Abstract
B-type cyclins are rapidly degraded at the transition between metaphase and anaphase and their ubiquitin-mediated proteolysis is required for cells to exit mitosis. We used a novel enrichment to isolate new budding mutants that arrest the cell cycle in mitosis. Most of these mutants lie in the CDC16, CDC23, and CDC27 genes, which have already been shown to play a role in cyclin proteolysis and encode components of a 20S complex (called the cyclosome or anaphase promoting complex) that ubiquitinates mitotic cyclins. We show that mutations in CDC26 and a novel gene, DOC1, also prevent mitotic cyclin proteolysis. Mutants in either gene arrest as large budded cells with high levels of the major mitotic cyclin (Clb2) protein at 37°C and cannot degrade Clb2 in G1-arrested cells. Cdc26 associates in vivo with Doc1, Cdc16, Cdc23, and Cdc27. In addition, the majority of Doc1 cosediments at 20S with Cdc27 in a sucrose gradient, indicating that Cdc26 and Doc1 are components of the anaphase promoting complex.
INTRODUCTION
Oscillations in the activity of cyclin-dependent kinases (Cdk) control the eukaryotic cell cycle (reviewed in Morgan, 1995). Multicellular eukaryotes express a number of Cdks with different activities through the cell cycle. Yeasts, however, express a single Cdk, called Cdc28 in budding yeast (Saccharomyces cerevisiae) and Cdc2 in fission yeast (Schizosaccharomyces pombe). The oscillation of Cdk activity in all eukaryotes depends on phosphorylations of the kinase subunit and the cyclic expression and degradation of cyclins, the regulatory subunits of Cdks. Entry into mitosis requires the expression of B-type (mitotic) cyclins, and exit from mitosis requires their degradation (reviewed in Murray and Kirschner, 1989; Nurse, 1990). The Cdk activity associated with B-type cyclins is required for chromosome condensation and proper formation of the mitotic spindle. The degradation of B-type cyclins, which leads to inactivation of the mitotic Cdk, results in chromosome decondensation, breakdown of the mitotic spindle, and cell division.
A short N-terminal sequence found in A- and B-type cyclins, termed the destruction box, is required for their proteolysis (Glotzer et al., 1991). Deletion or mutation of this sequence produces a nondegradable cyclin, and cells or extracts expressing these nondegradable B-type cyclins are unable to exit mitosis and remain arrested in anaphase (Murray et al., 1989; Holloway et al., 1993; Surana et al., 1993; Sigrist et al., 1995). In contrast, inhibiting the machinery that is required for cyclin proteolysis arrests cells in metaphase with unseparated sister chromatids (Holloway et al., 1993). This observation suggests that additional proteins, whose destruction is required for the separation of sister chromatids, must also be targets of the machinery that degrades cyclins. Candidates for these regulators of sister cohesion have recently been identified (Funabiki et al., 1996; Yamamoto et al., 1996).
The degradation of cyclin is regulated by the assembly of multiubiquitin chains onto the cyclin substrate that targets it for proteolysis by the proteasome. In frog egg extracts, mutations in cyclins that block their degradation also block ubiquitination (Glotzer et al., 1991; King et al., 1995), and methylated ubiquitin, an inhibitor of multiubiquitin chain formation, inhibits the degradation of cyclins in clam egg extracts (Hershko et al., 1991). In yeast, a number of mutants that block cyclin degradation also block ubiquitination (Zachariae and Nasmyth, 1996).
Ubiquitination of proteins begins with the activation of ubiquitin by a ubiquitin-activating enzyme (E1). The E1 forms a thiol ester with ubiquitin and then transfers that ubiquitin onto a ubiquitin-carrier protein (E2). Some substrates are directly ubiquitinated by an E2. Other substrates, however, also require a ubiquitin-protein ligase (E3), which appears to provide a higher level of specificity to the degradation system. Finally, the 26S proteasome recognizes multiubiquitinated substrates and degrades them (reviewed in Ciechanover, 1994).
In budding yeast, clam, and frog, a nonspecific E1 is capable of activating ubiquitin. A large family of E2s (UBCs) exist in yeast, and the E2 required for Clb degradation has not yet been definitively identified. E2-C (clam), homologues of the budding yeast UBC4, and a novel UBC, UBCX (frog) are capable of acting as E2s for cyclin B in reconstituted cyclin ubiquitination systems derived from clam and frog egg extracts (King et al., 1995; Sudakin et al., 1995). The E3 for B-type cyclins was first identified as a 20S complex in clam and frog egg extracts, known as the cyclosome or anaphase-promoting complex (APC; King et al., 1995; Sudakin et al., 1995). In frog egg extracts, yeast homologues of CDC16 and CDC27 were identified as components of this E3 complex, which appears to be active only in mitosis in both clams and frogs.
CDC16, CDC23, and CDC27 are all members of a family of tetratricopeptide repeat proteins and form a complex in yeast that is required for the ubiquitination and degradation of Clb2, the major mitotic cyclin (Lamb et al., 1994; Irniger et al., 1995; Zachariae and Nasmyth, 1996). Temperature-sensitive mutants in any of these genes cause a metaphase arrest with high levels of Clb2 and unseparated sister chromosomes. Another gene, CSE1, has also been implicated in cyclin ubiquitination (Irniger et al., 1995). Mutations in CDC16, CDC23, and CSE1 all result in increased chromosome loss, suggesting that proper regulation of the cyclin proteolysis machinery is required for faithful chromosome segregation (Hartwell and Smith, 1985; Xiao et al., 1993).
We developed an enrichment for cell cycle arrest mutants in budding yeast and used this to screen for new mitotic arrest mutants. We isolated a large number of alleles of previously identified cdc (cell division cycle) mutants, and identified a novel mutant, doc1 (destruction of cyclin B). CDC26 and DOC1 are involved in the degradation of Clb2 and the products of both genes associate with the yeast APC.
MATERIALS AND METHODS
Yeast Strains, Media, and Genetic Methods
All yeast strains are derivatives of W303, except those used for complementation analysis. All strains used in this work are listed in Table 1. Yeast media and genetic manipulations were as described (Sherman et al., 1974). LH103 (mec1–1) was made by backcrossing AFS85 (MATa, mec1–1, GAL-CLN3::URA3::cln3Δ, ssd1-v1::LEU2, leu2–3,112, ura3, his3–11,15,ade2–1, trp1–1) four times into W303 and subsequently crossing in mad1Δ::HIS3 (from KH123, Hardwick and Murray, 1995) and bar1 (from AFS92, kindly provided by Aaron Straight, University of California, San Francisco). BglII-cut pLH17 (CLB2-lacZ) was integrated at CLB2, recombination and loss of the URA3 marker were selected for on 5-fluoroorotic acid medium, and retention of CLB2-lacZ was screened for by using β-galactosidase (β-gal) plate assays. Strains containing GAL-CLB2 were made by integrating pDK27 (kindly provided by Doug Kellogg, University of California, Santa Cruz) at URA3.
Table 1.
