Abstract
Objective
To develop and compare two new technologies for diagnosing a contiguous gene syndrome, the Williams‐Beuren syndrome (WBS).
Methods
The first proposed method, named paralogous sequence quantification (PSQ), is based on the use of paralogous sequences located on different chromosomes and quantification of specific mismatches present at these loci using pyrosequencing technology. The second exploits quantitative real time polymerase chain reaction (QPCR) to assess the relative quantity of an analysed locus.
Results
A correct and unambiguous diagnosis was obtained for 100% of the analysed samples with either technique (n = 165 and n = 155, respectively). These methods allowed the identification of two patients with atypical deletions in a cohort of 182 WBS patients. Both patients presented with mild facial anomalies, mild mental retardation with impaired visuospatial cognition, supravalvar aortic stenosis, and normal growth indices. These observations are consistent with the involvement of GTF2IRD1 or GTF2I in some of the WBS facial features.
Conclusions
Both PSQ and QPCR are robust, easy to interpret, and simple to set up. They represent a competitive alternative for the diagnosis of segmental aneuploidies in clinical laboratories. They have advantages over fluorescence in situ hybridisation or microsatellites/SNP genotyping for detecting short segmental aneuploidies as the former is costly and labour intensive while the latter depends on the informativeness of the polymorphisms.
Keywords: Williams‐Beuren syndrome, aneuploidy, genomics, real time PCR, pyrosequencing
Increasing numbers of human diseases are recognised to result from recurrent DNA rearrangements involving unstable genomic regions. Both interchromosomal and intrachromosomal rearrangements are facilitated by the presence of region specific low copy repeats (LCRs) and result from non‐allelic homologous recombination. These diseases are traditionally diagnosed by fluorescence in situ hybridisation (FISH)1 or microsatellite/single nucleotide polymorphism (SNP) genotyping, or both.2 FISH has the disadvantages of being labour intensive and expensive, whereas microsatellite/SNP genotyping depends on the relative informativeness of the polymorphisms used and on parental DNA to ascertain the deletion.
The completion of the human genome sequence3,4,5 has provided an extensive catalogue of sequence features that can be exploited for the design of new diagnostic strategies. Here we propose and validate two recently described polymerase chain reaction (PCR) based diagnostic approaches to identify contiguous gene syndromes. We compare their efficiency and practicality for detecting the 7q11.23 microdeletion in patients with Williams–Beuren syndrome (WBS).6 WBS has a prevalence estimated between 1/7500 and 1/20 000.7,8,9 Patients are characterised by mental retardation with unique cognitive and personality profile, distinctive facial features, supravalvar aortic stenosis, short stature, connective tissue anomalies, hypertension, infantile hypercalcaemia, dental and kidney abnormalities, premature aging of the skin, impaired glucose tolerance, and silent diabetes.10,11,12
The first method is based on the use of paralogous sequences located on different chromosomes. Because these sequences are under different selective constraints, they accumulate nucleotide substitutions in a locus specific manner. Thus each locus will contain specific internal sequence differences (paralogous sequence mismatches or PSMs), which can be exploited to estimate their relative quantity. Technically, we co‐amplify paralogous fragments of identical size that map to different autosomes and quantify the PSMs to detect the presence of chromosome number abnormalities. This technique—named paralogous sequence quantification (PSQ)—was recently successfully implemented to detect autosomal trisomies as well as numerical abnormalities of the sex chromosomes.13
The second method, real time quantitative polymerase chain reaction (QPCR), allows detection of a PCR product as it accumulates during the reaction.14,15 The higher the copy number of a target sequence, the earlier a significant increase in fluorescence will be detected. We can thus estimate the relative quantity of an analysed locus. By designing multiple assays within and next to a segmental aneuploidy, we should be able to map exactly the extension of an indel. We report the implementation of these technologies to diagnose WBS and to map the breakpoints of atypical rearrangements.
Methods
Samples
Samples were collected by the CSS‐Mendel Institute, Rome (140 Italian WBS patients and 43 Italian controls), the Aghia Sofia Children's Hospital, Athens (31 Greek WBS patients) and the Department of Genetic Medicine and Development, University of Geneva, Geneva (34 French controls from the Departments of Savoie and Haute‐Savoie, 11 WBS patients and 15 controls from Western Switzerland), with appropriate informed consent obtained by the physician in charge and approval of the local ethics committee. Genomic DNA was prepared with either the PUREGENE whole blood kit (Gentra Systems, Minneapolis, Minnesota, USA) or the QIAamp kit (Qiagen, Valencia, California, USA).
