Abstract
Genome-wide model free linkage analysis was conducted for nicotine dependence and tobacco use phenotypes in 607 members of 158 nuclear families consisting of at least two ever smokers (100 or more cigarettes smoked in lifetime). DNA from whole blood was genotyped for 739 autosomal microsatellite polymorphisms with an average inter-marker distance of 4.6 cM. A peak LOD score of 2.7 was observed on chromosome 6 for scores for the Fagerström Test for Nicotine Dependence. Exploratory analyses were conducted to determine whether sequence variation at other loci affected other measures of dependence or tobacco use. Four additional loci with LOD scores of 2.7 or more were associated with alternative measures of nicotine dependence, one with current frequency of use, and one with smoking cessation. Several of the corresponding support intervals were near putative loci reported previously (on chromosomes 6, 7, and 8) while others appear to be novel (on chromosomes 5, 16, and 19).
Keywords: nicotine, dependence, linkage, genetics, smoking
Introduction
Twin studies have identified a significant genetic component to several aspects of tobacco use initiation and persistence [Carmelli et al., 1992; Heath et al., 1995; Li et al., 2003a; Sullivan and Kendler, 1999; Swan and Carmelli, 1997; Tyndale, 2003] and, more recently, to clinically meaningful phenotypes such as indicators of nicotine dependence (ND) [Kendler et al., 1999; Lessov et al., 2004; McGue et al., 2000; True et al., 1999], “failed cessation” [Xian et al., 2003], and nicotine withdrawal [Xian et al., 2003]. While none of the published twin studies utilized the Fagerström Test for Nicotine Dependence questionnaire (FTND) [Heatherton et al., 1991], considered by many to be the “gold-standard” for measuring ND, one study [Lessov et al., 2004] has reported high heritability (71%) for the Heaviness of Smoking Index [Heatherton et al., 1989], which is composed of two of the six FTND items. The one published non-twin study of familial aggregation of ND assessed the FTND in sibling pairs and found evidence for significant aggregation for ND with a recurrence risk of 1.7 [Niu et al., 2000].
There have been several published reports involving linkage between a variety of tobacco use phenotypes and specific genomic regions. Collectively, these investigations have relied on four population cohorts: the Collaborative Studies on the Genetics of Alcoholism cohort [Bergen et al., 1999; Bierut et al., 2004; Duggirala et al., 1999], the Framingham Heart Study cohort [Goode et al., 2003; Li et al., 2003b; Saccone et al., 2003], the Christchurch, New Zealand cohort [Straub et al., 1999; Sullivan et al., 2004], and a family study of panic disorder [Gelernter et al., 2004]. The first attempt to map susceptibility loci for ND per se utilized the precursor of the FTND, the Fagerström Tolerance Questionnaire [Fagerstrom, 1978] (FTQ; ND defined as a score of 7 or more) in a convenience sample of 130 families from Christchurch, New Zealand [Straub et al., 1999]. While initial results by Straub et al. showed only modest evidence for linkage with specific regions (the strongest being a sharp peak at or near D2S1326), a subsequent reanalysis of the same data using different methods detected the same peak with an estimated Z score of about 2.5 [Sullivan et al., 2004].
The present study, based on a cohort that was originally constructed to identify predictors of incident smoking behavior among adolescents, was primarily designed to map loci for ND as well as for other tobacco-related phenotypes. A LOD score of 2.7 was detected on chromosome 6 for the FTND phenotype. Because we suspect that evidence for linkage depends on the specific tobacco-related phenotype that is assessed, we explored other measures of ND and tobacco use in this analysis. The analysis of these additional phenotypes could provide insight regarding other phenotypes that also segregate at ND loci, and such knowledge could facilitate replication in the future.
Subjects and Methods
Setting
An ongoing interdisciplinary study to identify environmental and genetic determinants of tobacco use collected data from families ascertained through index participants (probands) in the Smoking in Families Study [SMOFAM; Hyman Hops, PI, Oregon Research Institute, DA003706) [Swan et al., 2003a]. The SMOFAM study, initiated in 1981, is a comprehensive, repeated measures cohort study of environmental and psychosocial risk factors for adolescent and young adult substance use, including tobacco [Hops et al., 2000]. All methods and procedures described herein were reviewed and approved by the Institutional Review Boards of the collaborating institutions (SRI International, Oregon Research Institute, and the University of California at San Francisco) and are in accord with the Code of Ethics of the World Medical Association (Declaration of Helsinki) and the standards of the University of California Tobacco-Related Diseases Research Program.
Subjects
The procedures and design of this study are described elsewhere [Swan et al., 2003a]. Briefly, the sample for our analysis consisted of a subset of 481 SMOFAM members who had participated in the longitudinal study. Compared to the original SMOFAM sample, the volunteers for this genetic investigation were more likely to be female, younger, and less likely to have smoked more than once. The majority (84%) of the probands identified themselves as of Caucasian origin with the remainder reporting Hispanic (2%), Native American (1%), or mixed (13%) origin.
