Abstract
Direct localization of specific genes, RNAs and proteins has allowed the dissection of individual nuclear speckles in relation to the molecular biology of gene expression. Nuclear speckles (aka SC35 domains) are essentially ubiquitous structures enriched for most pre-mRNA metabolic factors, yet their relationship to gene expression has been poorly understood. Analyses of specific genes and their spliced or mature mRNA strongly support that SC35 domains are hubs of activity, not stores of inert factors detached from gene expression. We propose that SC35 domains are hubs that spatially link expression of specific pre-mRNAs to rapid recycling of copious RNA metabolic complexes, thereby facilitating expression of many highly active genes. In addition to increasing the efficiency of each step, sequential steps in gene expression are structurally integrated at each SC35 domain, consistent with other evidence that the biochemical machineries for transcription, splicing, and mRNA export are coupled. Transcription and splicing are subcompartmentalized at the periphery, with largely spliced mRNA entering the domain prior to export. In addition, new findings presented here begin to illuminate the structural underpinnings of a speckle, by defining specific perturbations of phosphorylation that promote disassembly or assembly of an SC35 domain, in relation to other components. Results thus far are consistent with the SC35 spliceosome assembly factor as an integral structural component. Conditions that disperse SC35 also disperse poly A RNA, whereas the splicing factor ASF/SF2 can be dispersed under conditions in which SC35 or SRm300 remain as intact components of a core domain.
INTRODUCTION
In the eukaryotic nucleus, the distribution of pre-mRNA splicing factors is not uniform, but is markedly concentrated at 10-30 sites, with lower levels of factors diffusely distributed throughout the nucleoplasm (figure 1A). Variously referred to as “speckles”, “SC35 domains” or “splicing factor compartments (SFCs)”, these irregular but discrete domains are most frequently visualized with an antibody directed against the spliceosome assembly factor SC35 (Fu and Maniatis, 1990). Nuclear speckles were first described as the pattern of staining using an Sm antibody, which labels both SC35-rich domains and a smaller number of coilin-rich Cajal Bodies which lack SC35. The term “SC35 domains” denotes the 10-30 more prominent SC35 rich “speckles”, 0.5-3.0 micrometers in diameter; these correspond largely if not entirely to ultrastructures termed interchromatin granule clusters (IGCs) (Fakan and Puvion, 1980; Visa et al., 1993); reviewed in: (Spector, 1993); Moen, 1995). Although SC35 domains typically reside between chromosome territories (Zirbel et al., 1993; Clemson et al., 1996), much evidence indicates that SC35 domains are not just factors trapped in interchromatin space; rather, they behave as discretely bordered compartments within insoluble nuclear structure (Carter et al., 1993; Blencowe et al., 1994; Huang et al., 1994; Shopland and Lawrence, 2000; Moen et al., 2004). In addition to the SR protein SC35, many different factors involved in the transcription, processing, and export of mRNAs have been demonstrated to be enriched in these domains (Table 1). Although concentrated in the SC35 domains, these components are also more diffusely distributed throughout the nucleus. Importantly, several studies show that factors within SC35 domains are in rapid flux, although the positions of the domains themselves are relatively immobile (Kruhlak, 2000; Misteli, 2000)]; discussed in [(Shopland and Lawrence, 2000)].
Figure 1. Overview: The Anatomy of an SC35 Domain as Relates to the Molecular Biology of Gene Expression.
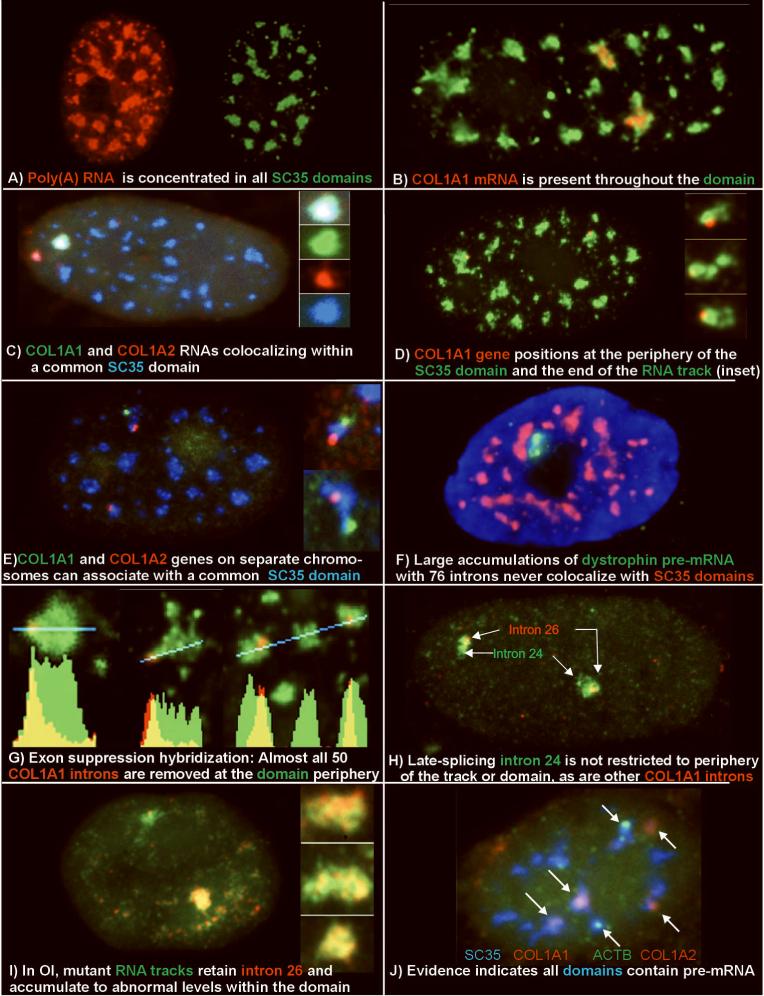
- A) Poly(A) RNA (red) is concentrated in all SC35 domains (green).