Paralogous sequence quantification and pyrosequencing
Paralogous sequence quantification and pyrosequencing were carried out as described.13 The forward and reverse primers and the internal sequencing primers were designed using Oligo 6.55 (Molecular Biology Insights, Cascade, Colorado, USA) and the manufacturer's site (http://www.pyrosequencing.com/index.html), respectively. Reverse and sequencing primers were desalted, while the biotinylated forward primers were desalted and purified by high performance liquid chromatography (HPLC) (Sigma‐Genosys, St Louis, Missouri, USA). PCR reactions with the selected primer pairs (supplementary table S1, which can be viewed on the journal website: http://www.jmedgenet.com/supplemental) were set up in a total volume of 25 μl containing 30 ng of genomic DNA, 400 μM of each primer and 200 μM dNTPs. We used 1.25 units of a standard Taq polymerase in the manufacturer provided buffer (Amersham Biosciences), or alternatively the ready‐made 2×Jumpstart PCR mastermix (Sigma‐Genosys) with varying levels of MgCl2 and DMSO depending on the assay (table S1). Three negative controls per plate were added and the PCR products were checked on agarose gel to ensure lack of PCR contamination and correct amplimer size. PCR products were purified and annealed to an internal sequencing primer close to the PSM site to be quantified (table S1; figs S1 and S2, http://www.jmedgenet.com/supplemental). The purification and pyrosequencing steps were carried out following the manufacturer's instructions (Biotage Inc, Charlottesville, Virginia, USA). The pyrosequencing software directly emits a quantitative value for the proportion of each PSM present in the PCR product (fig S3, http://www.jmedgenet.com/supplemental). We used the relative abundance of the PSM of the “query” chromosome as our statistic for all calculations. The methods used to determine the PSM relative abundance that could be confidently diagnosed (with 99% confidence interval), to determine the sensitivity and specificity of each assay, and to combine the assays have been described previously.13
Real time quantitative PCR
Oligos were designed using the PrimerExpress program (Applied Biosystems, Foster City, California, USA) with default parameters in every case (table S2, http://www.jmedgenet.com/supplemental) and ordered from Sigma‐Genosys. Amplicon sequences were checked by both BLAST and BLAT against the human genome to ensure specificity. All reverse transcriptase (RT)‐PCR reactions were carried out in two replicates in 9 μl final volume with concentrations of 0.96 ng/μl DNA, 0.42 μM of each primer, and 1×SybrGreen mastermix (Applied Biosystems). Reactions were set up in a 384 well plate format with a Biomek 2000 (Beckman) and run in an ABI Prism7900HT (Applied Biosystems) with the following amplification conditions: 50°C for two minutes, 95°C for 10 minutes, and 50 cycles of 95°C for 15 seconds/60°C for one minute. Raw Ct values were obtained using SDS2.0 (Applied Biosystems). We implemented the geNorm method16 to measure relative quantities. Four normalisation assays mapping to HSA21 (table S2, http://www.jmedgenet.com/supplemental) and four normalisation DNAs were systematically included in each run.
Results
Paralogous sequence quantification
Paralogous sequences were identified by comparing 10 kb long fragments covering the WBS deletion17 with the human genome sequence using both BLAT (http://genome.ucsc.edu/) and BLAST (http://www.ensembl.org/Multi/blastview). We identified two pairs of suitable paralogous sequences with one copy to chromosome 7q11.23 and only a single second copy elsewhere in the genome (figs S1 and S2, http://www.jmedgenet.com/supplemental). We aligned all matching sequences and built a consensus sequence to design three discriminating pyrosequencing assays (assay BAZ1B and assays CYLN2 and WBSCR3.4M, respectively) for WBS deletion detection. Each assay was tested with various PCR conditions on 10 control and 10 WBS samples. From this analysis we selected the conditions based on the following criteria: (1) the PSM quantification in control individuals should be close to 50%/50%, indicating that both “loci” amplify with equal efficiency; and (2) there should be a clear, non‐overlapping discrimination between control and WBS samples. For the CYLN2 and WBSCR3.4M assays we found conditions that showed PSM relative abundances close to the theoretical values of 50%/50% for control individuals and 66%/33% for patients (table S1, http://www.jmedgenet.com/supplemental). In contrast for the BAZ1B assay, and despite testing multiple triads of amplification and sequencing primers (data not shown), we were unable to identify conditions that produced PSM relative abundances close to the theoretical values.
To validate the method on a larger cohort, we tested 128 WBS patient, 16 probable WBS patients (see below), and 37 control samples collected by the CSS‐Mendel Institute. Typical results and distribution of PSM relative abundance of normal and affected samples for each assay are shown in fig S3 (http://www.jmedgenet.com/supplemental). We use the relative abundance of the PSM from the “query” chromosome—that is, the chromosome we want to quantify, here HSA7—as our statistic for all calculations. For both the CYLN2 and the WBSCR3.4M assay, the distribution of the “query” chromosome PSM relative abundance obtained for both cohorts show normal distributions, with medians close to the expected results of 33% for WBS patients (CYLN2 relative abundance: mean (SD), 33 (2)%; WBSCR3.4M: 37 (2)%) and 50% for control individuals (CYLN2 relative abundance: 47 (2)%; WBSCR3.4M: 50 (2)%) (table 1). The third assay recorded relative abundances of the “query” chromosome PSM under the expected value for both the control and patient samples (mean (SD), 41 (6)%, range 39 to 47, and 27 (5)%, range 12 to 44, respectively) and 3% of misdiagnoses (n = 160). It was therefore not retained further.