Ascertainment
SMOFAM study participants who completed at least seven of the first 10 repeat assessments were contacted for a Family History of Tobacco Use (FHTU) interview. Four hundred and eighty one SMOFAM study participants completed the interview. The FHTU interview was used to determine pedigree structure of the nuclear family, vital status, and lifetime “ever” smoking status (i.e., 100 or more cigarettes smoked) for each first-degree relative. Using these data, families were stratified based on “density” of the ever-smoking phenotype (i.e., the number of biological members who had smoked 100 or more cigarettes in their lifetime) within the nuclear family. An essential criterion for prioritizing families was that the family be composed of at least two living, first-degree, ever-smoking members. In anticipation of the genetic analysis, eligible families had to consist of a minimum “triad” configuration, defined as two biological parents plus an ever-smoking offspring, or a biological sibling pair plus a biological parent (with one ever-smoker among them). Regardless of their individual ever-smoking status, the proband and all first-degree relatives within these high priority families were invited to complete the Smoking History Questionnaire (SHQ) and to provide a blood sample [Swan et al., 2003a]. ND and tobacco use-related phenotypes examined in the linkage analyses were derived from the SHQ.
Phenotype Assessment
A consensus has emerged in which ND is viewed as multidimensional and, therefore, should be assessed and quantified accordingly [Colby et al., 2000; Hudmon et al., 2003; Lombardo et al., 1988; Moolchan et al., 2002; Pomerleau et al., 1993]. Our primary interest was to identify loci that segregate with ND as determined by the FTND. Additional measures were also included to capture the complexity of the tobacco dependence phenotype (Table 1). These included: (a) elements from the DSM-IV dependence criteria [American Psychiatric Association 1994]; (b) smoking frequency and quantity; and, (c) quitting history. Individuals who never tried even a puff of a cigarette are excluded from all definitions. Individuals who tried smoking and those who smoked 100 or more cigarettes lifetime but were never daily smokers are included in the zero category of DSM-IV related measures. All other measures included lifetime daily smokers only. The footnote to Table 1 provides explicit definitions for the phenotypes used in this study grouped into three main categories.
Table I.
Phenotype Means (SD) for Continuous Measures and Prevalence Estimates (%) for Categorical Measures in the Parental and Proband Generations and Among Probands Only.
| Phenotype (range) | Parental generation | Proband generation1 | Proband | |||
|---|---|---|---|---|---|---|
| N (range)2 | 178-275 | 213-336 | 105-160 | |||
| Dependence | ||||||
| FTND (0-10)3 | 4.7 (2.8) | 3.4 (2.4) | 3.2 (2.4) | |||
| DSM-IV-Like Severity: sum of 7 ND criteria (lifetime) (0-7)4 | 3.6 (2.2) | 3.3 (2.1) | 3.4 (1.9) | |||
| DSM-IV-Like Dependence: 3 or more of 7 ND criteria (lifetime) (% yes)5 | 74.0 | 70.3 | 73.9 | |||
| Withdrawal Severity: sum of 9 withdrawal symptom scores (0-27)6 | 10.4 (7.2) | 8.7 (6.5) | 8.5 (6.4) | |||
| Tobacco use | ||||||
| Quantity: number cigarettes smoked per day (past 3 months) (0-80)* | 4.7 (11.0) | 6.0 (9.0) | 6.5 (9.0) | |||
| Current Frequency: smoking one or more cigarettes on most days (% yes)* | 21.5 | 39.1 | 43.7 | |||
| Quitting history | ||||||
| Quit smoking cigarettes completely (% yes)* | 65.3 | 33.8 | 32.8 | |||
| Short-Term Quit: ever quit smoking for > 1 month but < 1 year (% yes)* | 54.1 | 56 | 49.5 | |||
The sample size for the proband generation includes the probands themselves.
The minimum and maximum sample size available for characterization of each generational grouping is given at the top of each column. The sample sizes vary due to the inclusion of data from lifetime smokers only for some of the phenotypes.
Fagerström Test for Nicotine Dependence summary score assessed during the heaviest period of smoking lifetime.
DSM-IV criteria were constructed using items from the DSM-based Nicotine Dependence Scale [Snedecor et al., 2004] in combination with other questionnaire items. DSM-IV ND was defined as a continuous severity summary score across seven ND criteria4 (tolerance, withdrawal, ever smoked more than meant to, difficulty quitting, great deal of time spent obtaining cigarettes, give up important social or occupational activities in order to smoke, continue smoking despite health problems caused or exacerbated by smoking), as well as a categorical measure to indicate the reporting of 3 or more of 7 ND criteria lifetime5.
Summation of nine withdrawal symptom scores [Hughes and Hatsukami, 1986; Jarvis et al., 1982]; depressed mood, trouble falling asleep, irritability/frustration/anger, anxiety, difficulty concentrating, restlessness, increased appetite/weight gain, cravings to smoke, decreased heart rate) rated on a 4-point scale, 0 = not at all, 3 = severe.
Definitions of these four phenotypes are self-explanatory.
Laboratory Analysis
Genotyping was done as described previously [Wilhelmsen et al., 2003]. DNA was isolated from frozen blood and DNA concentration was determined by optical density. Genotypes were determined for 739 autosomal microsatellite polymorphisms [Weber and May, 1989] (HD5, Applied Biosystems, 850 Lincoln Center Drive, Foster City, CA 94404). The average marker-to-marker distance was 4.6 cM with an average heterozygosity of greater than 77% in a Caucasian population. The sizes of marker amplimers were determined blind to pedigree structure and phenotype. Allele frequencies were estimated from the study population.
Pedigree Structure and Genotype Assessment
Genotypes were determined for 607 of the 867 participants who completed the SHQ assessment. Accuracy of pedigree structure and genotype determination was crucial for the planned genetic analyses, because it has been suggested elsewhere that a genotype error rate as small as 1% can significantly affect the results of a genomic linkage scan [Douglas et al., 2000]. A multistage data analysis approach was used to minimize errors in pedigree structure, sample identity, and genotypes. All available genotypes for all of the autosomal markers were analyzed for each family using PREST [McPeek and Sun, 2000] to detect sample and pedigree structure errors. Both phenotype and genotype data were excluded from further analysis when genotyping did not confirm the recorded pedigree structure (fewer than 1% of all participants). One hundred and fifty eight families had individuals with both phenotype and genotype information. In most families, siblings and some of their parents participated. An average of 3.8 (SD = 1.2) individuals participated per family; 269 sibling-sibling, 570 parent-child, 3 half-sibling and 112 spousal pairs with complete genotype and phenotype data were included in the analysis.