Yeast strains
| Strain | Relevant genotype | Source |
|---|---|---|
| LH103 | MATa mec1-1 mad1Δ::HIS3 clb2::CLB2-LacZ bar1 | This work |
| YCp50/MAD1 | ||
| AFS34 | MATa ade2-1 can1-100 ura3-1 leu2-3,112 his3-11,15 trp1-1 | R. Rothstein |
| (W303-1a) | ||
| YPH218 | MATα cdc16-1 | P. Hieter |
| YPH221 | MATα cdc23-1 | P. Hieter |
| LH127 | MATα cdc20-1 ura3-1 leu2-3,112 trp1-1 | This work |
| H160-3-3 | MATα cdc27-1 | L. Hartwell |
| H152-4-2 | MATα cdc26-1 | L. Hartwell |
| ELW65-93 | MATa cdc31-2 YCp50/CDC31 | M. Winey |
| CMY763 | MATα cim3-1 | C. Mann |
| CMY765 | MATα cim5-1 | C. Mann |
| LH202 | MATa cim5-1 bar1 | This work |
| LH103-15 | MATa doc1-1 mec101 mad1Δ::HIS3 clb2::CLB2-lacZ bar1 | This work |
| YCp50/MAD1 | ||
| LH103-66 | MATa cdc26-100 mec1-1 mad1Δ::HIS3 clb2::CLB2-lacZ bar1 | This work |
| YCp50/MAD1 | ||
| LH225 | MATa cdc26-100 clb2::CLB2-lacZ bar1 | This work |
| LH226 | MATa doc1-1 clb2::CLB2-lacZ bar1 | This work |
| ADR58 | MATa pDK27 (GAL-CLB2) | This work |
| ADR103 | MATa cdc16-1 pDK27 | This work |
| LH227 | MATa cdc26-100 bar1 pDK27 | This work |
| LH228 | MATa doc1-1 bar1 pDK27 | This work |
| LH229 | MATa cdc26-100 bar1 ura3-1::CDC26myc-URA3 pDK27 | This work |
| LH230 | MATa doc1-1 bar1 ura3-1::DOC1myc-URA3 pDK27 | This work |
| LH209 | MATa bar1 pUb-R-βgal | This work |
| LH210 | MATa bar1 pUbV76-V-eΔK-βgal | This work |
| LH211 | MATa doc1-1 bar1 pUb-R-βgal | This work |
| LH212 | MATa doc1-1 bar1 pUbV76-V-eΔK-βgal | This work |
| LH213 | MATa cdc26-100 bar1 pUb-R-βgal | This work |
| LH214 | MATa cdc26-100 bar1 pUbV76-V-eΔK-βgal | This work |
| LH215 | MATa cim3-1 bar1 pUb-R-βgal | This work |
| LH216 | MATa cim3-1 bar1 pUbV76-V-eΔK-βgal | This work |
| LH231 | MATa bar1 pWAM10 (CDC16HA) | This work |
| LH232 | MATa bar1 pRS239 (CDC23HA) | This work |
| LH233 | MATa bar1 pJL25 (CDC27HA) | This work |
| LH234 | MATa bar1 pLH32 (DOC1HA) | This work |
| LH235 | MATa bar1 cdc26Δ::URA3 pWAM10 | This work |
| LH236 | MATa bar1 cdc26Δ::URA3 pRS239 | This work |
| LH237 | MATa bar1 cdc26Δ::URA3 pJL25 | This work |
| LH238 | MATa bar1 cdc26Δ::URA3 pLH32 | This work |
| LH297 | MATa bar1 doc1Δ::URA3 leu2-3,112::DOC13XHA-LEU2 | This work |
| pJL25 |
All strains from this work are W303 (R. Rothstein).
Hydroxyurea (Sigma, St. Louis, MO) was used at 10 mg/ml, final concentration, in medium. α-Factor (Bio-synthesis, Lewisville, TX) was used at 1 μg/ml for bar1− strains and 10 μg/ml for BAR1 strains, from a stock solution at 10 mg/ml in dimethyl sulfoxide (Aldrich, Milwaukee, WI). Nocodazole (Sigma) was used at 15 μg/ml from a stock solution of 10 mg/ml in dimethyl sulfoxide. Cycloheximide (Sigma) was used at 10 μg/ml from a stock solution of 10 mg/ml. Nocodazole treatments were carried out at 23°C. The other treatments were performed at various temperatures.
Plasmid Constructions
For CLB2-lacZ (pLH17), CLB2 (including the open reading frame [ORF] and 301 bp upstream) was amplified by polymerase chain reaction (PCR) using oligomers containing BamHI and SalI sites at 5′ and 3′ ends, respectively. This was ligated into pRS304 (Sikorski and Hieter, 1989) and then cut with SalI and KpnI. The KpnI site was blunted with T4 polymerase. lacZ was cut from pAFS35 (kindly provided by Aaron Straight) by using SalI and BamHI. The BamHI site was blunted by using the Klenow fragment of DNA polymerase I. The lacZ fragment was then ligated into the CLB2 construct. A second BamHI site was created 3′ of lacZ by ligation of the blunted BamHI and KpnI sites. A 270-bp fragment of sequence immediately 3′ of the CLB2 ORF was amplified by PCR with BamHI and SalI sites 5′ and 3′, respectively. This was ligated into pRS306 (Sikorski and Hieter, 1989). The BamHI fragment containing CLB2-lacZ was then ligated into the pRS306 construct containing the 3′ flanking region of CLB2. The Clb2-LacZ protein is functional and capable of acting as the sole mitotic cyclin since pLH17 rescues the temperature sensitivity of a strain deleted for CLB1, CLB3, and CLB4 with CLB2 replaced with a temperature sensitive allele, K3080 (Amon et al., 1993).
pLH25 was made by PCR amplifying the ORF of DOC1 and 508 bp upstream with a SalI site 5′ and an NcoI site 3′ to the ORF. The PCR product was ligated into pKH511 (kindly provided by Kevin Hardwick, University of Edinburgh), to place a single myc tag at the C terminus of the protein. To make pLH23, pLH25 was cut with EcoRV and BamHI and the 1.0-kb fragment was ligated into pDK20 (kindly provided by Doug Kellogg) cut with SmaI and BamHI, to place the DOC1 ORF behind the GAL1–10 promoter. pLH24 was made by cutting pLH25 with SalI and BamHI and ligating the 1.5-kb fragment into YIplac211, cut with the same enzymes. pLH26–1B was made by cutting T1 (the original rescuing plasmid from the YCp50 library) with NdeI and blunting with Klenow. For pLH26–1A, URA3 was cut from pLH3 with XhoI and XbaI and also blunted with Klenow. The URA3 fragment was ligated into the blunted NdeI site in T1.
To make pLH32, the ORF of DOC1 with 508 bp upstream was amplified by PCR with a SalI site 5′ and a KpnI 3′ to the ORF. The KpnI site was designed so that the stop codon of DOC1 was excluded and the 3′ end of DOC1 would be in-frame with a triple hemaglutinin (3×HA) tag in YCplac111–3×HA. The DOC1 PCR product was cut with SalI, blunted with Klenow, and then cut with KpnI. YCplac111–3×HA was cut with EcoRI, blunted with Klenow, and then cut with KpnI. The PCR fragment was then ligated into YCplac111–3×HA. pLH59 (DOC1–3×HA-LEU2) was made by cutting pLH32 with SphI and SpeI and ligating the 1.8-kb fragment into YIplac128 cut with SphI and XbaI. All enzymes were from New England Biolabs (Beverly, MA) and used according to the manufacturer’s specifications.