- B) Collagen 1A1 RNA (red) is consistently seen within an SC35 domain (green) (from: (Johnson et al., 2000))
- C) Collagen 1A1 RNA (green) and collagen 1A2 RNA (red) can localize within a common SC35 (blue) (from: (Shopland et al., 2003))
- D) Collagen 1A1 DNA hybridization (red) positions at the periphery of an SC35 domain (green).
- E) Collagen 1A1 (green) and Col 1A2 (red) DNA hybridization shows multiple genes at the periphery of an SC35 domain (blue).
- F) Dystrophin nuclear RNA accumulation (green) detected here using a cDNA probe to exons 1-11 (L. Kunkel Harvard School of Medicine, Boston, MA) is never observed to colocalize with SC35 domains (red) (Smith et al., 1999).
- G) Exon suppression RNA hybridization with a labeled Collagen genomic probe (red) competed with excess cDNA shows that most introns are removed at the periphery of the SC35 domain (green) (from: (Johnson et al., 2000))
- H) Comparison of collagen intron 24 and intron 26 shows that intron 24 is spliced later, and is not restricted to the periphery of domain or RNA track (green) (from: (Johnson et al., 2000))
- I) In Osteogenesis Imperfecta, collagen mRNA from the mutant allele (lower signal and insets) retains intron 26 (green) throughout the mRNA track (red) and thus the Sc35 domain. The intron is removed from mRNA of the normal allele (upper signal) (from: (Johnson et al., 2000))
- J) Visualization of three different mRNAs(beta actin, green, and collagen 1A1 and collagen 1A2, red) within Sc35 domains (blue), illustrates that half of the domains in this cell label with probes to just three genes. (from: Shopland, 2002])
Table 1. Some Protein Components of SC35 Domains.
Proteomic analysis of biochemically purified ICGs have found at least 75 proteins enriched in these structures (Mintz et al., 1999). This table is not intended to be an exhaustive list of factors that are coincident with SC35 domains, nor is it practical to give a comprehensive accounting of the relevant references. It is meant to provide an impression of the number and variety of factors that appear to concentrate in these regions. Although the factors are organized by functional groups, new research often finds that a particular protein thought to have a particular function also is involved in another process. For instance, SRm160 functions as a splicing coactivator (Blencowe et al., 1998), 3′-end cleavage-stimulatory factor (McCracken et al., 2002) and is involved in mRNA export (Le Hir et al., 2000).
Proteins localized to Splicing Factor Domains
| Splicing factors | |
| snRNPs (U1 , U2, U4/U6, U5) | (Spector et al., 1983; Nyman et al.,1986) |
| SR proteins (SC35, ASF/SF2, 9G8, SRp20, SRp30c, SRp40, SRp55, Srp75, p54) |
(Fu and Maniatis, 1990; Fu, 1995);reviewed in: (Bourgeois et al.,2004) |
| SRm 160, SRm 300 | (Blencowe et al., 2000; Wagner et al., 2003) |
| U2AF | (Gama-Carvalho et al., 1997) |
| Pinin | (Zimowska et al., 2003) |
| RNPS1 | (Mayeda et al., 1999) |
| PAP-1 | (Maita et al., 2004) |
| Other mRNA processing | |
| Poly(A) polymerase | (Schul et al., 1998) |
| Poly(A) binding protein II | (Krause et al., 1994) |
| RNA helicase II/Gu (beta) | (Valdez et al., 2002) |
| Eukaryotic initiation factor 4E (eIF4E) | (Dostie et al., 2000) |
| Transcription | |
| Hyperphosphorylated RNA pol II | (Bregman et al., 1995) |
| WT1 | (Larsson et al., 1995) |
| FBI-1 | (Pendergrast et al., 2002) |
| HMG-17 | (Hock et al., 1998) |
| fibroblast growth factor receptor 1 | (Somanathan et al., 2003) |
| RNA helicase p68 | (Enukashvily et al., 2005) |
| PRCC | (Skalsky et al., 2001) |
| Structural proteins | |
| Lamin A | (Jagatheesan et al., 1999) |
| αB-crystallin and HSP27 | (van den et al., 2003) |
| Profilin | (Skare et al., 2003) |
| Phosphorylation/dephosphorylation | |
| Protein phosphatase 1 | (Trinkle-Mulcahy et al., 2001) |
| PSKH1 | (Brede et al., 2002) |
| PRP4 | (Kojima et al., 2001) |
| CLK/STY | (Colwill et al., 1996) |
| RNA Transport | |
| Aly/REF | (Zhou et al., 2000) |
| MLN51 | (Degot et al., 2004) |
| Magoh | (Le Hir et al., 2001) |
| Y14 | (Le Hir et al., 2001) |
A commonly perpetuated model of SC35 domains is straightforward, presenting these regions as merely stores of factors devoid of (pre)-mRNA, from which factors are “recruited” to active genes randomly dispersed in the nucleoplasm (Mattaj, 1994; Zhang et al., 1994)]. While such recruitment may take place to some degree, evidence now shows that much gene expression is structurally associated directly with these domains. The concept of domains as storage sites devoid of mRNA was rooted in the demonstration that tritiated-uridine incorporation labels the IGC very little relative to the surrounding nucleoplasm (Fakan and Bernhard, 1971)]; reviewed in: [(Spector, 1993; Fakan, 1994)], although some studies do find newly synthesized RNA concentrated within these domains (Wei et al., 1999). However, labeling methods such as 3H-uridine or bromo-UTP are wholly non-specific and do not necessarily reflect the distribution of pre-mRNA; these methods label mostly ill-defined hnRNA (Salditt-Georgieff et al., 1981)] and introns (most of pre-mRNA mass) that may be rapidly removed and dispersed during the labeling period (discussed in [(Moen et al., 1995). Recent genomic expression studies reveal widespread transcription of non-genic DNA throughout the transcriptome (Cheng et al., 2005). This is also suggested by the copious amounts of RNA throughout the nucleoplasm detected by RNA hybridization using Cot-1 DNA, composed largely of repetitive elements such as Alu and LINEs ((Hall et al., 2002); Clemson et al., submitted). Thus other sorts of RNA which do not necessarily exit the nucleus may constitute a major fraction of RNA labeled by uridine incorporation, and it could be this RNA that is excluded from SC35 domains.