Pedcheck was used to detect non-Mendelian inheritance patterns [O'Connell and Weeks, 1998]. Relevant genotypes were reviewed blind to phenotypic status. For each Mendelian inconsistency, genotypes for the nuclear family were removed. To further reduce errors, the probability that each genotype was correct was assessed in the context of all other available genotypes using the maximum-likelihood error-checking algorithm implemented in Merlin [Abecasis et al., 2002]. Genotypes with a probability of less than 0.025 of being correct were removed from further consideration. Less than 0.5% of all genotypes were excluded.
Linkage Analysis
Multipoint LOD scores were determined using the QTL statistic implemented in Merlin (Abecasis et al. 2002). Merlin uses sparse binary trees to represent patterns of gene flow in general pedigrees to calculate exact multipoint estimates of IBD. The Merlin QTL statistic is a non-parametric statistic implemented in the general framework of Whittemore and Halpern (1994) as extended by Kong and Cox (1997). Discrete traits were coded as 2 for affected individuals and 1 for unaffected individuals but otherwise treated as continuous variables. In this approach a score, based on one of many possible functions, is computed for each family for the observed inheritance vector as well as all possible inheritance vectors. A family specific Z score is calculated based on the observed score and the mean and variance of scores for all possible inheritance vectors, and the weighted family specific Z scores are combined to produce an overall score. Merlin calculates a founder allele score for each family, which is the sum of the phenotype (less the sample mean) for each family member that carries the founder allele. The score function used by the QTL statistic is the sum of the square of the founder allele scores. As with all scores, the family scores are compared to the alternative scores for that family for all possible inheritance vectors. The contribution to the QTL statistic of individual families is weighted by family size. The QTL statistic is less likely to be affected by extreme phenotypes and the distribution of the phenotypes since the scores for each family are normalized by the mean of all possible scores for that family. In contrast, the maximum-likelihood based variance-component (Blangero et al. 2000) method is more likely to be biased by outliers. The use of the QTL statistic in this case allowed for a greater proportion of the phenotypic information than the more commonly used affected sib pair methods (e.g. NPL and ALL, Kruglyak et al. 1996) because information from both affected and unaffected individuals could be used.
The robustness of the QTL statistic to violations of multivariate normality, as implemented in Merlin (Abecasis et al. 2002) has not been reported. To assess the statistical significance of the observed QTL LOD scores, 10000 genomes of genotype data were constructed independently of the measured phenotypes using the simulation option in Merlin. The algorithm constructed haplotypes for the founders based on the estimated allele frequencies in the original sample and then allowed them to segregate through the families. Only genotypes consistent with the measured sample were retained for analysis. The Merlin QTL option normalizes phenotypes to the population mean and constructs a score statistic, which is robust to non-normality. Genome-wide multipoint LOD scores were then calculated using the original measured phenotypes. All LOD score peaks were identified and used to estimate the number of times that a peak was observed above a specific LOD score/per simulated random genome.
The frequency peaks above specific thresholds per simulated random genome were calculated. It was observed that with this set of families and markers, independent of phenotype, the number of times a peak above a threshold was observed per random genome was equivalent across phenotypes. This was true for both continuous variables as well as for dichotomous traits. LOD scores of 2.7 and 3.4 occurred an average of 1.0 and 0.1 times per random simulated genome, respectively. The uniformity of the number of peaks above specific thresholds for all phenotypes suggested that the QTL statistic is robust to deviations from multivariate normality in a moderately large set of nuclear families with dense genotyping data.
Results
Description of Members of the Proband and Parental Generations
Participating SMOFAM members of the proband generation were 28.2 years of age (SD = 4.4), while members of the parental generation averaged 54.3 years of age (SD = 4.9). Approximately 51% of the proband generation was female. The prevalence of education beyond high school was 63.3% among the proband generation and 76.9% among parents. Twenty percent of the proband generation and 41% of parents earned $50,000 or more annually. Consistent with the initial construction of the SMOFAM cohort and subsequent recruitment of families, there was an overall high prevalence of current smoking among both generations with a higher prevalence among probands and siblings (39.1%) than among their parents (21.5%). The study sample exhibited a higher prevalence of lifetime DSM-IV-like ND, 74.0% and 70.3% for members of the parental and proband generations, respectively, than does the general U.S. population (24.1%); [Kandel et al., 1997; Kandel and Chen, 2000].
Table 1 provides descriptive statistics for the phenotypes used in this analysis for both parental and proband generations as well as for the probands themselves. The minimum and maximum sample size available for characterization of each generational grouping is given at the top of each column. The sample sizes vary due to the inclusion of data from lifetime smokers only for some of the phenotypes. The table reveals higher FTND scores for parents (M = 4.7) than for members of the proband generation (M = 3.4).
Primary Linkage Analysis
For the FTND summary score a maximum LOD score of 2.7 was seen at 178 cM on chromosome 6 (Figure 1 and Table 2). The marker closest to the peak was D6S446. The support interval (defined as the region in which LOD scores are greater than the maximum LOD score – 1) included 156 to 191 cM (D6S1581-D6S281). The next largest LOD score peak had a maximum of 2.4 at 88 cM on chromosome 20 (data not shown).