Table 1 Specificity and sensitivity of paralogous sequence quantification assays.
| Assay | CYLN2 | WBSCR3.4M | Combined | |||
|---|---|---|---|---|---|---|
| Mean control | 46.7 | 50.3 | 50.0 | |||
| SD of control | 1.9 | 2.2 | 1.6 | |||
| Mean WBS | 32.9 | 37.3 | 36.1 | |||
| SD of WBS | 2.2 | 1.7 | 1.6 | |||
| Number of samples | 163 | 160 | 160 | |||
| Number of uncertain samples | 6 | 4 | 3 | |||
| Sensitivity | 0.95 | 0.97 | 0.98 | |||
| Specificity | 1.00 | 1.00 | 1.00 |
WBS, Williams‐Beuren syndrome samples.
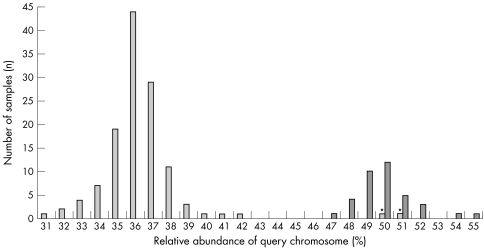
Both retained assays were characterised by no false negative or false positive calls, but some samples fell outside of the 99% confidence interval and could therefore not be unambiguously diagnosed. They were treated either as false positives or as false negatives according to the known FISH result to estimate sensitivity and specificity (table 1). Combining both assays resulted in a clearer separation between the control and the patient samples (fig 1) and a significant improvement in the sensitivity (table 1).
Figure 1 Diagnosing Williams‐Beuren syndrome (WBS) by paralogous sequence quantification. Distribution of the relative abundances of the query chromosome upon combination of the CYLN2 and the WBSCR3.4M assays. Control samples are in dark grey and WBS patients in light grey. The positions of samples corresponding to the two patients carrying an atypical deletion (CO3 and IM3)—that is, euploid for the CYLN2 and WBSCR3.4M assays—are indicated by asterisks.
Six subjects of the “probable WBS” cohort, who were clinically diagnosed as WBS but were not analysed by FISH or Southern blotting, showed “query” chromosome relative abundance that classified them in the 99% confidence interval of the control cohort with all three assays (not shown). Consistently, for two of these patients there was only a suspicion of WBS, while a third showed facial features, but no other characteristic phenotypes that can be attributed to the disease (see below).
Quantitative real time PCR
A quantitative real time PCR (QPCR) method has recently been successfully implemented to diagnose aneuploid mice.18 To assess if the technology could be used to diagnose human segmental aneuploidies and to evaluate its qualities/disadvantages v the PSQ technology, we applied this second high throughput technique to diagnose the WBS segmental aneuploidy. Implementation of this second procedure should allow clarification of ambiguous cases and atypical deletions and, eventually, exact mapping of the breakpoints of unusual rearrangements.
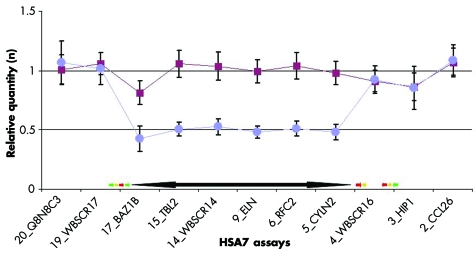
We designed 20 QPCR assays spread from HSA7 base pair (bp) 70012994 to bp 75057642 (table S2, http://www.jmedgenet.com/supplemental) and preliminarily tested them on DNA samples from four controls and three WBS patients. A set of 11 assays was retained and used to analyse 94 WBS patients, 16 probable WBS, and 61 controls, collected by the CSS‐Mendel Institute (55 WBS, 16 probable WBS, 14 controls), the Department of Genetic Medicine, Geneva (8 patients, 47 controls), and the Aghia Sofia Children's Hospital, Athens (31 patients). These assays map either centromerically (two assays), inside (six assays), or telomerically (two assays) to the commonly deleted region. A further assay, named 4_WBSCR16, maps between the two blocks of repeats that flank the telomeric side of the WBS‐critical region inside the WBSCR16 gene.19 Mean results are presented in fig 2 and typical examples in fig S4 (http://www.jmedgenet.com/supplemental). These assays show mean results close to the expected value of 1 for controls. Consistently, the mean results obtained with the patients samples are close to 1 for assays outside the common deletion and to 0.5 for assays inside the deletion (table 2). There were no false negative or false positive calls in any of the six diagnostic assays, but a few samples (0.7–4.6%) fell outside the 99% confidence intervals for one (12 samples), three (two samples), or even four assays (one sample; table 2). However, even these samples can be unambiguously diagnosed upon combination of the six assays (pairwise t test: all individual p values <0.0016, n = 153, atypical deletions not included; see table 2). To further validate this method, we carried out a blind test on 24 DNA samples collected by the Department of Genetic Medicine, Geneva and previously analysed by FISH. We confirmed the clinical diagnosis of WBS (11 cases) and normal samples (15 cases) for the 7q11.23 region in all cases (all individual p values <0.0009).
Figure 2 Diagnosing Williams‐Beuren syndrome (WBS) by real time quantitative polymerase chain reaction (QPCR). Means and standard deviations of real time quantitative PCR results obtained for 11 assays from the 7q11.23 region with 61 control (red squares) and 92 WBS patient (blue circles) DNA samples. The extension of the classical deletion observed in WBS (double head black arrow) and the location of the low copy repeats (LCRs; green arrow, BLOCK C; yellow arrow, BLOCK A; red arrow, BLOCK B20) are indicated.