Plasmids expressing Ub-R-βgal and UbV76-V-eΔK-βgal were kindly provided by Dr. Erica Johnson (Rockefeller University, NY). Plasmids containing HA-tagged CDC16 (pWAM10), CDC23 (pRS239), and CDC27 (pRS248) were kindly provided by Dr. Phillip Heiter (Johns Hopkins University, MD).
Mutant Isolation
Strain LH103 (8 × 106 cells) was mutagenized with ethyl methanesulfonate (Sigma) to 50% killing. The mutagenized cells were diluted 1:25 and allowed to recover for 12 h at room temperature in YPD. They were shifted to 37°C, to prearrest potential G1 and mitotic arrest mutants, for 2 h, then hydroxyurea was added to 10 mg/ml, and cells were incubated at 37°C for an additional 5 h. This culture was then plated on YPD plates and incubated at 23°C. Surviving colonies were replica plated onto YPD plates in duplicate with one set at 23°C and the other at 37°C to test for temperature sensitivity.
Clb2-LacZ and Visual Screen
Temperature-sensitive mutants were patched onto YPD plates and allowed to grow overnight at 23°C. They were then replica plated to YPD and placed at 37°C for 4 h, replica plated again to Whatman filters (VWR, San Francisco, CA) on YPD plates that contained 1 μg/ml α-factor, and returned to 37°C for 5 h. The filters were assayed for β-gal activity by freezing them in liquid nitrogen, thawing them, and incubating them on Whatman paper soaked in Z buffer plus 5-bromo-4-chloro-3-indolyl β-d-galactoside, US Biological, Swampscott, MA; 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 0.03% 5-bromo-4-chloro-3-indolyl β-d-galactoside) at 30°C overnight.
Strains that produced bright blue patches were further analyzed by microscopy. The strains were grown in liquid medium to logarithmic phase at 23°C and then shifted to 37°C. Cells were taken and fixed in 3.7% formaldehyde (Fisher, Santa Clara, CA) after 3 and 6 h at 37°C. They were stained with 4,6-diamidino-2-phenylindole (1 μg/ml) and anti-tubulin (YOL1/34, Accurate Chemical and Scientific, Westbury, NY) diluted 1:200, and their arrest phenotype was determined by light microscopy.
Mitotic arrest mutants were tested for complementation against a collection of known mitotic mutants. Only two mutants were not identified as previously isolated mutants. One was cloned and named DOC1, and the other (two alleles) has not been cloned and appears to arrest in mitosis with pleiotropic defects. One allele of cdc14 was also isolated; however, it was isolated as a double mutant with a cdc16 mutant. CDC14 codes for a phosphatase, and mutants in this gene arrest in anaphase of the cell cycle (Wan et al., 1992).
Cloning of DOC1
A YCp50 library (described in Hardwick and Murray, 1995) was transformed into doc1–1 by lithium acetate transformation and plated on −Ura medium at 37°C. Plasmids that were capable of rescuing the temperature sensitivity were recovered and the inserts were sequenced at both 5′ and 3′ ends. These sequences were used to probe the yeast genome database.
Preparation of Antibodies against Cdc26, Western Blot Analysis, and Immunoprecipitations
The ORF of CDC26 was amplified by PCR with oligomers containing BamHI and EcoRI sites 5′ and 3′ to the ORF, respectively. This fragment was cloned into pGEX-1 cut with BamHI and EcoRI (Smith and Johnson, 1988), forming a glutathione S-transferase (GST) fusion construct (pLH29). pLH29 was transformed into Escherichia coli strain TG1 (Maniatis et al., 1982) and induced for expression with 0.1 mM isopropyl β-d-thiogalactoside (US Biologicals) for 2 h at 37°C. Cells were pelleted, washed with PBS (140 mM Na2HPO4, 1.8 mM KH2PO4, 138 mM NaCl, 2.7 mM KCl, pH 7.2), pelleted again, and frozen in liquid nitrogen. The frozen pellet was resuspended in five volumes of PBS containing 1 mM EDTA, 1 mM EGTA, 1 mM phenylmethylsulfonyl fluoride, and 200 μg/ml lysozyme and then briefly sonicated. Triton X-100 was added to 0.5%, dithiothreitol was added to 15 mM, and cells were sonicated again. The lysate was spun at 15,000 rpm in an SS34 rotor (Sorvall, Burbank, CA) for 30 min, and then the supernatant was transferred to a new tube and spun again for 20 min. The cleared supernatant was then loaded onto a 5-ml glutathione-agarose column (Sigma) that was then washed with PBS containing 0.5 M NaCl, 1 mM EDTA, and 1 mM EGTA. The Cdc26-GST fusion protein was eluted with 5 mM reduced glutathione (Sigma) in 50 mM Tris(hydroxymethyl)aminomethane, pH 8.1. The peak fractions were pooled and dialyzed into 50 mM HEPES, pH 7.6, 50 mM KCl, and 30% glycerol. The protein was sent to Berkeley Antibody (Berkeley, CA) where it was used to immunize a rabbit. The rabbit serum was passed over a 50-ml column of GST protein coupled to Affi-Gel 10 (Bio-Rad Labs, Hercules, CA) to remove anti-GST antibodies and then affinity purified by using a 5-ml column of the Cdc26-GST fusion protein coupled to Affi-Gel 10 (Bio-Rad).
Yeast extracts for immunoblotting and immunoprecipitations were made by bead beating cells for two 90 s periods, in lysis buffer A (50 mM HEPES, pH 7.6, 75 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.1% Nonidet P-40, 50 mM NaF, 100 μM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride, and LPC (10 μg/ml each leupeptin, pepstatin, and chymostatin; Boehringer, Indianapolis, IN). The lysates were spun briefly to separate beads from the lysate and then cleared by centrifugation for 5 min in an Eppendorf microcentrifuge at 4°C. This step was repeated three times. Protein concentrations of extracts were determined with the Bradford assay using bovine serum albumin as a standard (Bradford, 1976). For immunoblotting, the extracts were adjusted to the same protein concentration and diluted with 2× SDS sample buffer (1× SDS sample buffer: 80 mM Tris(hydroxymethyl)aminomethane, pH 6.8, 2% SDS, 10% glycerol, 10 mM EDTA, 0.0013% bromophenol blue, 5% 2-mercaptoethanol). Standard methods were used for SDS-PAGE and protein transfer to nitrocellulose (Harlow and Lane, 1988). Blots were stained with Ponceau S (Fisher) to confirm transfer and equal protein loading and then blocked for 30 min with blotto (4% dried milk, PBS, 0.2% Tween 20). Antibodies were used at a 1:1200 dilution for anti-Clb2, a 1:1000 dilution for 12CA5 (BABCO, Berkeley, CA), a 1:1000 dilution for anti-β-gal (Cappel, Durham, NC), and a 1:1000 dilution for anti-Cdc26 at either room temperature for 1 h or 4°C overnight. Blots were washed three times 10 min in PBS with 0.2% Tween 20 (PBST) and then incubated with horseradish peroxidase-conjugated anti-rabbit or anti-mouse antibodies (Amersham, Buckinghamshire, United Kingdom) at a 1:5000 dilution in PBST. They were washed again and developed by using Amersham ECL detection reagents following manufacturer’s instructions. Cdc26 protein is not detectable by Western blot, likely due to an inability to bind to nitrocellulose.