Fig. 1.
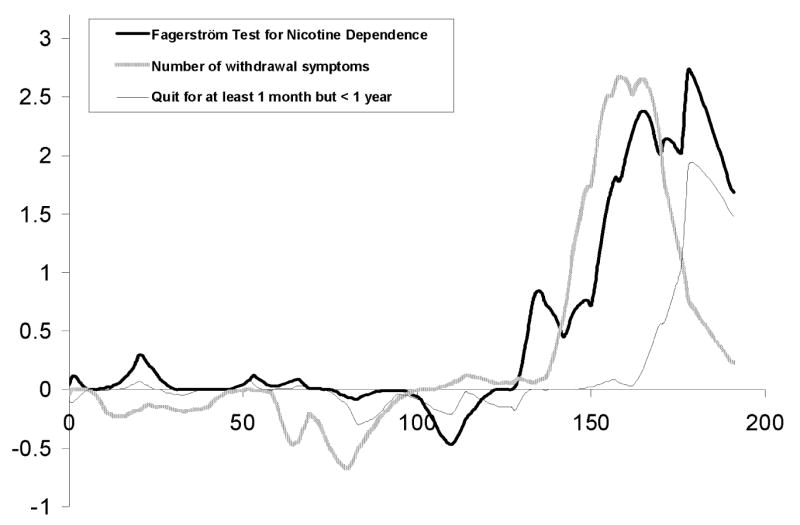
Multipoint LOD scores are shown for chromosome 6 FTND (thick solid black line), Withdrawal Severity (thick gray line), and Short-Term Quit (gray thin line) phenotypes.
Table II.
Multipoint Linkage Results for Nicotine Dependence Phenotypes.
| Chr | Phenotype (Category) | Peak cM (Support Interval) | Max LOD | Closest marker to peak |
|---|---|---|---|---|
| 5 | Quantity: number cigarettes smoked per day (past 3 months) | 62 (57-70) | 2.9 | D5S1969 |
| 6 | FTND | 178 (156-190) | 2.7 | D6S446 |
| Withdrawal Severity: sum of 9 withdrawal symptom scores | 159 (148-173) | 2.7 | ||
| 7 | DSM-IV-Like Severity: sum of 7 ND criteria (lifetime) | 164 (159-167) | 3.0 | D7S636 |
| 8 | DSM-IV-Like Dependence: 3 or more of 7 ND criteria (lifetime) | 31 (23-39) | 2.7 | D8S258 |
| 16 | Short-Term Quit: ever quit smoking for > 1 month but < 1 year | 70 (52-80) | 4.0 | D16S415 |
| 19 | Current Frequency: smoking one or more cigarettes on most days | 90 (83-97) | 2.9 | D19S572 |
Secondary Linkage Analysis
In subsequent analyses we examined additional tobacco use phenotypes for evidence of linkage. To minimize the reporting of results that may be due to chance, we report individual LOD scores of 2.7 or greater (see Table 2).
Other measures of ND
The support interval for Withdrawal Severity overlapped the FTND support interval on chromosome 6 (Figure 1) with a peak LOD score of 2.7. Also shown in Figure 1 is a quitting history phenotype, Short-Term Quit that had a peak LOD score of 1.9 in the same region.
The largest LOD score for any ND phenotype (LOD score = 3.0) was observed for DSM-IV-like ND Severity (see footnotes 4 and 5, Table 1 for a list of the criteria) near D7S636 (164 cM; support interval 159-167 cM; Table 2). For the dichotomous DSM-IV-like ND measure, a maximum LOD score of 2.7 was observed on chromosome 8 at 31 and 35 cM (near marker D8S258).
Tobacco use
Cigarette Quantity produced a peak LOD score of 2.9 on chromosome 5 near marker D5S1969. A peak LOD score of 2.9 was observed on chromosome 19 (at 90 cM near D19S572) for Current Smoking Frequency (Table 2).
Quitting history
The largest linkage peak (LOD score = 4.0) in the entire study was observed for Short-Term Quit at D16S415 (Table 2).
The primary and secondary linkage analyses were re-run after limiting the sample to only those individuals of Caucasian or Hispanic origin (a total of 86% of the entire sample). The primary LOD score for the FTND on chromosome 6 remained unchanged (2.7). The linkage peaks observed on chromosomes 5 (Cigarette Quantity) and 16 (Short-Term Quit) were somewhat reduced but remained above 2.7 and were 2.8 and 3.1, respectively. In the restricted sample, the remaining four secondary linkage peaks fell below the 2.7 cutoff (Withdrawal Severity, DSM-ND Severity, dichotomous DSM-ND, and Current Smoking Frequency).
Discussion
A search for autosomal loci affecting ND and a variety of tobacco-related phenotypes was conducted in nuclear families with a high prevalence of smoking. In discussing these results, we indicate, where possible, their proximity to other reported linkage peaks or candidate genes. Potential candidate genes were selected on the basis of previously reported significant association with tobacco use phenotypes [Tyndale, 2003] or their location within relevant receptor, metabolic, or signal transduction pathways [Sullivan et al., 2004]. Linkage peaks that may not have been reported previously are noted as novel.