An approach which avoids the limitations of non-specific labeling of all nuclear RNAs is to use high-resolution, sensitive methods to detect specific RNAs and individual endogenous genes directly within nuclear structure. This allows direct demonstration of the organization, relative to SC35 domains, of specific protein-coding genes, in the transcribed or non-transcribed state, as well as the sites of splicing and post-splicing maturation of the cognate RNA. Based on observations from our lab for >25 genes and a number of individual chromosome bands, a picture has emerged of a highly non-random distribution of sequences relative to this compartment (e.g. Smith et al., 1999; Moen et al., 2004). Notably, the inactive or non-pre-mRNA coding sequences studied thus far show no preferential association with these domains but rather localize to peripheral or nucleolar heterochromatin (Xing et al., 1995; Zink et al., 2004). In contrast, somewhat more than half of the active genes studied to date associate at a level well above random (70-99% association). This has led us to propose that each SC35 domain lies at the functional center of a local euchromatic neighborhood, around which many but not all active genes tend to cluster (Shopland et al., 2003).
This association of genes with SC35 domains has far-reaching and complex implications for genomic organization, which will largely be discussed elsewhere; here we focus on the detailed structural and functional analysis of single SC35 domains in precise relationship to distinct steps in the molecular biology of specific gene expression. A number of findings presented here illuminate the molecular anatomy of an individual speckle and show that these structures have far more functional and structural complexity that initially envisioned. Results strongly support a surprising model of each SC35 domain as a structurally organized hub that facilitates the efficiency and integration of distinct steps in gene expression, ranging from transcription to mRNA export.
While the first part of this paper deals with the relationship of SC35 speckles to the molecular biology of gene expression based primarily on our analyses of specific genes and RNAs, the second part presents results of ongoing studies to investigate the structural underpinnings of these nuclear domains. A host of prior observations from other labs shows that the localization and function of SR proteins in speckles is controlled by phosphorylation events (Reviewed in: Ma and He, 2003; Bourgeois et al., 2004; Huang and Steitz, 2005). We have thus defined conditions for specific perturbations of phosphorylation that promote the breakdown or maintenance of SC35 defined domains to determine the impact on other components of these structures. Results thus far support that the SC35 protein likely provides a structural component that is integral to the formation and maintenance of these domains of RNA metabolic activity, in contrast to components such as ASF/SF2 which has a more tentative relationship to domain structure.
Dissecting Molecular Anatomy of Individual SC35 Domains by Precise Localization of Specific Genes and RNAs
Through relatively unique analysis of precise spatial relationships of DNA, RNA, and protein within a single domain we have conducted a “molecular dissection of a speckle” as exemplified in the images in Figure 1. Since such experiments provide a wealth of specific information that is complicated to present, we focus here on six unanticipated findings that derive from these experiments, which are then integrated into an overall model for domain structure and function. While each of these findings brought some element of surprise and complexity, together they fit into an internally consistent view of SC35 domains as intricately linked steps in specific gene expression. For consistency, Figure 1 presents detailed analysis of distinct steps in nuclear metabolism of the collagen 1A1 gene (COL1A1), and to a lesser extent the COL1A2 gene located on a different chromosome. However, observations on a number of other genes indicate that the structural organization shown here also applies to the many other active genes which spatially associate with SC35 domains.
Finding #1: SC35 domains are consistently enriched in poly(A) RNA
The finding that poly(A) RNA, comprised largely of messenger RNA, concentrates at sites co-localized with snRNP and SC35 rich domains was the first challenge to suggest that pre-mRNA metabolism associates with the SC35 compartment (Carter et al., 1991; Carter et al., 1993; Xing et al., 1993)]. As shown in figure 1A and 2, poly(A) RNA is found throughout the nucleoplasm, but concentrates in regions coincident with the prominent accumulations or “domains” of SC35. These SC35 and Poly(A) RNA domains position within regions sharply depleted of DAPI staining, however they do not fill the whole interchromatin space as shown, even that immediately in the environs of the domains (Figure 2G). This indicates that the structure of the SC35 domain is not entirely determined by the surrounding chromatin. Also, it is apparent that the poly(A) signal consistently demarcates a slightly larger “speckle” region than SC35 (fig. 2E-F and Carter et al. 1993). This suggests that the SC35 signal defines a “core” domain which is surrounded by a larger shell. This shell may be, at least in part, coincident with the “paraspeckle” described by Fox et al (Fox et al., 2002) which is a nuclear domain marked by several RNA binding proteins. Other results presented below that define specific sites of gene transcription suggest that on an ultrastructural level, the speckle shell may correspond to perichromatin fibrils, which label strongly with uridine and have been noted to surround IGCs (Fakan and Puvion, 1980; Visa et al., 1993).