For immunoprecipitations, antibodies were used at a 1:33 dilution for 12CA5 and a 1:50 dilution for Cdc26. Lysates with primary antibody were rotated for 1 h at 4°C, then transferred to a tube containing protein A-Sepharose beads (Pharmacia, Pleasant Hill, CA), and rotated at 4°C for an additional hour. The beads were washed twice with lysis buffer A, transferred to a new tube, washed again with PBST, and then resuspended in SDS sample buffer.
Sucrose Gradient Analysis
Solutions with lysis buffer A were prepared containing either 5% or 40% sucrose. Additional solutions with 13.75%, 22.5%, and 31.25% sucrose were made by combining various amounts of the 5% and 40% solutions. Nine hundred fifty microliters of each solution were layered into Beckman Ultra-Clear Centrifuge tubes (Beckman, Palo Alto, CA), size 0.5 × 2 inches. The gradient was incubated 12–16 h at 4°C. Yeast extract (100 μl), made as described above, was layered onto the gradient and spun at 50,000 rpm in a SW-55 rotor (Sorvall) for 4 h at 4°C. Sixteen 300-μl fractions were taken, the fractions were precipitated with 10% trichloroacetic acid, loaded onto SDS-Page gels, and analyzed by immunoblotting. Molecular weight markers were analyzed by Coomassie blue staining (Fisher).
RESULTS
Screening for New Mitotic cdc Mutants
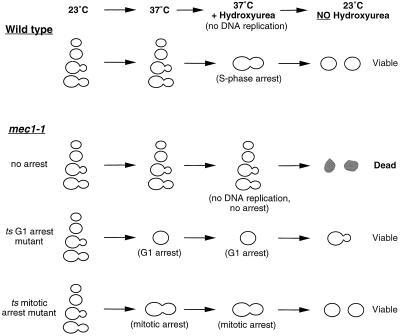
We designed a strategy that enabled us to enrich for temperature-sensitive budding yeast mutants that arrest in G1 or mitosis (Figure 1). We used mec1–1, a cell cycle checkpoint mutant, to devise conditions under which cycling cells would die and arrested ones would survive. mec1–1 mutants lack the checkpoint that detects unreplicated DNA and DNA damage and cannot arrest their cell cycle in the presence of unreplicated DNA (Weinert, 1992). As a result, treating cycling mec1–1 cells with DNA replication inhibitors, such as hydroxyurea, induces rapid cell death. In contrast, treating G1 or mitotically arrested mec1–1 cells with hydroxyurea will not kill them. We exploited this difference to enrich for mutants that caused a temperature-sensitive arrest in G1 or mitosis. This protocol does not enrich for most mutants arrested in S or G2 phase, because their arrest requires the MEC1-dependent checkpoint. A mec1–1 strain was mutagenized and the mutagenized cells were shifted to 37°C to allow potential cell division cycle mutants to prearrest and then treated with hydroxyurea to kill the majority of cells that continued to progress through the cell cycle. Cells that arrest in either G1 or mitosis remain viable during the hydroxyurea treatment and form colonies on plating at the permissive temperature of 23°C.
Figure 1.
Schematic representation of the mutant enrichment strategy. Hydroxyurea, a drug that inhibits DNA synthesis, arrests wild-type cells in S phase. Mutants that are defective in the checkpoint gene MEC1 are unable to arrest in response to unreplicated DNA and die in the presence of hydroxyurea. If mec1–1 cells are prearrested in G1 or mitosis, they remain viable after exposure to hydroxyurea.
We mutagenized 8 × 106 cells to 50% survival with ethyl methanesulfonate, and 40,000 survived the mec1–1 enrichment. Of those, 1968 (5%) were temperature sensitive. To distinguish mutants that arrested in mitosis from those that arrested in G1, we exploited the observation that mitotic cyclins are strongly expressed in mitosis but rapidly degraded as cells exit mitosis and enter G1 (Amon et al., 1994). The temperature-sensitive mutants were tested for expression of Clb2-lacZ, a protein fusion between the major mitotic cyclin (Clb2) and β-gal, whose activity can be easily monitored by exposing cells to a chromogenic substrate. Cells from mutants that expressed Clb2-lacZ strongly at 37°C (see MATERIALS AND METHODS) were screened visually for their arrest phenotypes. We found 32 mutants that arrested with large budded cells at 37°C, a phenotype that is consistent with a mitotic arrest. These mutants fell into 11 complementation groups. Each group was tested against known mitotic mutants by complementation of temperature sensitivity and for rescue of temperature sensitivity by transformation with plasmids containing wild-type copies of genes known to function in mitosis.
We isolated mitotic arrest mutations in nine genes (Table 2). One, CDC31 (one allele) is involved in spindle pole body duplication and cdc31ts mutants arrest in mitosis with only one spindle pole body (Byers, 1981; Baum et al., 1986). Another, PRP22 (two alleles) codes for an RNA helicase involved in mRNA splicing (Company et al., 1991). The alleles of PRP22 isolated in this screen arrests in mitosis with no microtubules (our unpublished results). TUB1 and TUB3, the genes that code for α-tubulin, each contain an intron. It is likely that the tubulin defect seen in these mutants is due to a failure to splice the mRNAs from these genes that leads to a deficiency in α-tubulin.
Table 2.
Mitotic-arrest mutants isolated by mec1-1 enrichment
| Gene | No. of alleles isolated | Biochemical function | Reference |
|---|---|---|---|
| CDC16 | 16 | APC component | Irniger et al. (1995), King et al. (1995) |
| CDC23 | 3 | APC component | Irniger et al. (1995) |
| CDC27 | 4 | APC component | King et al. (1995) |
| CDC26 | 1 | APC component | This work |
| DOC1 | 1 | APC associated | This work |
| CIM3 | 1 | Proteasome component | Ghislain et al. (1993) |
| CDC20 | 1 | Unknown | Sethi et al. (1991) |
| CDC31 | 1 | SPB component | Baum et al. (1986) |
| PRP22 | 2 | Splicing | Company et al. (1991) |
| Uncloned | 2 | Unknown |
Mutants were identified based on complementation of temperature sensitivity with previously identified mutants. They were also tested for rescue of temperature sensitivity by wild-type copies of known genes on plasmids.
Five of the genes we identified are involved in the proteolysis of cyclin that triggers the exit from mitosis. CDC16 (16 alleles), CDC23 (3 alleles), and CDC27 (4 alleles) encode proteins that are involved in cyclin proteolysis and have been shown to form a multiprotein complex in yeast, frogs, and clams (Lamb et al., 1994; Irniger et al., 1995; King et al., 1995; Sudakin et al., 1995). CDC20 (1 allele) has also been implicated in cyclin proteolysis. Its homologue in Drosophila, fizzy, appears to be required for the proteolysis of cyclin A and B (Dawson et al., 1995; Sigrist et al., 1995). In yeast, cdc20 mutants arrest in metaphase and appear to have microtubule abnormalities (Byers and Goetsch, 1974; Palmer et al., 1989; Sethi et al., 1991). CIM3 (1 allele) is a subunit of the 26S proteasome, the multiprotein complex that degrades ubiquitinated proteins (Ghislain et al., 1993).