A maximum LOD peak on chromosome 6 was detected for the FTND summary score, and this peak was supported by a nearby peak for Withdrawal Severity. Previous work has identified linkage peaks at or near the support interval reported here for FTND [Bergen et al., 1999; Sullivan et al., 2004]. Moreover, the support interval is very close to the OPRM1 gene (mu 1 opioid receptor) and contains MAP3K4 (mitogen-activated protein kinase 4) and LPAAT-delta (lysophosphatidic acid acyltransferase, delta), both candidate genes for ND [Sullivan et al., 2004]. We therefore report this finding with increased confidence.
We are encouraged that several loci reported here have been detected in other linkage studies as well. The support interval on chromosome 7 observed here for DSM-IV-like ND Severity is near the linkage peak, D7S1804, reported previously for the FTQ [Sullivan et al., 2004], and is near the candidate gene HTR5A (5-hydroxytryptamine [serotonin] receptor 5A). The support interval seen in the present study on chromosome 8 for DSM-IV-like ND is near previously reported linkage peaks for the ever smoking phenotype [Bergen et al., 1999] and is close to the candidate genes CHRNA2 (alpha 2 nicotinic acetylcholine receptor) and ADRA1A (alpha 1A adrenergic receptor).
Heterogeneity inherent in ND is apparent when viewing these results. DSM-IV and FTND definitions of ND are poorly correlated with a kappa value of agreement of 0.2 [Moolchan et al., 2002]. The development of the DSM classification of drug dependence was largely based on evidence from the alcohol field and may not be specific to ND. The parent instrument of the FTND, the FTQ, on the other hand, was specifically developed to measure ND. In our study, the FTND was assessed for the heaviest period of smoking in the respondent's life [Hudmon et al., 2005] and, therefore, can be thought of as a lifetime assessment of ND severity that may have a physiological basis. Whether the heterogeneity across chromosomes for indices of ND derives from genetic or measurement sources cannot be determined from the present study and will need to be addressed in future research.
The few previous biometric or measured genetic studies that were designed to evaluate the role of genetic influences on the ability to stop smoking and/or to maintain abstinence support the notion of a genetic substrate. Carmelli et al. [Carmelli et al., 1992] found epidemiological evidence suggestive of genetic involvement in relapse following cessation. Significant heritability estimates have been reported for “failed cessation” (54%) [Xian et al., 2003] and for difficulty quitting (54-68%) [Lessov et al., 2004]. Previous work has determined that variation in dopaminergic (DRD2), opioidergic (OPRM1), and cytochrome P450 metabolic (CYP2B6) genes is associated with the ability to stop smoking following pharmacological treatment [Lerman et al., 2003; Lerman et al., 2004; Miksys et al., 2003; Swan et al., 2005]. The support interval reported on chromosome 16 for Short-Term Quit may be novel and is near several candidate genes proposed recently [Sullivan et al., 2004] including PRKCB1 (Protein kinase C, beta 1), CBFB (core binding factor beta subunit), and ATP6V0D1 (vacuolar H+-ATPase, V0 subunit d isoform 1).
The support interval on chromosome 19 for Current Smoking Frequency also appears to be new and is not near any previously reported peaks. Several candidate genes map near the interval including CYP2A6 (cytochrome P450 2A6), CYP2B6 (cytochrome P450 2B6), and PTPRH (protein tyrosine phosphatase, receptor type, H).
Heritability estimates for most or all of these phenotypes have been reported previously, including: the number of cigarettes smoked per day (range of estimates, 49-72%); [Carmelli et al., 1990; Hettema et al., 1999; Kaprio et al., 1984; Lessov et al., 2004; Swan et al., 1990], FTND (71% for a composite of two items from the scale) [Lessov et al., 2004], DSM-IV ND (present/absent; range of estimates, 56-60%) [Lessov et al., 2004; True et al., 1999], and current smoking (range of estimates 27-53%) [Heath and Martin, 1993; Madden et al., 1999, 2004].
The LOD scores reported here are among the highest reported for ND and related phenotypes. The higher LOD scores observed were in part due to the sample size and ascertainment strategy used. In addition, the linkage analysis was conducted with the QTL score statistic to use all of the available information for each trait. Simulation analysis indicated that the QTL score statistic is robust to significant deviations from normality even to the extent that it can be used for dichotomous traits. Unlike other commonly used methods, the QTL score statistic allows information to be used from both affected and unaffected individuals. Implicit in our use of the QTL score statistic is the assumption that affected and unaffected individuals are equally informative. We think it is likely that non-genetic factors have as big an impact on not smoking as they do on smoking [Heath and Martin, 1993; Madden et al., 1999; Swan et al., 2003a; Swan et al., 2003b; True et al., 1999].
In summary, the present results are encouraging and provide support for the importance of taking into account the phenotypic complexity of ND in genetic investigations. Rather than one global measure of ND as used in previous studies, the use of multiple measures more accurately reflects the current state-of-the-art of the measurement of ND. The genetic contributions to different components of ND appear to be due to different loci. This conclusion is supported by previously published studies. That different loci are associated with different components of ND may be important to the eventual understanding of the underlying genetic architecture of ND.
Acknowledgments
We thank the participants of this study for their time and patience. We acknowledge the effort of James Lee, Samantha Segal, Kymberli Hemberger, Dorit Carmelli, Taline V. Khroyan, Dale McBride, Chris Webster, Barbara Koenig, Lorraine Caron, Judy Illes, and Lisa M. Jack in the conduct of this work. We thank Gonçalo Abecasis for helpful discussions. The authors also wish to thank Ovide and Cynthia Pomerleau, Ray Niaura, and Harold Javitz for assistance in the selection of measurement tools for tobacco-related phenotypes and with data management and analysis. Research supported by grants 7PT2000, 2001, 2002, 2003, and 2004 from the University of California Tobacco-Related Diseases Research Program and by funds from the State of California for medical research on alcohol and substance abuse through the Department of Neurology of University of California at San Francisco.