Figure 2. Relationship of Poly(A) containing RNA to SC35.
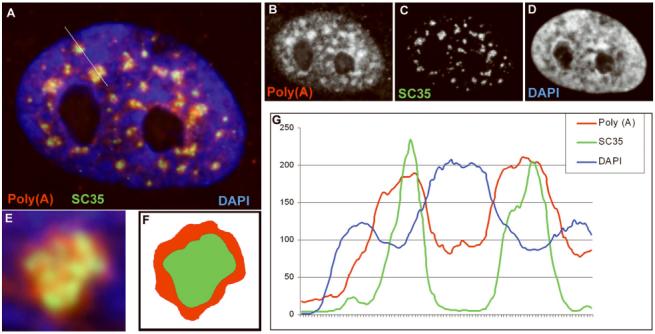
(A) Three color images of Poly(A) RNA (red) detected by hybridization with an oligo dT probe and SC35 (green), detected with an anti-SC35 monoclonal antibody (Sigma) in human fibroblasts. DNA (blue) is detected by DAPI stain. Grayscale images of each color are represented in B, C and D. (E) Blowup of a single SC35 domain with poly(A). (F) The image in E was masked by color thresholding of red and green signals to more clearly show that poly(A) signal extends beyond the borders of the SC35 signal. (G) Histogram of signal intensities along a line shown in (A) demonstrates that poly(A) (red line) defines regions larger than those seen with SC35 staining (green line) and that these domains lie within regions of lower DNA density (blue line).
Finding #2: Each SC35 domain can contain multiple specific pre-mRNAs, including some of those most highly expressed in the cell
While poly(A) RNA is suggestive of mRNA distribution, it also includes some less defined nuclear RNAs that could have a different role (discussed in: Lawrence et al., 1993; Huang et al., 1994), such as a long-lived structural RNA like Xist (Clemson et al., 1996). Because global transcriptional inhibition impacts stability and export of many mRNAs, the retention of RNAs in domains under these conditions does not necessarily indicate they have a structural role (Shopland et al., 2002). While it is likely that some poly(A) RNA in the nucleus and potentially in SC35 domains has other functions, the localization of specific (pre)-mRNAs not only adjacent to (Xing et al., 1993), but within SC35 domains (Xing et al., 1995) (and fig. 1B) indicate that a significant portion of the poly(A) RNAs present in domains represent transcripts of protein coding genes. Since this initial report, numerous mRNAs have been shown to associate with SC35 domains (Smith et al., 1999; Shopland et al., 2002; Moen et al., 2004) clearly demonstrating that some, likely all, of the prominent SC35 “speckles” indeed contain transcripts from individual protein coding genes. Not all (pre)-mRNAs that associate with (contact) SC35 domains are necessarily seen within them. However, we found that pre-mRNAs which appeared to accumulate at just the domain periphery using a genomic probe, are found enriched within the domain when a more sensitive detection of spliced mRNA was achieved using a cDNA probe (Shopland et al., 2002). The reasons for this are related to the localization of splicing events relative to the domain, as will be explained in Finding #5.
That the domain is not just the site of one highly abundant pre-mRNA, is an important point is which will be evidenced by several findings below. Recent analyses of multiple nuclear RNAs from different genes in the same nuclei (Shopland et al., 2003) show that an individual domain contains multiple mRNAs, likely many. For example, as shown in Fig. 3, COL1A1 and COL1A2 RNA accumulations which are transcribed from different chromosomes can intermingle within the same domain, as viewed in 3-D or by optical sectioning. Similarly, other transcripts can be found there, supporting the domain as a structure which contains multiple mRNAs rather than splicing factors with just a single (pre)-mRNA.
Figure 3. A Model of SC35 Domain Function.
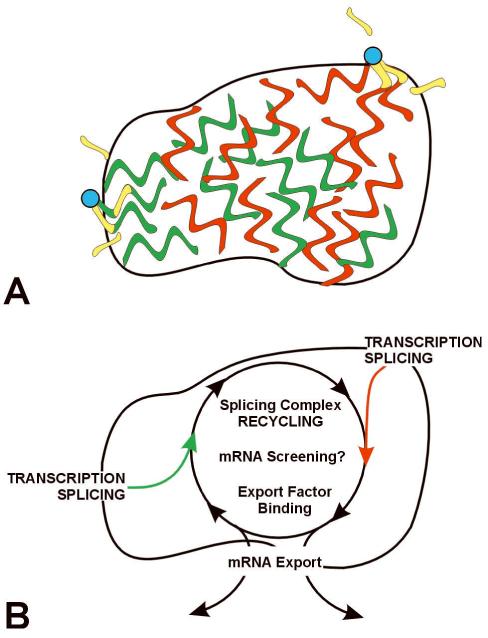
(A) Multiple genes (blue) associate with a single domain. Transcription occurs at the outer edge of the domain, with all or almost all introns removed in the periphery at or near the gene. Essentially spliced mRNA distributes throughout the domain. (B) Model suggesting a functional rationale for coupling the completion of mRNA maturation and release for mRNA export with the recycling/preassembly of factors within SC35 domains. Because the RNA metabolic machinery requires interaction of such a large number of different factors, their concentration at a site would facilitate recycling and a rapid rate of reuse for expression of adjacent genes. The closer the completion of maturation (and release for export) occurs to putative sites of recycling, the more efficient the total process. In this model, mRNAs enter the domains, where splicing complex recycling and binding of export factors occurs. If mRNAs are not properly processed, transit through the domain may be impeded, thus domains may also be the site of mRNA “screening” functions.