Finally, we isolated one allele each of CDC26 and a novel gene that we named DOC1 (destruction of cyclin B). CDC26 is a mitotic arrest mutant, identified in the original Hartwell screen for cdc mutants (Hartwell et al., 1970). The CDC26 gene is essential only at 37°C; cdc26Δ cells grow well at 23°C (Araki et al.., 1992), a finding that we confirmed in the W303 strain background.
CDC26 and DOC1 Are Required for Clb2 Proteolysis
Because the majority of mutants isolated in our screen identify genes involved in mitotic cyclin proteolysis, we tested the cdc26 and doc1 mutants for defects in this process. To determine whether CDC26 and DOC1 were involved in cyclin proteolysis, we exploited the observation that cyclin proteolysis begins in late mitosis and persists well into G1 (Amon et al., 1994). Haploid yeast cells can be arrested in G1 by treatment with mating pheromone. Thus in MATa cells arrested in G1 by α-factor treatment, the B-type cyclin proteolysis machinery is active and ectopic expression of the CLB2 gene does not lead to Clb2 protein accumulation. In a cdc16–1 mutant, however, Clb2 protein does accumulate in G1-arrested cells as a result of a defect in the ubiquitination of Clb2 (Irniger et al., 1995; Zachariae and Nasmyth, 1996).
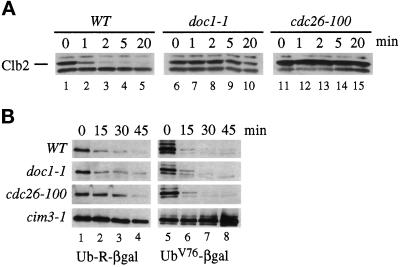
To determine the half-life of mitotic cyclins, we integrated a copy of CLB2 under the control of the inducible GAL promoter into wild-type and mutant strains. Wild-type and the doc1–1 and cdc26–100 mutants isolated in our screen were arrested with α-factor and shifted to 37°C for 20 min, and CLB2 expression was induced with galactose. Cycloheximide and glucose were added after a 30-min induction to stop Clb2 protein synthesis and CLB2 transcription. Clb2 protein levels were monitored by Western blotting using anti-Clb2 antibodies. Figure 2A shows that the half-life of Clb2 in wild-type cells is less than 1 min. In doc1–1 and cdc26–100 mutants, the half-life of Clb2 protein increases to greater than 20 min. This defect is rescued in doc1–1 and cdc26–100 by the integration of a wild-type copy of the corresponding gene at the LEU2 locus (our unpublished results). Others have also identified CDC26 as playing a role in cyclin proteolysis (Zachariae et al., 1996).
Figure 2.
Phenotype of cdc26 and doc1 mutants. (A) Stability of Clb2 in proteolysis mutants. The indicated strains contained an integrated copy of pGAL-CLB2, were grown in YEP + 2% raffinose medium, and arrested in G1 by exposure to α-factor for 3 h at 23°C. The cells were shifted to 37°C for 20 min before CLB2 expression was induced by the addition of 2% galactose while maintaining the α-factor arrest. Cycloheximide (10 μg/ml) and glucose (2%) were added after a 30-min induction and time points were taken at 0, 1, 2, 5, and 20 min. An exposure of a Western blot probed with polyclonal anti-Clb2 antibodies is shown. Wild-type (lanes 1–5), doc1–1 (lanes 6–10), and cdc26–100 (lanes 11–15). (B) Stability of β-gal derivatives in proteolysis mutants. Wild-type, doc1–1, cdc26–100, and cim3–1 strains containing plasmids expressing either Ub-R-β-gal (lanes 1–4) or UbV76-V-eΔK-βgal (UbV76-βgal; lanes 5–8) under the control of the GAL promoter were grown in YEP + 2% raffinose and then arrested in G1 with α-factor for 3 h. Expression from the plasmids was induced by the addition of 2% galactose for 2 h. At time zero, cycloheximide was added to 10 μg/ml and samples were taken at 15, 30, and 45 min. An exposure of a Western blot probed with anti-β-gal antibodies is shown.
The Clb2 destruction defect in cdc26–100 and doc1–1 appears to be specific to Clb2 and not a general defect in the proteolysis of ubiquitinated substrates. Figure 2B shows that cdc26–100 and doc1–1 cells have no defect in the degradation of two substrates that require ubiquitination for destruction: Ub-R-βgal, which generates β-gal with an N-terminal arginine, a substrate that is degraded by the N-end rule (Figure 2B, Ub-R-βgal; Bachmair et al., 1986), and UbV76-V-eΔK-βgal, a fusion with a noncleavable N-terminal ubiquitin (Figure 2B, UBV76-βgal; Johnson et al., 1992). Cycloheximide was added to cells expressing Ub-R-βgal or UbV76-V-eΔK-βgal to inhibit protein synthesis, and stability of the substrates was determined by Western blotting with anti-β-gal antibodies at different times after inhibiting protein synthesis. cim3–1, a mutant in a subunit of the 26S proteasome (Ghislain et al., 1993) that degrades all ubiquitinated proteins, is defective in the degradation of both substrates (Figure 2B, lanes 1–8).
To see if the mutant phenotype of cdc26–100 and doc1–1 cells reflected defects in mitotic proteolysis, we overexpressed a mitotic cyclin in these mutants. cdc26–100 and doc1–1 strains containing an integrated copy of GAL-CLB2 are unable to grow on galactose at their permissive temperature (data not shown). We suggest that these strains are already crippled in their Clb2 destruction machinery at the permissive temperature and that excess Clb2 overloads this machinery and results in a cell cycle arrest.
Cloning and Disruption of DOC1
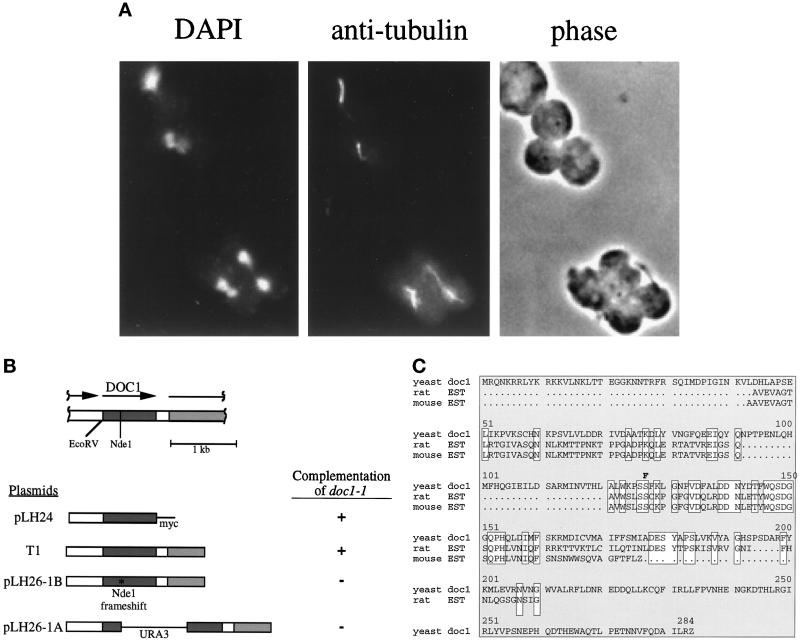
Like previously identified mitotic cyclin proteolysis mutants, doc1–1 mutants arrest as large budded cells at 37°C with a single nucleus and a short spindle (Figure 3A and Table 3).