Support from grants 7PT2000, 2001, 2002, 2003, and 2004 from the University of California Tobacco-Related Diseases Research Program, grant DA03706 from the National Institute of Drug Abuse, and by funds from the State of California for medical research on alcohol and substance abuse through the Department of Neurology of University of California at San Francisco.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders: DSM-IV. 4th 1994. [Google Scholar]
- Bergen AW, Korczak JF, Weissbecker KA, Goldstein AM. A genome-wide search for loci contributing to smoking and alcoholism. Genet Epidemiol. 1999;17 Suppl 1:S55–60. doi: 10.1002/gepi.1370170710. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Rice JP, Goate A, Hinrichs AL, Saccone NL, Foroud T, Edenberg HJ, Cloninger CR, Begleiter H, Conneally PM, Crowe RR, Hesselbrock V, Li TK, Nurnberger JI, Jr, Porjesz B, Schuckit MA, Reich T. A genomic scan for habitual smoking in families of alcoholics: common and specific genetic factors in substance dependence. Am J Med Genet. 2004;124A:19–27. doi: 10.1002/ajmg.a.20329. [DOI] [PubMed] [Google Scholar]
- Blangero J, Williams JT, Almasy L. Robust LOD scores for variance component-based linkage analysis. Genet Epidemiol. 2000;19 Suppl 1:S8–14. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI2>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Carmelli D, Swan GE, Robinette D, Fabsitz R. Genetic influence on smoking--a study of male twins. N Engl J Med. 1992;327:829–833. doi: 10.1056/NEJM199209173271201. [DOI] [PubMed] [Google Scholar]
- Carmelli D, Swan GE, Robinette D, Fabsitz RR. Heritability of substance use in the NAS-NRC Twin Registry. Acta Genet Med Gemellol (Roma) 1990;39:91–98. doi: 10.1017/s0001566000005602. [DOI] [PubMed] [Google Scholar]
- Colby SM, Tiffany ST, Shiffman S, Niaura RS. Measuring nicotine dependence among youth: a review of available approaches and instruments. Drug Alcohol Depend. 2000;59 Suppl 1:S23–39. doi: 10.1016/s0376-8716(99)00163-5. [DOI] [PubMed] [Google Scholar]
- Douglas JA, Boehnke M, Lange K. A multipoint method for detecting genotyping errors and mutations in sibling-pair linkage data. Am J Hum Genet. 2000;66:1287–1297. doi: 10.1086/302861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggirala R, Almasy L, Blangero J. Smoking behavior is under the influence of a major quantitative trait locus on human chromosome 5q. Genet Epidemiol. 1999;17 Suppl 1:S139–144. doi: 10.1002/gepi.1370170724. [DOI] [PubMed] [Google Scholar]
- Fagerstrom KO. Measuring degree of physical dependence to tobacco smoking with reference to individualization of treatment. Addict Behav. 1978;3:235–241. doi: 10.1016/0306-4603(78)90024-2. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Liu X, Hesselbrock V, Page GP, Goddard A, Zhang H. Results of a genomewide linkage scan: support for chromosomes 9 and 11 loci increasing risk for cigarette smoking. Am J Med Genet. 2004;128B:94–101. doi: 10.1002/ajmg.b.30019. [DOI] [PubMed] [Google Scholar]
- Goode EL, Badzioch MD, Kim H, Gagnon F, Rozek LS, Edwards KL, Jarvik GP. Multiple genome-wide analyses of smoking behavior in the Framingham Heart Study. BMC Genet. 2003;4 Suppl 1:S102. doi: 10.1186/1471-2156-4-S1-S102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Madden PA, Slutske WS, Martin NG. Personality and the inheritance of smoking behavior: a genetic perspective. Behav Genet. 1995;25:103–117. doi: 10.1007/BF02196921. [DOI] [PubMed] [Google Scholar]
- Heath AC, Martin NG. Genetic models for the natural history of smoking: evidence for a genetic influence on smoking persistence. Addict Behav. 1993;18:19–34. doi: 10.1016/0306-4603(93)90005-t. [DOI] [PubMed] [Google Scholar]
- Heatherton TF, Kozlowski LT, Frecker RC, Fagerström KO. The Fagerström Test for Nicotine Dependence: a revision of the Fagerström Tolerance Questionnaire. Br J Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- Heatherton TF, Kozlowski LT, Frecker RC, Rickert W, Robinson J. Measuring the heaviness of smoking: using self-reported time to the first cigarette of the day and number of cigarettes smoked per day. Br J Addict. 1989;84:791–799. doi: 10.1111/j.1360-0443.1989.tb03059.x. [DOI] [PubMed] [Google Scholar]
- Hettema JM, Corey LA, Kendler KS. A multivariate genetic analysis of the use of tobacco, alcohol, and caffeine in a population based sample of male and female twins. Drug Alcohol Depend. 1999;57:69–78. doi: 10.1016/s0376-8716(99)00053-8. [DOI] [PubMed] [Google Scholar]
- Hops H, Andrews JA, Duncan SC, Duncan TE, Tildsley E. Adolescent drug use development: A social interactional and contextual perspective. In: Samerhoff A, Lewis M, Miller S, editors. Handbook of Developmental Psychopathology. New York: Kluwer Academic/Plenum Publishers; 2000. pp. 589–605. [Google Scholar]
