poznanski A K, Nagy J M. Metacarpophalangeal length in the evaluation of skeletal malformation. Radiology 1972105375–381. [DOI] [PubMed] [Google Scholar]
- 7.Poznanski A K.The hand in radiologic diagnosis with gamuts and pattern profiles. Philadelphia: WB Saunders, 198446–54.
- 8.Ott J. A computer programme for linkage analysis of general human pedigree. Am J Hum Genet 197328528–529. [PMC free article] [PubMed] [Google Scholar]
- 9.Schäffer A A, Gupta S K, Shriram K, Cottingham R W. Avoiding recomputation in linkage analysis. Hum Hered 199444225–237. [DOI] [PubMed] [Google Scholar]
- 10.Schreuder H, Liesum A, Pohl J, Kruse M, Koyama M. Crystal structure of recombinant human growth and differentiation factor 5: evidence for interaction of the type I and type II receptor‐binding sites. Biochem Biophys Res Commun 20053291076–1086. [DOI] [PubMed] [Google Scholar]
- 11.Galjaard R J, van der Ham L I, Posch N A, Dijkstra P F, Oostra B A, Hovius S E, Timmenga E J, Sonneveld G J, Hoogeboom A J, Heutink P. Differences in complexity of isolated brachydactyly type C cannot be attributed to locus heterogeneity alone. Am J Med Genet 200198256–262. [DOI] [PubMed] [Google Scholar]
- 12.Everman D B, Bartels C F, Yang Y, Yanamandra N, Goodman F R, Mendoza‐Londono J R, Savarirayan R, White S M, Graham J M, Jr, Gale R P, Svarch E, Newman W G, Kleckers A R, Francomano C A, Govindaiah V, Singh L, Morrison S, Thomas J T, Warman M L. The mutational spectrum of brachydactyly type C. Am J Med Genet 2002112291–296. [DOI] [PubMed] [Google Scholar]
- 13.Keller S, Nickel J, Zhang J L, Sebald W, Mueller T D. Molecular recognition of BMP‐2 and BMP receptor IA. Nat Struct Mol Biol 200411481–488. [DOI] [PubMed] [Google Scholar]
- 14.Storm E E, Huynh T V, Copeland N G, Jenkins N A, Kingsley D M, Lee S J. Limb alterations in brachypodism mice due to mutations in a new member of the TGF beta‐superfamily. Nature 1994368639–643. [DOI] [PubMed] [Google Scholar]
- 15.Nishitoh H, Ichijo H, Kimura M, Matsumoto T, Makishima F, Yamaguchi A, Yamashita H, Enomoto S, Miyazono K. Identification of type I and type II serine/threonine kinase receptors for growth/differentiation factor‐5. J Biol Chem 199627121345–21352. [DOI] [PubMed] [Google Scholar]
- 16.Ahmad M, Abbas H, Wahab A, Haque S. Fibular hypoplasia and complex brachydactyly (Du Pan syndrome) in an inbred Pakistani kindred. Am J Med Genet 199036292–296. [DOI] [PubMed] [Google Scholar]
- 17.Faiyaz‐Ul‐Haque M, Ahmad W, Zaidi S H, Haque S, Teebi A S, Ahmad M, Cohn D H, Tsui L C. Mutation in the cartilage‐derived morphogenetic protein‐1 (CDMP1) gene in a kindred affected with fibular hypoplasia and complex brachydactyly (DuPan syndrome). Clin Genet 200261454–458. [DOI] [PubMed] [Google Scholar]
- 18.Thomas J T, Lin K, Nandedkar M, Camargo M, Cervenka J, Luyten F P. A human chondrodysplasia due to a mutation in a TGF‐beta superfamily member. Nat Genet 199612315–317. [DOI] [PubMed] [Google Scholar]