Figure 3.
(A) doc1–1 arrest phenotype. The top two large-budded cells arrested with short spindles and a single DNA mass. The bottom two adjacent cells are large-budded. One has arrested with a short spindle and single DNA mass, the other appears to have undergone anaphase. doc1–1 cells were grown to logarithmic phase at room temperature and then shifted to 37°C for 3 h. The cells were fixed and stained with 4,6-diamidino-2-phenylindole and anti-tubulin. (B) Structure of the DOC1 locus and complementation of doc1–1 temperature sensitivity with various plasmids. The shaded region represents the DOC1 ORF and the hatched region represents the 5′ end of the CSE1 ORF. The arrows show the direction of transcription. The DOC1 ORF with 508 bp upstream is sufficient to complement the temperature sensitivity of doc1–1 at 37°C. TI, the rescuing plasmid from a YCp50-based library, contains approximately 640 bp upstream of the ORF of DOC1 and approximately 1150 bp of the CSE1 ORF. T1 was cut with NdeI and blunted with the Klenow fragment of DNA polymerase I (New England Biolabs), producing a frameshift in the DOC1 ORF (pLH26–1B). The frameshifted construct was unable to rescue doc1–1. pLH26–1A, in which URA3 is inserted into the NdeI-blunted site, was also unable to rescue doc1–1. (C) Amino acid sequence of DOC1. This sequence corresponds to ORF YGL240W in the yeast genome database (the GenBank accession number for DOC1 is Z72762). The DOC1 amino acid sequence is aligned with rat and mouse expressed sequence tag sequences (accession numbers H33761 and W49295, respectively). Identities are boxed. The doc1–1 mutation is indicated above the sequence. The mutation in doc1–1 (S137 → F137) is indicated in boldface type.
Table 3.
doc1-1 arrest phenotype
| Strain | % of total
|
|||||
|---|---|---|---|---|---|---|
| Wild type | 75 | 10 | 11 | 4 | ||
| doc1-1 | 16 | 1 | 79 | 4 | ||
Wild-type and doc1-1 strains were grown to logarithmic phase and then shifted to 37°C for 3 h. The cells were fixed and then stained for DNA using 4,6-diamino-2-phenylindole and tubulin by using immunofluorescence. Percentage of total cell morphologies of unbudded with a single nucleus and a G1 microtubule array, small budded with a single nucleus and short spindle, large budded with a single nucleus and short spindle, and large budded with separated DNA masses and long spindle are indicated from left to right. Two hundred cells were counted for each strain.
DOC1 was cloned by complementing the temperature sensitivity of doc1–1 with a genomic yeast library on a centromeric vector (Figure 3B, plasmid T1). The minimal complementing region contains a single ORF coding for a protein of 283 amino acids. A construct containing the ORF of DOC1 and 508 bp upstream, with a single myc tag on the C terminus and no sequences 3′ to the ORF, complements the temperature sensitivity and Clb2 proteolysis defect of the doc1–1 mutant (Figure 3B, plasmid pLH24). In addition, a construct containing a frameshift at an NdeI site 297 bp into the DOC1 ORF, will not rescue the temperature sensitivity of doc1–1 (Figure 3B, plasmid pLH26–1B). Disruption of DOC1 by insertion of the URA3 gene at the NdeI site (Figure 3B, plasmid pLH26–1A) results in viable cells that grow poorly at 23°C, forming colonies in which most of the cells have large buds and cannot grow at 37°C. Coincidentally, DOC1 lies directly upstream of the CSE1 gene, which is also implicated in cyclin proteolysis (Xiao et al., 1993; Irniger et al., 1995). These genes have distinct functions in proteolysis because plasmids that contain only the CSE1 gene do not complement doc1–1. The Doc1 amino acid sequence is shown in Figure 3C. The Doc1 sequence lacks obvious motifs and shows no homology to proteins of known function; however, it does show homology to rat (Lee et al., 1995) and mouse expressed sequence tag sequences (GenBank accession numbers H33761 and W49295, respectively). The doc1–1 mutation was recovered and sequenced. The mutation changes a conserved serine (Ser-137) to phenylalanine.
Cdc26 and Doc1 Associate with APC Components (Cdc16, Cdc23, and Cdc27)
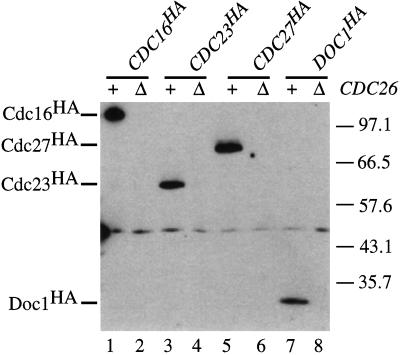
Cdc16, Cdc23, and Cdc27 form a complex in yeast (Lamb et al., 1994) and their homologues are components of a 20S complex in frog egg extracts (King et al., 1995). This complex behaves as a ubiquitin ligase for B-type cyclins, and CDC16, CDC23, and CDC27 are required for the ubiquitination of Clb2 in yeast (Zachariae and Nasmyth, 1996). We therefore tested whether Cdc26 or Doc1 associated with Cdc16, Cdc23, or Cdc27. Extracts were made from exponentially growing wild-type or cdc26Δ strains that had been transformed with centromeric plasmids containing HA-tagged versions of CDC16, CDC23, CDC27, and DOC1. Cdc26 was immunoprecipitated with polyclonal antibodies against Cdc26. The bound proteins were probed by Western blotting with mouse monoclonal antibodies against HA (12CA5) to test whether the tagged proteins coimmunoprecipitate with Cdc26.
Figure 4 shows that immunoprecipitation of Cdc26 in a wild-type strain results in the coimmunoprecipitation of HA-tagged Cdc16, Cdc23, Cdc27, and Doc1. No HA-tagged Cdc16, Cdc23, Cdc27, or Doc1 was detected when immunoprecipitations were performed on extracts made from a cdc26Δ strain. These experiments suggest that Cdc26 is a functional component of the APC, and this suggestion is strengthened by the observation that the Xenopus homologue of Cdc26 is a component of the purified APC (M. Kirschner, personal communication).
Figure 4.