- Hudmon KS, Marks JL, Pomerleau CS, Bolt DM, Brigham J, Swan GE. A multidimensional model for characterizing tobacco dependence. Nicotine Tob Res. 2003;5:655–664. doi: 10.1080/1462220031000158672. [DOI] [PubMed] [Google Scholar]
- Hudmon KS, Pomerleau CS, Brigham J, Javitz H, Swan GE. Validity of retrospective assessments of tobacco dependence: A preliminary report. Addict Behav. 2005;30:613–617. doi: 10.1016/j.addbeh.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Hughes JR, Hatsukami D. Signs and symptoms of tobacco withdrawal. Arch Gen Psychiatry. 1986;43:289–294. doi: 10.1001/archpsyc.1986.01800030107013. [DOI] [PubMed] [Google Scholar]
- Jarvis MJ, Raw M, Russell MA, Feyerabend C. Randomised controlled trial of nicotine chewing-gum. Br Med J (Clin Res Ed) 1982;285:537–540. doi: 10.1136/bmj.285.6341.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel D, Chen K, Warner LA, Kessler RC, Grant B. Prevalence and demographic correlates of symptoms of last year dependence on alcohol, nicotine, marijuana and cocaine in the U.S. population. Drug Alcohol Depend. 1997;44:11–29. doi: 10.1016/s0376-8716(96)01315-4. [DOI] [PubMed] [Google Scholar]
- Kandel DB, Chen K. Extent of smoking and nicotine dependence in the United States: 1991-1993. Nicotine Tob Res. 2000;2:263–274. doi: 10.1080/14622200050147538. [DOI] [PubMed] [Google Scholar]
- Kaprio J, Koskenvuo M, Langinvainio H. Finnish twins reared apart. IV: Smoking and drinking habits. A preliminary analysis of the effect of heredity and environment. Acta Genet Med Gemellol (Roma) 1984;33:425–433. doi: 10.1017/s0001566000005870. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Neale MC, Sullivan P, Corey LA, Gardner CO, Prescott CA. A population-based twin study in women of smoking initiation and nicotine dependence. Psychol Med. 1999;29:299–308. doi: 10.1017/s0033291798008022. [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ. Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- Lerman C, Shields PG, Wileyto EP, Audrain J, Hawk LH, Jr, Pinto A, Kucharski S, Krishnan S, Niaura R, Epstein LH. Effects of dopamine transporter and receptor polymorphisms on smoking cessation in a bupropion clinical trial. Health Psychol. 2003;22:541–548. doi: 10.1037/0278-6133.22.5.541. [DOI] [PubMed] [Google Scholar]
- Lerman C, Wileyto EP, Patterson F, Rukstalis M, Audrain-McGovern J, Restine S, Shields PG, Kaufmann V, Redden D, Benowitz N, Berrettini WH. The functional mu opioid receptor (OPRM1) Asn40Asp variant predicts short-term response to nicotine replacement therapy in a clinical trial. Pharmacogenomics J. 2004;4:184–192. doi: 10.1038/sj.tpj.6500238. [DOI] [PubMed] [Google Scholar]
- Lessov CN, Martin NG, Statham DJ, Todorov AA, Slutske WS, Bucholz KK, Heath AC, Madden PA. Defining nicotine dependence for genetic research: evidence from Australian twins. Psychol Med. 2004;34:865–879. doi: 10.1017/s0033291703001582. [DOI] [PubMed] [Google Scholar]
- Li MD, Cheng R, Ma JZ, Swan GE. A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction. 2003a;98:23–31. doi: 10.1046/j.1360-0443.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- Li MD, Ma JZ, Cheng R, Dupont RT, Williams NJ, Crews KM, Payne TJ, Elston RC. A genome-wide scan to identify loci for smoking rate in the Framingham Heart Study population. BMC Genet. 2003b;4 Suppl 1:S103. doi: 10.1186/1471-2156-4-S1-S103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo TW, Hughes JR, Fross JD. Failure to support the validity of the Fagerström Tolerance Questionnaire as a measure of physiological tolerance to nicotine. Addict Behav. 1988;13:87–90. doi: 10.1016/0306-4603(88)90030-5. [DOI] [PubMed] [Google Scholar]
- Madden PA, Heath AC, Pedersen NL, Kaprio J, Koskenvuo MJ, Martin NG. The genetics of smoking persistence in men and women: a multicultural study. Behav Genet. 1999;29:423–431. doi: 10.1023/a:1021674804714. [DOI] [PubMed] [Google Scholar]
- Madden PA, Pedersen NL, Kaprio J, Koskenvuo MJ, Martin NG. The epidemiology and genetics of smoking initiation and persistence: crosscultural comparisons of twin study results. Twin Res. 2004;7:82–97. doi: 10.1375/13690520460741471. [DOI] [PubMed] [Google Scholar]
- McGue M, Elkins I, Iacono WG. Genetic and environmental influences on adolescent substance use and abuse. Am J Med Genet. 2000;96:671–677. doi: 10.1002/1096-8628(20001009)96:5<671::aid-ajmg14>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66:1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miksys S, Lerman C, Shields PG, Mash DC, Tyndale RF. Smoking, alcoholism and genetic polymorphisms alter CYP2B6 levels in human brain. Neuropharmacology. 2003;45:122–132. doi: 10.1016/s0028-3908(03)00136-9. [DOI] [PubMed] [Google Scholar]