Table 2 Specificity and sensitivity of real time quantitative polymerase chain reaction assays.
| Assay | 20_Q8N8C3 | 19_WBSCR17 | 17_BAZ1B | 15_TBL2 | 14_WBSCR14 | 9_ELN | 6_RFC2 | 5_CYLN2 | 4_WBSCR16 | 3_HIP1 | 2_CCL26 | 6 Dx assays combined | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean control | 1.01 | 1.06 | 0.81 | 1.06 | 1.04 | 0.99 | 1.04 | 0.98 | 0.91 | 0.86 | 1.07 | 0.99 | ||||||||||||
| SD of control | 0.12 | 0.09 | 0.10 | 0.11 | 0.12 | 0.10 | 0.11 | 0.10 | 0.09 | 0.12 | 0.12 | 0.13 | ||||||||||||
| Mean WBS | 1.07 | 1.02 | 0.43 | 0.51 | 0.53 | 0.48 | 0.51 | 0.48 | 0.92 | 0.86 | 1.09 | 0.49 | ||||||||||||
| SD of WBS | 0.18 | 0.13 | 0.11 | 0.06 | 0.07 | 0.05 | 0.06 | 0.06 | 0.12 | 0.18 | 0.13 | 0.08 | ||||||||||||
| Number of samples | 153 | 153 | 152 | 153 | 153 | 152 | 152 | 152 | 153 | 153 | 153 | 153 | ||||||||||||
| Number of uncertain samples | 2 | 2 | 1 | 7 | 4 | 6 | 0 | |||||||||||||||||
| Sensitivity | 0.98 | 0.99 | 0.99 | 0.96 | 0.97 | 0.96 | 1.00 | |||||||||||||||||
| Specificity | 1.00 | 0.98 | 1.00 | 0.96 | 0.98 | 0.97 | 1.00 |
WBS, Williams‐Beuren syndrome samples.
The six patients of the “probable WBS” group, with PSM relative abundances that place them in the control group (see above), showed no evidence of deletion in the 7q11.23 region with the QPCR method either (combination of six diagnostic assays, pairwise t test: all individual p values <0.006). Thus the results obtained with the two independent methods (PSQ and QPCR) are consistent with the interpretation that these six patients were clinically misdiagnosed. As no FISH or Southern blot results were available for these, we considered that they were clinically misdiagnosed and reclassified them as non‐WBS. However, we cannot exclude the possibility that these individuals carry an inversion or a subtle deletion that maps between the assays we used (maximum size 415 kb between WBSCR14 and ELN).
Atypical deletions
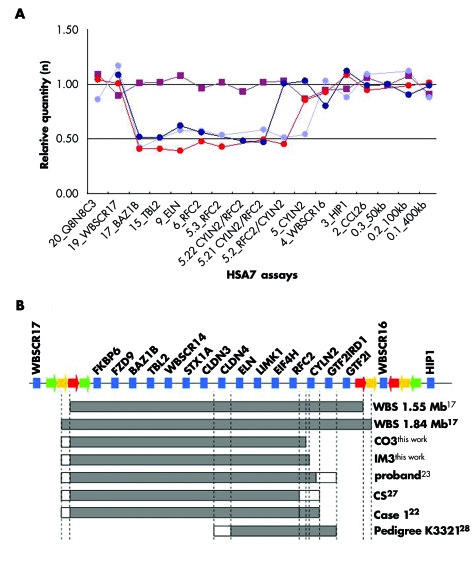
Two patients, CO3 and IM3, presented an atypical deletion on the telomeric side as measured by both PSQ and QPCR. Their DNAs contained one copy of the BAZ1B (diagnosed with PSQ and QPCR), TBL2 (QPCR), ELN (QPCR), and RFC2 (QPCR) loci, but were unaffected in the WBSCR3.4M (PSQ) and CYLN2 (PSQ and QPCR) assays (figs 1 and 3). To further narrow down the breakpoints, we designed more QPCR assays between RFC2 and CYLN2 (table S2, http://www.jmedgenet.com/supplemental). These additional assays allowed mapping of the CO3 and IM3 breakpoints to 13 011 and 66 180 bp long intervals, respectively. Thus these patients present atypical deletions smaller by more than 400 kb, when compared to the extent of the classical WBS deletion (fig 3).17 The breakpoints of IM3 and CO3 map in the region between the RFC2 and CYLN2 genes. All the WBSCR genes centromerically positioned from this point, starting with the RFC2 gene, are deleted, while the two telomeric genes, GTF2IRD1 and GTF2I, are preserved. The situation is more ambiguous for the CYLN2 gene: 12.3 to 25.3 kb separate the mapped CO3 breakpoint from the first known exon of CYLN2. Similarly, the IM3 breakpoint was narrowed down to a region that encompasses the promoter and the three first exons of that gene. Thus both cases display genomic organisations that might impair the normal expression of CYLN2.