Other analyses show that in a metabolically active cell it is not just a few SC35 domains that contain mRNAs, but that all of them do (Shopland et al., 2002). For example, fig. 1J shows that simultaneous hybridization to nuclear RNAs from just three genes (beta actin, COL1A1, and COL1A2) can label half (6 of 12) of the domains in a cell. Since some lower abundance transcripts have also been found within domains (eg. Moen, 2004), clearly these structures contain transcripts from many genes.
Finding # 3: For RNAs that accumulate within the SC35 domain, the corresponding genes position just outside the domain, to one side of their RNA accumulations
If some mRNAs associate with SC35 domains, what about the gene loci from which they are produced? An important though often overlooked observation is that genes consistently associate with the outer periphery of the domain, not the interior (fig. 1D). This peripheral association results in a reproducible orientation of the gene relative to an RNA focus within the domain. As seen in fig. 1D (inset), as opposed to surrounding the site of transcription, the RNA forms a larger accumulation that extends away from the gene locus and into the domain. In some cases it has been directly demonstrated that this is a post-transcriptional accumulation of transcripts that have detached and begun to move away from the gene (Smith et al., 1999; Johnson et al., 2000). Such a polarized distribution of released transcripts relative to the gene we refer to loosely as an “RNA track”. In contrast, the dystrophin gene, which does not associate with SC35 domains (see below), produces foci of nascent transcripts still attached to this large gene which overlies and surrounds the gene signal, rather than a polarized “RNA track” of detached transcripts (Smith et al., 1999).
In keeping with Finding #2, that multiple mRNAs are within a domain (fig. 1C), multiple genes are found to cluster around their periphery. In fact, it is not uncommon for two non-syntenic genes, COL1A1 and COL1A2, to associate with a common domain or for two alleles of the same gene to be at the same domain (fig. 1E). While the implications of this for genome organization are discussed elsewhere (Shopland et al., 2003), this demonstrates that an individual SC35 domain can serve as a metabolic center for multiple specific transcripts, with genes at the periphery. The positioning of many active genes at the domain periphery, with corresponding RNA within the domain, supports our suggestion that each splicing factor rich domain, as viewed by light microscopy, is a structural entity comprised of functionally distinct zones, with transcription at the periphery. This detail is important because it is compatible with other evidence (discussed above) that transcription does not occur within IGC (interchromatin granule clusters), which are ultrastructures that may correspond to the interior zone of “speckles”.
Finding #4: Splicing factor distribution is not proportional to the amount of spliced pre-mRNA at a given site, but rather reflects a locus-specific organization of certain active genes with a larger domain of metabolic factors
Some abundant nuclear accumulations of highly spliced pre-mRNAs never associate with SC35 domains. This finding argues against the co-localization of pre-mRNA and SC35 accumulations being just the consequence of factors accumulated on splice sites of intron-containing pre-mRNA. Whether a pre-mRNA accumulation is associated with a domain of splicing factors is not simply explained by the amount or complexity of transcripts present. For example, although the dystrophin gene has 76 introns, there is no visible increase in the concentration of splicing factors co-localized with a large molar accumulation of dystrophin pre-mRNA (fig. 1F) which we showed was undergoing splicing. In contrast, a similar molar accumulation of myosin heavy chain (MyHC) transcripts was always associated with a “domain” of splicing factors (Smith et al., 1999). This tells us that there are sufficient quantities of splicing factors in the nucleoplasm to splice at least some RNAs without any obvious accumulation. Similarly, mRNAs like myogenin have a high (70-95%) but not invariant association with domains, but usually are concentrated more at the periphery of a much larger domain. This indicates that there is a much larger accumulation of factors in the domain than are needed to process the myogenin mRNA (Moen et al., 2004).
Finding #5: Splicing occurs with the gene at the outer periphery of the domain, yet essentially spliced mRNA still passes into the domain of splicing factors
The simplest interpretation of a focus of RNA overlapping a focus rich in splicing factors is that splicing is occurring throughout the RNA accumulation. However, it has been shown that normal COL1A1 mRNA is almost entirely spliced before it enters into the splicing factor rich domain. Hybridization of specific intron and cDNA sequences showed that the intron RNA consistently concentrated at only one end of a larger RNA “track” (delimited by the cDNA) and at the periphery of an SC35 domain (Johnson et al., 2000). Moreover, we used an “exon suppression” approach which demonstrated that most introns are removed co-transcriptionally before mRNA enters the domain (Fig. 1G). Since introns comprise >80% of most pre-mRNAs this also helps to explain the lower levels of uridine seen within SC35 domains.
Although most splicing is accomplished at the edge of the domain, complete splicing may be rate limiting for some transcripts to exit SC35 domains: While most of the 50 COL1A1 introns are absent from pre-mRNA within SC35 domains, intron 24, which is spliced later than others, was present throughout most of the RNA track (within the domain interior), indicating post-transcriptional splicing of this intron (fig. 1H) (Johnson et al., 2000). Hence, retention of one or a few introns could explain why the mRNA may “pause” within this compartment prior to export. However, based on other evidence under Finding #6, we suggest that other rate limiting steps in preparation for mRNA export may also occur within the domain.