- Moolchan ET, Radzius A, Epstein DH, Uhl G, Gorelick DA, Cadet JL, Henningfield JE. The Fagerström Test for Nicotine Dependence and the Diagnostic Interview Schedule: do they diagnose the same smokers? Addict Behav. 2002;27:101–113. doi: 10.1016/s0306-4603(00)00171-4. [DOI] [PubMed] [Google Scholar]
- Niu T, Chen C, Ni J, Wang B, Fang Z, Shao H, Xu X. Nicotine dependence and its familial aggregation in Chinese. Int J Epidemiol. 2000;29:248–252. doi: 10.1093/ije/29.2.248. [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerleau OF, Collins AC, Shiffman S, Pomerleau CS. Why some people smoke and others do not: new perspectives. J Consult Clin Psychol. 1993;61:723–731. doi: 10.1037//0022-006x.61.5.723. [DOI] [PubMed] [Google Scholar]
- Saccone NL, Neuman RJ, Saccone SF, Rice JP. Genetic analysis of maximum cigarette-use phenotypes. BMC Genet. 2003;4 Suppl 1:S105. doi: 10.1186/1471-2156-4-S1-S105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedecor SM, Mehringer AM, Marks JL, Pomerleau CS. Development and validation of a self-rated DSM-based scale for assessing nicotine dependence. Annual Meeting of the Society for Research on Nicotine and Tobacco; Scottsdale, AZ. February.2004. [Google Scholar]
- Straub RE, Sullivan PF, Ma Y, Myakishev MV, Harris-Kerr C, Wormley B, Kadambi B, Sadek H, Silverman MA, Webb BT, Neale MC, Bulik CM, Joyce PR, Kendler KS. Susceptibility genes for nicotine dependence: a genome scan and followup in an independent sample suggest that regions on chromosomes 2, 4, 10, 16, 17 and 18 merit further study. Mol Psychiatry. 1999;4:129–144. doi: 10.1038/sj.mp.4000518. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Kendler KS. The genetic epidemiology of smoking. Nicotine Tob Res. 1999;1 Suppl 2:S51–7. doi: 10.1080/14622299050011811. discussion S69-70. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Neale BM, van den Oord E, Miles MF, Neale MC, Bulik CM, Joyce PR, Straub RE, Kendler KS. Candidate genes for nicotine dependence via linkage, epistasis, and bioinformatics. Am J Med Genet. 2004;126B:23–36. doi: 10.1002/ajmg.b.20138. [DOI] [PubMed] [Google Scholar]
- Swan GE, Carmelli D. Behavior genetic investigations of cigarette smoking and related issues in twins. In: Blum K, Noble EP, editors. Handbook of Psychiatric Genetics. Boca Raton: CRC Press, Inc.; 1997. pp. 387–405. [Google Scholar]
- Swan GE, Carmelli D, Rosenman RH, Fabsitz RR, Christian JC. Smoking and alcohol consumption in adult male twins: genetic heritability and shared environmental influences. J Subst Abuse. 1990;2:39–50. doi: 10.1016/s0899-3289(05)80044-6. [DOI] [PubMed] [Google Scholar]
- Swan GE, Hudmon KS, Jack LM, Hemberger K, Carmelli D, Khroyan TV, Ring HZ, Hops H, Andrews JA, Tildesley E, McBride D, Benowitz N, Webster C, Wilhelmsen KC, Feiler HS, Koenig B, Caron L, Illes J, Cheng LS. Environmental and genetic determinants of tobacco use: methodology for a multidisciplinary, longitudinal family-based investigation. Cancer Epidemiol Biomarkers Prev. 2003a;12:994–1005. [PMC free article] [PubMed] [Google Scholar]
- Swan GE, Hudmon KS, Khroyan TV. Tobacco dependence. In: Nezu A, Nezu C, editors. Health Psychology. New York: Wiley; 2003b. pp. 147–168. [Google Scholar]
- Swan GE, Valdes A, Ring HZ, Khroyan TV, Jack LM, Ton C, Curry SJ, McAfee T. Dopamine receptor DRD2 genotype and smoking cessation outcome following treatment with bupropion SR. Pharmacogenomics. 2005;5:21–29. doi: 10.1038/sj.tpj.6500281. [DOI] [PubMed] [Google Scholar]
- True WR, Xian H, Scherrer JF, Madden PA, Bucholz KK, Heath AC, Eisen SA, Lyons MJ, Goldberg J, Tsuang M. Common genetic vulnerability for nicotine and alcohol dependence in men. Arch Gen Psychiatry. 1999;56:655–661. doi: 10.1001/archpsyc.56.7.655. [DOI] [PubMed] [Google Scholar]
- Tyndale RF. Genetics of alcohol and tobacco use in humans. Ann Med. 2003;35:94–121. doi: 10.1080/07853890310010014. [DOI] [PubMed] [Google Scholar]
- Weber JL, May PE. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet. 1989;44:388–396. [PMC free article] [PubMed] [Google Scholar]
- Whittemore AS, Halpern J. A class of tests for linkage using affected pedigree members. Biometrics. 1994;50:118–127. [PubMed] [Google Scholar]
- Wilhelmsen KC, Schuckit M, Smith TL, Lee JV, Segall SK, Feiler HS, Kalmijn J. The search for genes related to a low-level response to alcohol determined by alcohol challenges. Alcohol Clin Exp Res. 2003;27:1041–1047. doi: 10.1097/01.ALC.0000075551.02714.63. [DOI] [PubMed] [Google Scholar]
- Xian H, Scherrer JF, Madden PA, Lyons MJ, Tsuang M, True WR, Eisen SA. The heritability of failed smoking cessation and nicotine withdrawal in twins who smoked and attempted to quit. Nicotine Tob Res. 2003;5:245–254. [PubMed] [Google Scholar]