Figure 3 Atypical deletions. (A) Typical real time quantitative polymerase chain reaction (QPCR) results obtained for samples from a control (red squares) and a typical WBS patient (light blue circles) besides the samples of atypical patients CO3 (dark blue circles) and IM3 (red circles). (B) Schematic partial transcript map of the WBS critical region. Single copy transcripts mapping to the WBS critical region are represented by blue boxes, while duplicons within the low‐copy repeats are depicted by coloured arrows (BLOCK A: yellow; BLOCK B: red; BLOCK C: green20). The names of the hemizygous and normal copy genes are printed diagonally and vertically, respectively. The extent of the deletion observed in classical WBS (short and long deletions), as well as in the atypical patients discussed in this paper, are denoted by grey boxes below the transcript map. Unclear deletion statuses are represented by white boxes. Patient symbols and references are indicated on the right. WBS, Williams‐Beuren syndrome.
Patient CO3, a male, was born by vaginal delivery after an uneventful pregnancy of 32 weeks. Birth weight was 1850 g (50th–75th centile), length 43 cm (50th centile), and head circumference 29 cm (25th centile). A congenital heart defect was diagnosed at five months of age and echocardiography showed supravalvar aortic stenosis. Patient IM3, a female, was born by vaginal delivery at term of an uneventful pregnancy. Birth weight was 3100 g, length 49 cm, and head circumference 34 cm. A congenital heart defect was diagnosed shortly after birth. Echocardiography showed supravalvar aortic and peripheral pulmonary artery stenoses which were surgically corrected at five years. Both patients had motor developmental milestones in the normal range, though mild cognitive deficit was evident at four years in CO3. Both CO3 and IM3 had difficulties with non‐linguistic and visuo‐motor abilities, overfriendliness, and preservation of linguistic abilities. Wechsler intelligence scale testing performed at 15 and 10 years of age showed mild mental retardation with IQs of 72 (verbal, 74; performance, 68) and 71 (verbal, 73; performance, 66), respectively. Clinical examination showed mild facial anomalies, including periorbital fullness (mild in CO3) and thick lower lip, normal size and number of teeth, and blood glucose levels in the average range.
At the time of writing, CO3 was 30 years old; his weight was 77 kg (90th to 97th centile), height 168 cm (10th centile), head circumference 53 cm (3). His irises were normal and no signs of premature aging were noticeable. Ophthalmological and audiological evaluations were normal. Renal ultrasonography showed no malformations. Measurements of blood pressure at rest and during 24 hour ambulatory monitoring were in the normal range.
The auxological variables in IM3 at 11 years of age were normal. Her weight was 38.5 kg (50th to 75th centile), height 140 cm (50th centile), head circumference 50 cm (3rd centile). She presented a hoarse voice, a stellate iris, and mild malar hypoplasia. Strabismus was operated on at seven years of age. Audiological evaluation and renal ultrasonography were normal. Measurements of blood pressure at rest and during 24 hour ambulatory monitoring showed mild systolic hypertension.
Discussion
We report the setup and validation of two recently described methods for diagnosing the recurrent DNA deletion present in WBS patients.6,17 The first technique, paralogous sequence quantification (PSQ), is based on the quantification of two paralogous sequences by pyrosequencing.13,21 PSQ allowed us to correctly segregate 161 of the 163 tested samples with no false positives or false positive calls. The diagnosis of the remaining two samples was inconclusive as they fell outside of the confidence interval of the combined assay (table 1). We conclude that the PSQ technology is a reliable, effective, and inexpensive tool for diagnosing segmental aneuploidies, but is not suitable for measuring the extent of the deletion in WBS because of the lack of exploitable paralogous sequences at the centromeric end of the deletion.
The second method takes advantage of quantitative real time PCR to evaluate the relative abundance of a DNA fragment. We designed a set of 11 assays mapping to the 7q11.23 region and tested them on 92 WBS patient and 61 control DNA samples. They allowed the correct segregation of the two analysed cohorts in all the tested genomes with 100% sensitivity and specificity (table 2).
The PSQ method is less expensive than the QPCR technology for diagnosing segmental aneuploidies. However, PSQ has the weakness of being dependent on the presence of suitable sequences—that is, sequences paralogous to sequences somewhere else on the genome. This is not a major problem when the goal is to diagnose aneuploidies of an entire chromosome or a large chromosomal region,13 but it becomes important in partial aneuploidies of small size (<5 Mb) such as WBS. In contrast, assays for the QPCR technology can be designed on any genomic fragment that is uniquely represented in the human genome. This allows a large number of assays to be designed along the region to be tested—for example, a segmental aneuploidy—decreasing the risk of false positive and false negative results and increasing the resolution. Moreover, atypical indels can then be characterised further by designing more assays close to the supposed breakpoints, as we did to characterise WBS patients who carry two copies of the CYLN2, GTF2IRD1, and GTF2I genes.
Genetic diagnostic laboratories attached to genetic counselling clinics routinely use FISH, microsatellite/SNP genotyping, and Southern blotting to diagnose segmental aneuploidies, including the WBS. FISH and Southern blotting have success rates reaching 99%, but they have the disadvantage of being costly and labour intensive. Microsatellite/SNP genotyping requires the analysis of multiple assays of both the proband and their parents to ensure the informativeness of the genotyped polymorphisms. Another drawback inherent in this method is the unwanted prospect of identifying non‐paternity. Advantages and disadvantages of these different methods for diagnosing segmental aneuploidies are summarised in table 3.