Finding #6: Links between SC35 domains, quality control and mRNA export
Why does the largely spliced RNA enter into the SC35 domain enriched in such a diversity of mRNA metabolic factors (Table I)? The finding that most splicing of COL1A1 mRNA occurs at the outer edge of the domain first led us to suggest that post-splicing steps linked to export may occur within the domain. We had shown that intron 26 was removed prior to entry of mRNA into the domain, but what would happen if this intron was not properly removed? A finding that links the SC35 domain to mRNA export is that a splice-defective mutant of COL1A1 in Osteogenesis Imperfecta (OI) fails to export mutant mRNA to the cytoplasm, but instead accumulates excess COL1A1 mRNA within SC35 domains. Both mutant and normal COL1A1 pre-mRNAs form post-transcriptional “tracks” that enter SC35 domains, showing that mutant RNA can begin transport from the gene. However, mutant transcripts retaining an intron accumulate to abnormal levels within domains (fig. 1I) and are not found in the cytoplasm, indicating that further transport out of the domain is impeded. This demonstrates that the SC35 domain is the intranuclear site at which mutant COL1A1 pre-mRNAs are detained (Johnson et al., 2000, Appendix).
Related to this, over the past few years several labs have reported the localization of various factors implicated in mRNA export, such as REF/Aly (Zhou et al., 2000) and the breast cancer linked shuttling protein MLN51 (Degot et al., 2004), to the SC35 domains. There are also examples in the literature linking export of poly(A) containing RNA to the SC35 domain. Molenaar et al (Molenaar et al., 2004) tracked poly(A) containing RNA in live cells, finding that a significant amount of the poly(A) containing RNA in the nucleus transits through domains prior to export out of the nucleus. Tpr, a nuclear protein localized to the nuclear periphery, is part of a filament that extends from the nuclear pore complex into the nucleoplasm and is thought to be involved in mRNA export (Bangs et al., 1998). Interestingly, loss of Tpr results in the accumulation of poly(A) RNA in SC35 speckles (Shibata et al., 2002), suggesting that, as seen in the OI example above, mRNAs accumulate in domains when their export is blocked.
The results with mutant splice-defective COL1A1 mRNA indicate that an unknown step in “screening” for defective mRNAs may occur within domains and is linked to the mRNA’s export, with improper mRNA being detained. Similarly, it has been suggested that speckles may function as checkpoints in which inappropriately processed mRNAs are screened out (Molenaar et al., 2004). Related to this idea, several factors now known to be involved in nonsense mediated decay (NMD) of RNA, such as Y14 (Kataoka et al., 2000) and RPNS1 (Mayeda et al., 1999) localize to speckles in the nucleus. Thus, it is possible that some portion of transcripts within domains become detained there, possibly for NMD.
Model of SC35 Domain Relationship to Molecular Steps in Gene Expression (Fig. 3)
We propose that SC35 domains function as nuclear hubs; structures that spatially link specific pre-mRNA transcription, splicing and export, to rapid recycling of RNA metabolic complexes, thereby facilitating efficient expression of many highly active genes. In addition to increasing the efficiency of each step, we propose that sequential steps of gene expression are structurally integrated at each SC35 domain, consistent with other evidence (Maniatis and Reed, 2002) that the biochemical machineries for transcription, splicing, and mRNA export are coupled.
While pol II transcription and splicing clearly occurs throughout the nucleoplasm, our evidence shows that a substantial fraction of specific protein-coding genes are expressed in intimate association with SC35 domains. Specific gene loci position at the immediate periphery of the SC35 domain, and include, but are not restricted to, many of the cell’s most highly expressed genes. While associated genes are at the periphery, the RNA accumulations emanating from them often extend into the domain interior (fig. 1D). For the pre-mRNAs studied in detail, most splicing occurs at or near the gene and hence is essentially completed before the mRNA enters the domain interior (fig. 1G). We thus postulate that the first step in export of these RNAs involves movement from the gene into the domain, after which the RNA might disperse to the nuclear pores. Hence, as shown in figure 3, we hypothesize that SC35 domains generally, as viewed by light microscopy, are sub-compartmentalized, with transcription and splicing at the periphery, and post-transcriptional accumulations of more mature mRNA appearing transiently within them, perhaps as a necessary step in screening or maturation prior to export.
In keeping with earlier uridine labeling studies, our results support the concept that the concentration of splicing factors within the domain interior is in great excess to splicing factors bound to even the most abundant individual pre-mRNAs found there. Therefore, we hypothesize that the concentration of various mRNA metabolic factors in domains facilitates a rapid recycling and/or reassembly of macromolecular complexes required for efficient expression of highly active genes (fig. 3). Rather than stores of inert factors, our evidence indicates that in metabolically active cells SC35 domains are actually “hubs”, comprising some of the most actively utilized factors in the cell.
Dissecting the Structure of Speckle by Perturbation of SR Protein Phosphorylation
The work described above addresses the spatial relationship of individual SC35 domains to the molecular biology of gene expression. Ultimately we also want to understand the structural underpinnings of a speckle: what induces it to form, which components provide the structural foundation, and which factors are more transient? One approach to this is the identification of conditions in which the SC35 domain is induced to disassemble or assemble. We have used specific inhibitors of phosphorylation to define conditions that promote either assembly or disassembly of the SC35 defined speckles, and examined how this impacts both other SR proteins and poly(A) RNA or specific RNA within the domains. Here we describe our initial progress towards this goal.