Coimmunoprecipitation of Cdc26 with epitope-tagged forms of Cdc16, Cdc23, Cdc27, and Doc1 (Cdc16HA, Cdc23HA, Cdc27HA, and Doc1HA). Wild-type (+) and cdc26Δ (Δ) strains containing centromeric plasmids expressing Cdc16HA (pWAM10; lanes 1 and 2), Cdc23HA (pRS239; lanes 3 and 4), Cdc27HA (pRS248; lanes 5 and 6), or Doc1HA (pLH32; lanes 7 and 8) were grown to logarithmic phase in selective medium. Cdc26 was immunoprecipitated from those cell lysates with polyclonal anti-Cdc26 antibodies. Immunoprecipitates were analyzed by immunoblotting with 12CA5. The positions of molecular mass markers and Cdc16HA, Cdc23HA, Cdc27HA, and Doc1HA are indicated in kilodaltons.
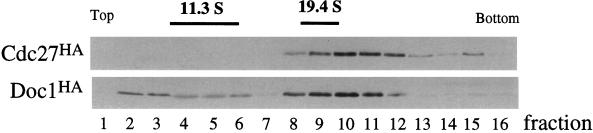
The association of Doc1 with Cdc26 could be interpreted in two ways. Either Doc1 is a component of the APC or there are two pools of Cdc26, one associated with the APC and one bound to Doc1. To distinguish these possibilities, we analyzed the sedimentation of Doc1 and Cdc27 in a sucrose gradient. Extracts from exponentially growing cells expressing both HA-tagged Doc1 and HA-tagged Cdc27 were separated on a 5–40% sucrose gradient (Figure 5). The majority of Doc1 sediments in a peak at approximately 20S, coinciding with the sedimentation of Cdc27. The sedimentation coefficient of the frog and clam APC is 20S (King et al.., 1995; Sudakin et al., 1995).
Figure 5.
Cosedimentation of Doc1 with Cdc27. A lysate from exponentially growing cells expressing both Doc1 and Cdc27 tagged with the HA epitope (Doc1HA and Cdc27HA) was separated through a 5–40% sucrose gradient. Fractions were analyzed by immunoblotting with 12CA5.
Genetic Interactions among CDC26, DOC1, and Other Mitotic Mutants
Genetic interactions between mutants often indicate that they function in the same pathway. We found that double mutants of cdc26–100 and doc1–1 are synthetically lethal (Table 4), suggesting that the functions of Cdc26 and Doc1 overlap. In addition, cdc26–100 is synthetically lethal with cdc16–1 and cdc23–1, and cdc26–100 cdc27–1 double mutants have a lower permissive temperature than either single mutant. doc1–1 double mutants with cdc16–1 or cdc23–1 have a lower permissive temperature than any of the single mutants. The interactions of cdc26–100 and doc1–1 with mutants involved in the ubiquitination of mitotic cyclins do not simply reflect synthetic interactions between any pair of mitotic mutants. Double mutants between cdc26–100 or doc1–1 and cim5–1 (a subunit of the 26S proteasome, required for the destruction of ubiquitinated proteins; Ghislain et al., 1993) have the same nonpermissive temperature as the cdc26–100 or doc1–1 single mutants.
Table 4.
Genetic interactions between cdc26, doc1, and other mitotic mutants
| cdc26-100 | doc1-1 | cdc16-1 | cdc23-1 | cdc27-1 | cim5-1 | |
|---|---|---|---|---|---|---|
| Maximum permissive temperature (°C) | 33 | 33 | 30 | 30 | 33 | 37 |
| Maximum permissive temperature of double mutant with cdc26-100 (°C) | 33 | Dead | Dead | Dead | 30 | 33 |
| Maximum permissive temperature of double mutant with doc1-1 (°C) | Dead | 33 | 23 | 23 | 30 | 33 |
Synthetic interactions among cdc26-100, doc1-1, and various mitotic mutants. cdc26-100 and doc1-1 were mated to each other or to cdc16-1, cdc23-1, cdc27-1, or cim5-1. The resultant diploids were sporulated and tetrads were dissected and germinated at 23°C. They were then tested for growth at 23°C, 30°C, 33°C, 35°C, and 37°C. The genotype of viable spores was determined by complementation testing with appropriate mutant tester strains. The maximum permissive temperatures of the cdc26-100 and doc1-1 mutants are shown in boldface type.
DISCUSSION
We have identified two new components of the cyclin proteolysis machinery, Cdc26 and Doc1, and show that Cdc26 and Doc1 are physically associated with the APC/cyclosome, the multiprotein complex that acts as an E3 for cyclin ubiquitination (King et al., 1995; Sudakin et al., 1995; Zachariae and Nasmyth, 1996). Similar results were recently obtained for CDC26 by Zachariae et al. (1996).
We isolated temperature-sensitive mitotic mutants by using an enrichment that kills the majority of cells passing through S phase at the nonpermissive temperature. Of the 32 temperature-sensitive mitotic arrest mutations we isolated, 27 are in genes involved in the cyclin proteolysis machinery; 1 is in CDC31, a gene required for spindle pole body duplication; and 2 are in PRP22, which encodes a splicing function that is probably required to ensure adequate levels of α-tubulin synthesis. Why do proteolysis mutants dominate the metaphase arrest mutants isolated in both this screen and the original Hartwell screen for cdc mutants? We believe that three factors account for this observation. First, although defects in the mitotic spindle can arrest cells in mitosis by transiently activating the spindle assembly checkpoint, the checkpoint eventually adapts to persistent defects thus allowing cells to exit mitosis (Hardwick, Rudner, Wiess, Winey, and Murray, unpublished results). Because cells that adapt and leave mitosis with a defective spindle are likely to suffer lethal errors in chromosome segregation, mutants that cause spindle defects may have died during the prolonged incubation at 37°C that was used during the mutant enrichment. Second, there appears to be considerable functional overlap among the components that assemble the mitotic spindle, so that mutations that inactivate single components do not cause a mitotic arrest. For example, strains lacking the Kar3 microtubule motor are viable, although mitosis in these cells is clearly abnormal (Meluh and Rose, 1990). Finally, the integrity of some spindle functions, such as the protein kinase Mps1 (Hardwick et al., 1996), are required for the spindle assembly checkpoint to detect defects in the spindle. As a result, lesions in these components would not arrest cells in mitosis.
cdc26 and doc1 mutants arrest in metaphase and cannot destroy mitotic cyclins that are expressed in G1-arrested cells. Unlike mutations in the proteasome, cdc26 and doc1 mutants do not suffer general defects in ubiquitin-mediated proteolysis. A substrate that is recognized by a destabilizing N-terminal amino acid or a noncleavable N-terminal ubiquitin is degraded normally in cdc26 and doc1 mutants. Thus, these observations suggest that the mitotic arrest of cdc26 and doc1 reflects their inability to degrade mitotic cyclins and proteins, such as Cut2 and Pds1, whose destruction is required for sister chromatid separation (Funabiki et al., 1996; Yamamoto et al., 1996). This conclusion is supported by both genetic and biochemical evidence for interactions of Cdc26 and Doc1 with the APC. Immunoprecipitates prepared with anti-Cdc26 antibodies contain Cdc16, Cdc23, and Cdc27, all of which are characterized components of the APC, as well as Doc1. In addition, a population of Doc1 cosediments with Cdc27. This evidence suggests that both Cdc26 and Doc1 are associated with the APC. Because we cannot easily measure the amounts of Cdc26 and Doc1 relative to other components of the APC, it is unclear whether Cdc26 and Doc1 are stoichiometric components of the APC. The observation that neither Cdc26 or Doc1 show any homology to cloned subunits of the biochemically purified Xenopus APC suggests that Cdc26 and Doc1 cannot be tightly associated with all APC complexes (King et al., 1995). The observation that doc1–1 is synthetically lethal with cdc26–100, but not with mutants in other components of the APC, suggests that Doc1 and Cdc26 may play partially overlapping roles in cyclin proteolysis. Although cdc26Δ mutants grow well at 23°C, they are synthetically lethal with cdc16–1 and cdc23–1, suggesting that Cdc26 may play a role in stabilizing the APC and that this role is dispensable under optimal conditions but becomes essential under environmental stress or in the presence of defects in other APC components.