Table 3 Advantages and disadvantages of the multiple techniques used to diagnose segmental aneuploidies.
| Method | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| QPCR | PSQ | FISH | Microsatellites | Southern | ||||||
| Speed (days) | 1 | 1 | 5 (2)* | 1 | 6 | |||||
| Cost per assay (€) | 30 | 10 | 80 | 15/30/45†‡ | 80 | |||||
| Advantages | Great adaptability to uncommon rearrangement§ | Possibility of diagnosing inversion¶ | Possibility of diagnosing inversion | |||||||
| Disadvantages | Inversion impossible to diagnose | Relies on paralogous sequences; impossible to diagnose inversion | Labour intensive | Relative informativeness of SNPs; discovery of non‐paternity; impossible to diagnose inversion | Requires working with radioactivity**; labour intensive |
*A preliminary result can be given after two days.
†Price for QF‐PCR.
‡€45/assay includes both parents.
§See, for example, the mapping of breakpoints of atypical deletion patient detailed in this report.
¶By three colour FISH (see, for example, Bayes et al, 200317).
**Possibility of working with non‐radioactive digoxygenin, but with a diminished sensitivity.
FISH, fluorescence in situ hybridisation; PSQ, paralogous sequence quantification; QPCR, quantitative real time polymerase chain reaction; SNP, single nucleotide polymorphism.
We also report the detection of two patients who carry partial deletions that do not extend telomerically further than the RFC2 gene (fig 3). The status of the relative expression of the neighbouring CYLN2 gene remains uncertain, as we can hypothesise that the hemizygosity of the sequences preceding that gene might play an important role in the regulation of its expression. These atypical patients can provide important information for genotype and phenotype correlations. A few WBS individuals with atypical deletions, as well as patients with isolated SVAS and partial deletions, have been identified previously.22,23,24,25,26,27,28,29,30 In particular an individual was reported who carried an atypical deletion extending from the centromeric LCR to the CYLN2 or the GTF2IRD1 gene (fig 3C) and presented with an unusual cognitive profile, supravalvar aortic stenosis, milder facial features, short stature, and an IQ of 83, but not the specific visuospatial impairment of WBS patients.24 Likewise, Tassabehji et al28 describe a seven year old girl with a deletion from the centromeric LCR to RFC2 (status of CYLN2 unclear; fig 3B) with no dysmorphic features and verbal and performance scores slightly above full deletion WBS. In addition, a Japanese patient with a deletion spanning most of the WBS region with the exception of GTF2IRD1 and GTF2I had abilities to visualise at spatial and global levels that outperformed typical WBS cases, despite having a verbal IQ of only 6423 (fig 3B). Finally, the affected members of a kindred with supravalvar aortic stenosis but without typical Williams facies, and with visuo‐spatial abilities above that of full deletion WBS, were shown to carry a smaller deletion from ELN to GTF2IRD1, thus maintaining euploidy of GTF2I29 (fig 3B). These combined observations suggest that the GTF2I or GTF2IRD1 genes, or both, are likely to be linked to the characteristic WBS visual spatial deficits.23,29 The milder cognitive deficit evidenced in patients with atypical deletions from the present series is consistent with this hypothesis. However, the presence of some aspects of the characteristic WBS cognitive profile indicates that one should be cautious in drawing firm conclusions on genotype–phenotype correlations from a limited number of subjects all bearing different microdeletions (fig 3B). This is further emphasised by the identification of an unusually highly functioning patient with the classical deletion26 and by a family with a balanced translocation t(7;16)(q11.23;q13) within the elastin gene associated with variable expression of the WBS phenotype.31 Interestingly, both of our atypical patients and none of the previously described patients with similar partial deletions23,24,28,29 showed the facial features characteristic of WBS, suggesting that hemizygosity of genes mapping to the telomeric end of the common deletion might be involved. An additional aspect which can be putatively linked to hemizygosity of telomeric genes is premature aging, as this feature was absent in IM3 at the age of 30.
Phenotype–genotype correlations are one of the major challenges in the clinical human genetics of contiguous gene syndromes. The techniques used in this report, in particular QPCR, are suitable for the precise mapping of any aneuploidy, thus providing a reliable means of large scale diagnosis in uncommon conditions, with a rapid report time and at a reasonable cost.
The supplementary tables and figures are available on the journal website (http://www.jmedgenet.com/supplemental)
Acknowledgements
We thank P Descombes, M Docquier, M Gagnebin, C Ucla, and C Wyss for assistance and discussion. This work was supported by grants from the Jérôme Lejeune Foundation, Telethon Action Suisse, the Désirée and Niels Yde Foundation and the Swiss National Science Foundation to AR and from the Italian Minister of Health “Ricerca Corrente” 2005 to GM.