SR proteins have a characteristic arginine/serine-rich C-terminal domain that undergoes dynamic phosphorylation changes. These cycles of phosphorylation and dephosphorylation have been reported to mediate localization of specific SR proteins to the nucleus and to intranuclear domains, as well as mediate the protein/protein interactions necessary for spliceosome assembly, splicing and mRNA export (Reviewed in: Ma and He, 2003; Bourgeois et al., 2004; Huang and Steitz, 2005). Of particular interest are recent findings that only hypophosphorylated SR proteins preferentially bind mRNA export proteins (Gilbert and Guthrie, 2004; Huang et al., 2004; Lai and Tarn, 2004) ensuring that only spliced mRNAs are exported. Subsequently, rephosphorylation of the SR protein in the cytoplasm relocates it to the nucleus.
While a number of studies have reported the impact of perturbing phosphorylation in specific systems, such impact may have been oversimplified in the initial studies, as there are many different SR proteins and a number of kinases responsible for their phosphorylation (reviewed in: Bourgeois et al., 2004). Not surprisingly, several reports suggested that the affect of phosphorylation on the RS domain and SR function may be substrate-specific (Zhu and Krainer, 2000), and can have positive, negative or no effect on its splicing function (Xiao and Manley, 1998). Similarly, localization of the different SR proteins to intra-nuclear domains may be differently impacted.
For these reasons, we have taken an empirical approach to identifying specific inhibitors and conditions that assemble or disassemble the SC35 defined “speckle”. While the SR protein ASF/SF2 has been the focus of many of the studies on phosphorylation effects, we have focused on the SC35 protein first because we hypothesize that SC35 may be a core structural component of the domain, unlike many SR components such as ASF/SF2. SC35 is more concentrated in a tightly defined domain than are snRNP antigens or ASF/SF2, which are more diffusely distributed in the nucleoplasm as well as somewhat concentrated in speckles. SC35 also defines a slightly smaller “core” domain than does poly(A) RNA (fig. 2E-F), and its localization is highly resistant to detergent and nuclear extraction. We have also previously noted conditions (transcriptional inhibition) in which the SC35 defined domains are still present, but the other snRNP proteins disperse, indicating that the snRNP localization is not required for the SC35 domain structure (Moen et al., 1995).
While aggregation of SC35 would predictably be controlled by phosphorylation events (Gui et al., 1994; Zhu et al., 2003; reviewed in: Lin et al., 2005), we sought to define conditions that easily manipulate assembly or disassembly of SC35 domains. To do this we surveyed a number of reagents that perturb phosphatases in specific ways. Okadaic acid (OKA), cantharidin and tautomycin are potent and cell-permeating inhibitors of serine/threonine phosphatases which have been shown to differentially target PP2A or PP1. At the concentrations used here in order to impact domain formation, okadaic acid and cantharidin inhibit PP2A (Favre et al., 1997), while tautomycin inhibits PP1 (Li and Casida, 1992).
Both okadaic acid (0.3-1uM) and cantharidin (10-15uM) cause SC35 domains to coalesce into larger domains (fig. 4A), suggesting that the hyperphosphorylation caused by this inhibition (likely of PP2A) promotes SC35 aggregation. On the other hand, inhibition of PP1 with the phosphatase inhibitor tautomycin (2-3uM) causes SC35 domains to disassemble, as shown in figure 4B. This breakdown of SC35 domains is similar to that seen with increased activity of Clk/Sty kinase and SRPK1 involved in SR protein phosphorylation (Gui et al., 1994; Colwill et al., 1996), while the aggregation of the domains was seen with increased activity of protein kinase C (Zhu et al., 2003) and may suggest epitope-specific consequences of hyperphosphorylation.
Figure 4. Perturbation of phosphorylation provides a means to promote assembly or disassembly of SC35 domains and investigate their structural underpinnings.
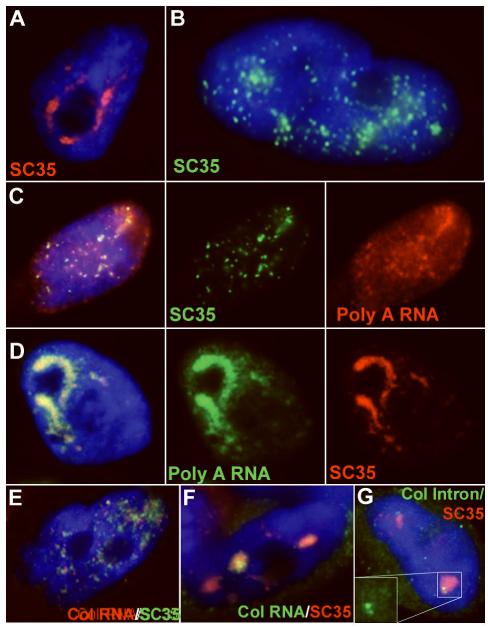
A) Cantharidin (PP2A inhibitor) treatment of TIG-1 cells results in the accumulation of SC35 (red) into larger domains. Compare to untreated cells in Fig 1 and 2. B) Tautomycin (PP1 inhibitor) treatment results in the dispersion of SC35 (green) from domains into the nucleoplasm as small punctuate dots. C) Poly(A) RNA (red) localization to domains is similarly affected by tautomycin treatment, following the pattern of SC35 ( green). D) Poly(A) RNA (green) accumulates into the larger domains along with the SC35 (red) after cantharidin treatment. E) When the SC35 (green) domains break down, collagen RNA (red) is no longer found accumulated in tracks or domains. F) Collagen RNA (green) is found in very large tracks in the large SC35 (red) domains found in cantharidin treated TIG-1 cells. G) Collagen mRNA appears to be efficiently spliced in cantharidin treated cells because the signal for intron 26 (green) is as small as those seen in control cells and intron-containing RNA does not accumulate in SC35 domains (red).