Defects in the mitotic spindle activate a spindle assembly checkpoint that arrests eukaryotic cells in mitosis (Hoyt et al., 1991; Li and Murray, 1991). Activation of the checkpoint prevents the destruction of mitotic cyclins and sister chromatid separation, suggesting that the principal action of the checkpoint is to inhibit the proteolysis of mitotic cyclins and proteins that play a role in maintaining the linkage between sisters. It is currently unclear whether this inhibition is due to inactivation of the APC itself or to reactions that protect specific substrates from a normally active APC in checkpoint-arrested cells. Further biochemical comparison between APC components in anaphase- and checkpoint-arrested extracts should help to resolve this issue.
ACKNOWLEDGMENTS
We thank Phil Hieter, Lee Hartwell, Mark Winey, Carl Mann, Erica Johnson, Kevin Hardwick, Doug Kellogg, and Aaron Straight for yeast strains and plasmids; Cathy Mistrot for technical assistance; the members of the Murray and Morgan labs for invaluable discussions and critical reading of the manuscript, especially Sue Biggins, Rey-Huei Chen, Hironori Funabiki, Marion Shonn, Dana Smith, and Alex Szidon. L.H.H. is supported by a National Science Foundation Fellowship. This work was supported by grants to A.W.M. from the National Institutes of Health, the March of Dimes, and the David and Lucile Packard Foundation.
REFERENCES
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. doi: 10.1016/0092-8674(94)90443-x. [DOI] [PubMed] [Google Scholar]
- Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- Araki H, Awane K, Ogawa N, Oshima Y. The CDC26 gene of Saccharomyces cerevisiae is required for cell growth only at high temperature. Mol Gen Genet. 1992;231:329–331. doi: 10.1007/BF00279807. [DOI] [PubMed] [Google Scholar]
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–185. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Baum P, Furlong C, Byers B. Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc Natl Acad Sci USA. 1986;83:5512–5516. doi: 10.1073/pnas.83.15.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Byers B. Multiple roles of the spindle pole bodies in the life cycle of Saccharomyces cerevisiae. Mol Genet Yeast. 1981;16:119–133. [Google Scholar]
- Byers B, Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harbor Symp Quant Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- Company N, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- Dawson IA, Roth S, Artavanis-Tsakonas S. The Drosophila cell cycle gene fizzy is required for normal degradation of cyclins A and B during mitosis and has homology to the CDC20 gene of Saccharomyces cerevisiae. J Cell Biol. 1995;129:725–737. doi: 10.1083/jcb.129.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yangida M. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366:358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Hardwick K, Murray AW. Mad1p, a phosphoprotein component of the spindle assembly checkpoint in budding yeast. J Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies. A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hartwell LH, Culotti J, Reid B. Genetic control of cell division in yeast, I. Detection of mutants. Proc Natl Acad Sci USA. 1970;66:352–359. doi: 10.1073/pnas.66.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985;110:381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ganoth D, Pehrson J, Palazzo RE, Cohen LH. Methylated ubiquitin inhibits cyclin degradation in clam embryo extracts. J Biol Chem. 1991;266:16376–16379. [PubMed] [Google Scholar]
- Holloway SL, Glotzer M, King RW, Murray AW. Anaphase is initiated by proteolysis rather than by the inactivation of MPF. Cell. 1993;73:1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Trotis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;77:1037–1050. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Bartel B, Seufert W, Varshavsky A. Ubiquitin as a degradation signal. EMBO J. 1992;11:497–505. doi: 10.1002/j.1460-2075.1992.tb05080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Lamb JR, Michaud WA, Sikorski RS, Hieter PA. Cdc16p, Cdc23p and Cdc27p form a complex essential for mitosis. EMBO J. 1994;13:4321–4328. doi: 10.1002/j.1460-2075.1994.tb06752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NH, et al. Comparative expressed sequence tag analysis of differential gene expression profiles in PC-12 cells before and after nerve growth factor treatment. Proc Natl Acad Sci USA. 1995;92:8303–8307. doi: 10.1073/pnas.92.18.8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- Meluh PB, Rose MD. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Morgan DO. Principles of CDK regulation. Nature. 1995;374:131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- Murray AW, Kirschner MW. Dominoes and clocks: the union of two views of cell cycle regulation. Science. 1989;246:614–621. doi: 10.1126/science.2683077. [DOI] [PubMed] [Google Scholar]
- Murray AW, Solomon MJ, Kirschner MW. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989;339:280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- Nurse P. Universal control mechanism regulating cell cycle timing of M-phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- Palmer RE, Koval M, Koshland D. The dynamics of chromosome movement in the budding yeast Saccharomyces cerevisiae. J Cell Biol. 1989;109:3355–3366. doi: 10.1083/jcb.109.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi N, Monteagudo MC, Koshland D, Hogan E, Burke DJ. The CDC20 gene product of Saccharomyces cerevisiae, a β-transducin homolog, is required for a subset of microtubule-dependent cellular processes. Mol Cell Biol. 1991;11:5592–5602. doi: 10.1128/mcb.11.11.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink G, Lawrence C. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1974. [Google Scholar]
- Sigrist S, Jacobs H, Stratmann R, Lehner C. Exit from mitosis is regulated by Drosophila fizzy and the sequential destruction of cyclins A, B, and B3. EMBO J. 1995;14:4827–4838. doi: 10.1002/j.1460-2075.1995.tb00164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitination ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–198. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Xu H, Grunstein M. CDC14 of Saccharomyces cerevisiae. Cloning, sequence analysis, and transcription during the cell cycle. J Biol Chem. 1992;267:11274–11280. [PubMed] [Google Scholar]
- Weinert TA. Dual cell cycle checkpoints sensitive to chromosome replication and DNA damage in the budding yeast Saccharomyces cerevisiae. Radiat Res. 1992;132:141–143. [PubMed] [Google Scholar]
- Xiao Z, McGrew JT, Schroeder AJ, Fitzgerald-Hayes M. CSE1 and CSE2, two new genes required for accurate mitotic chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:4691–4702. doi: 10.1128/mcb.13.8.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YA, Guacci V, Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. TPR proteins required for anaphase progression mediate ubiquitination of mitotic B type cyclins in yeast. Mol Biol Cell. 1996;7:791–801. doi: 10.1091/mbc.7.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Shin TH, Galova M, Obermaier B, Nasmyth K. Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science. 1996;274:1201–1204. doi: 10.1126/science.274.5290.1201. [DOI] [PubMed] [Google Scholar]