Abbreviations
FISH - fluorescence in situ hybridisation
LCR - low copy repeat
PSM - paralogous sequence mismatch
PSQ - paralogous sequence quantification
QPCR - quantitative real time polymerase chain reaction
SNP - single nucleotide polymorphism
WBS - Williams‐Beuren syndrome
Footnotes
Conflicts of interest: none declared
The supplementary tables and figures are available on the journal website (http://www.jmedgenet.com/supplemental)
References
- 1.Lowery M C, Morris C A, Ewart A, Brothman L J, Zhu X L, Leonard C O, Carey J C, Keating M, Brothman A R. pp. 49–53. [PMC free article] [PubMed]
- 2.Bonnet D, Cormier‐Daire V, Kachaner J, Szezepanski I, Souillard P, Sidi D, Munnich A, Lyonnet S. Microsatellite DNA markers detects 95% of chromosome 22q11 deletions. Am J Med Genet 199768182–184. [PubMed] [Google Scholar]
- 3.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov J P, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange‐Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin J C, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston R H, Wilson R K, Hillier L W, McPherson J D, Marra M A, Mardis E R, Fulton L A, Chinwalla A T, Pepin K H, Gish W R, Chissoe S L, Wendl M C, Delehaunty K D, Miner T L, Delehaunty A, Kramer J B, Cook L L, Fulton R S, Johnson D L, Minx P J, Clifton S W, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng J F, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs R A, Muzny D M, Scherer S E, Bouck J B, Sodergren E J, Worley K C, Rives C M, Gorrell J H, Metzker M L, Naylor S L, Kucherlapati R S, Nelson D L, Weinstock G M, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith D R, Doucette‐Stamm L, Rubenfield M, Weinstock K, Lee H M, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis R W, Federspiel N A, Abola A P, Proctor M J, Myers R M, Schmutz J, Dickson M, Grimwood J, Cox D R, Olson M V, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans G A, Athanasiou M, Schultz R, Roe B A, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie W R, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey J A, Bateman A, Batzoglou S, Birney E, Bork P, Brown D G, Burge C B, Cerutti L, Chen H C, Church D, Clamp M, Copley R R, Doerks T, Eddy S R, Eichler E E, Furey T S, Galagan J, Gilbert J G, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson L S, Jones T A, Kasif S, Kaspryzk A, Kennedy S, Kent W J, Kitts P, Koonin E V, Korf I, Kulp D, Lancet D, Lowe T M, McLysaght A, Mikkelsen T, Moran J V, Mulder N, Pollara V J, Ponting C P, Schuler G, Schultz J, Slater G, Smit A F, Stupka E, Szustakowski J, Thierry‐Mieg D, Thierry‐Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf Y I, Wolfe K H, Yang S P, Yeh R F, Collins F, Guyer M S, Peterson J, Felsenfeld A, Wetterstrand K A, Patrinos A, Morgan M J, de Jong P, Catanese J J, Osoegawa K, Shizuya H, Choi S, Chen YJ; International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature 2001409860–921. [DOI] [PubMed] [Google Scholar]
- 4.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, Gocayne J D, Amanatides P, Ballew R M, Huson D H, Wortman J R, Zhang Q, Kodira C D, Zheng X H, Chen L, Skupski M, Subramanian G, Thomas P D, Zhang J, Gabor Miklos G L, Nelson C, Broder S, Clark A G, Nadeau J, McKusick V A, Zinder N, Levine A J, Roberts R J, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu‐Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian A E, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman T J, Higgins M E, Ji R R, Ke Z, Ketchum K A, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov G V, Milshina N, Moore H M, Naik A K, Narayan V A, Neelam B, Nusskern D, Rusch D B, Salzberg S, Shao W, hue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng M L, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers Y H, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint N N, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn‐Deen E, Wolfe K, Zaveri J, Zaveri K, Abril J F, Guigo R, Campbell M J, Sjolander K V, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes‐Stine J, Caulk P, Chiang Y H, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science 20012911304–1351. [DOI] [PubMed] [Google Scholar]
- 5.Consortium IHG Finishing the euchromatic sequence of the human genome. Nature 2004431931–945. [DOI] [PubMed] [Google Scholar]
- 6.Ewart A K, Morris C A, Atkinson D, Jin W, Sternes K, Spallone P, Stock A D, Leppert M, Keating M T. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet 1993511–16. [DOI] [PubMed] [Google Scholar]
- 7.Stromme P, Bjornstad P G, Ramstad K. Prevalence estimation of Williams syndrome. J Child Neurol 200217269–271. [DOI] [PubMed] [Google Scholar]
- 8.Francke U. Williams‐Beuren syndrome: genes and mechanisms. Hum Mol Genet 199981947–1954. [DOI] [PubMed] [Google Scholar]
- 9.Osborne L R. Williams‐Beuren syndrome: unraveling the mysteries of a microdeletion disorder. Mol Genet Metab 1999671–10. [DOI] [PubMed] [Google Scholar]
- 10.Morris C A, Demsey S A, Leonard C O, Dilts C, Blackburn B L. Natural history of Williams syndrome: physical characteristics. J Pediatr 1988113318–326. [DOI] [PubMed] [Google Scholar]
- 11.Bellugi U, Bihrle A, Jernigan T, Trauner D, Doherty S. Neuropsychological, neurological, and neuroanatomical profile of Williams syndrome. Am J Med Genet Suppl 19906115–125. [DOI] [PubMed] [Google Scholar]
- 12.Cherniske E M, Carpenter T O, Klaiman C, Young E, Bregman J, Insogna K, Schultz R T, Pober B R. Multisystem study of 20 older adults with Williams syndrome. Am J Med Genet 2004131A255–264. [DOI] [PubMed] [Google Scholar]
- 13.Deutsch S, Choudhury U, Merla G, Howald C, Sylvan A, Antonarakis S E. Detection of aneuploidies by paralogous sequence quantification. J Med Genet 200441908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real‐time monitoring of DNA amplification reactions. Biotechnology (NY) 1993111026–1030. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi R, Dollinger G, Walsh P S, Griffith R. Simultaneous amplification and detection of specific DNA sequences. Biotechnology (NY) 199210413–417. [DOI] [PubMed] [Google Scholar]
- 16.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real‐time quantitative RT‐PCR data by geometric averaging of multiple internal control genes. Genome Biol 20023RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayes M, Magano L F, Rivera N, Flores R, Perez Jurado L A. Mutational mechanisms of Williams‐Beuren syndrome deletions. Am J Hum Genet 200373131–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu D P, Schmidt C, Billings T, Davisson M T. Quantitative PCR genotyping assay for the Ts65Dn mouse model of Down syndrome. Biotechniques. 2003;35: 1170–4, 1176, 1178 passim, [DOI] [PubMed]
- 19.Merla G, Ucla C, Guipponi M, Reymond A. Identification of additional transcripts in the Williams‐Beuren syndrome critical region. Hum Genet 2002110429–438. [DOI] [PubMed] [Google Scholar]
- 20.Valero M C, de Luis O, Cruces J, Perez Jurado L A. Fine‐scale comparative mapping of the human 7q11. 23 region and the orthologous region on mouse chromosome 5G: the low‐copy repeats that flank the Williams‐Beuren syndrome deletion arose at breakpoint sites of an evolutionary inversion(s), Genomics 2000691–13. [DOI] [PubMed] [Google Scholar]
- 21.Antonarakis S E, Lyle R, Dermitzakis E T, Reymond A, Deutsch S. Chromosome 21 and Down syndrome: from genomics to pathophysiology. Nat Rev Genet 20045725–738. [DOI] [PubMed] [Google Scholar]
- 22.Botta A, Novelli G, Mari A, Novelli A, Sabani M, Korenberg J, Osborne L R, Digilio M C, Giannotti A, Dallapiccola B. Detection of an atypical 7q11.23 deletion in Williams syndrome patients which does not include the STX1A and FZD3 genes. J Med Genet 199936478–480. [PMC free article] [PubMed] [Google Scholar]
- 23.Hirota H, Matsuoka R, Chen X N, Salandanan L S, Lincoln A, Rose F E, Sunahara M, Osawa M, Bellugi U, Korenberg J R. Williams syndrome deficits in visual spatial processing linked to GTF2IRD1 and GTF2I on chromosome 7q11.23. Genet Med 20035311–321. [DOI] [PubMed] [Google Scholar]
- 24.Gagliardi C, Bonaglia M C, Selicorni A, Borgatti R, Giorda R. Unusual cognitive and behavioural profile in a Williams syndrome patient with atypical 7q11.23 deletion. J Med Genet 200340526–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heller R, Rauch A, Luttgen S, Schroder B, Winterpacht A. Partial deletion of the critical 1.5 Mb interval in Williams‐Beuren syndrome. J Med Genet 200340e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karmiloff‐Smith A, Grant J, Ewing S, Carette M J, Metcalfe K, Donnai D, Read A P, Tassabehji M. Using case study comparisons to explore genotype‐phenotype correlations in Williams‐Beuren syndrome. J Med Genet 200340136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korenberg J R, Chen X N, Hirota H, Lai Z, Bellugi U, Burian D, Roe B, Matsuoka R. VI. Genome structure and cognitive map of Williams syndrome. J Cogn Neurosci 200012(suppl 1)89–107. [DOI] [PubMed] [Google Scholar]
- 28.Tassabehji M, Metcalfe K, Karmiloff‐Smith A, Carette M J, Grant J, Dennis N, Reardon W, Splitt M, Read A P, Donnai D. Williams syndrome: use of chromosomal microdeletions as a tool to dissect cognitive and physical phenotypes. Am J Hum Genet 199964118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris C A, Mervis C B, Hobart H H, Gregg R G, Bertrand J, Ensing G J, Sommer A, Moore C A, Hopkin R J, Spallone P A, Keating M T, Osborne L, Kimberley K W, Stock A D. GTF2I hemizygosity implicated in mental retardation in Williams syndrome: genotype‐phenotype analysis of five families with deletions in the Williams syndrome region. Am J Med Genet 2003123A45–59. [DOI] [PubMed] [Google Scholar]
- 30.Tassabehji M. Williams‐Beuren syndrome: a challenge for genotype‐phenotype correlations. Hum Mol Genet 200312(Spec No 2)R229–R237. [DOI] [PubMed] [Google Scholar]
- 31.Duba H C, Doll A, Neyer M, Erdel M, Mann C, Hammerer I, Utermann G, Grzeschik K H. The elastin gene is disrupted in a family with balanced translocation t(7;16)(q11.23; q13) associated with a variable expression of the Williams‐Beuren syndrome. Eur J Hum Genet 200210351–361. [DOI] [PubMed] [Google Scholar]