Having identified these conditions for assembly/disassembly of SC35 defined domains, we have begun to examine the impact upon and relationship to both RNA and other SR proteins. One important question concerns whether an SC35 domain (or Sm speckle) requires poly(A) or mRNA for its formation. We previously showed that as the nucleus is reconstructed after mitosis, the Sm speckle defined by an antibody to snRNPs does not form until poly(A) RNA “patches” are re-formed in the nucleoplasm (Carter et al., 1991). Thus, in addition to examining the relationship between SC35 domain association and other SR proteins, we can also address whether the localization of poly(A) RNA or specific mRNAs is dependent upon SC35 localization, and vice versa.
Results here demonstrate that disassembly of SC35 domains by tautomycin causes the poly A RNA within domains to disperse in a distribution similar to that of SC35 (fig. 4C). Similar to the Poly(A) distribution, no collagen mRNA accumulations or “tracks” are found in tautomycin treated cells without a corresponding SC35 accumulation (fig. 4E). Cantharidin treated cells on the other hand, in which SC35 combines into larger domains, exhibit a similar aggregation of poly(A) RNA (fig. 4D). Similarly, COL1A1 mRNA “tracks” look very robust in these cells, with large accumulations visible within the aggregate domains (fig. 4F). These RNA tracks were similar to, but appeared more robust than, untreated controls. To rule out that these large domain RNA accumulations were not due to phosphorylation-induced perturbations of splicing, we used a probe to intron 26 of COL1A1 RNA, which verified that the RNA within domains was still primarily spliced (fig. 4G). This suggests that at least for the collagen gene, the hyperphosphorylation induced by inhibition of PP2A, which results in large domain aggregation, does not necessarily block splicing. However, we cannot rule out that mRNA export is inhibited, thereby causing mature mRNA to accumulate in the domains.
Consistent with the idea that the SC35 protein may be a more structural component of the domain, SC35 does not appear to shuttle as do many other domain factors such as ASF/SF2 (discussed in: Lin et al., 2005). Staining for ASF/SF2 demonstrates that, while it does concentrate to some degree in domains, it has a much more diffuse nucleoplasmic component than either SC35 or SRm300. SRm300 is another speckle component that appears to define a more discrete domain (fig. 5A), and, both SRm300 (Blencowe et al., 2000) and SC35 have been shown to be highly resistant to nuclear matrix extraction (Bisotto et al., 1995). Thus, we hypothesize that SRm300 may also be a structural component of a “core” domain, and therefore examined the relative behaviors of SRm300 and ASF/SF2 under conditions defined above to impact SC35 domains. We speculated that we would find conditions under which ASF/SF2 would be induced to leave domains while the core structural components of the domain remained intact.
Figure 5. Tautamycin treatment (phosphatase 1A inhibition) results in dispersal of ASF/SF2 from the SC35 domain before SRM-300.
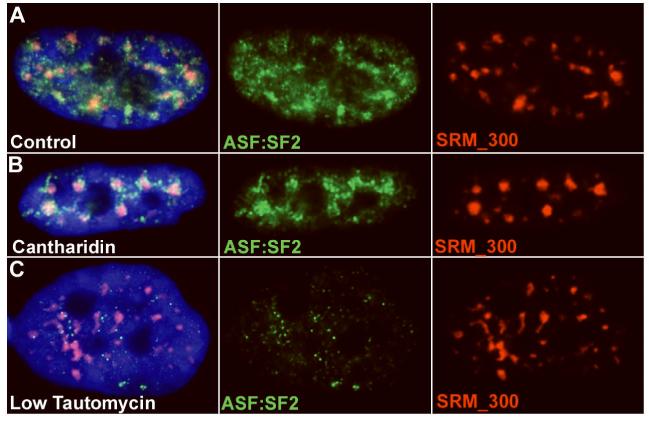
A) In untreated TIG-1 cells ASF/SF2 (green) is present throughout the nucleoplasm but somewhat more concentrated within the SC35 domain (speckle), defined by SRm300 staining (red) B) Upon cantharidin treatment, the ASF/SF2 protein (green) appears to increase its presence within the SRm300 (red) domains. C) In low tautomycin concentrations, where SC35 domains have yet to break down and SRm300 (red) is still concentrated in domains, ASF/SF2 (green) is observed to disperse from the speckle (SRm300 - red).
As with SC35, cantharidin treatment (10-15uM), results in SRm300 aggregation into larger domains, as shown in fig. 5B. Interestingly, cantharidin also caused the ASF/SF2 protein to concentrate more in these SRm300 defined domains, with a suggestion that the ASF/SF2 was accumulating both within the domain, and in the “shell” around the domain core (fig. 5B). Not surprisingly, concentrations of tautomycin (2-3uM) that cause disassociation of SC35, COL1A1 RNA and poly(A) also resulted in the dispersion of ASF/SF2 and SRm300. However, lower concentrations of tautomycin (1uM) that did not result in the breakdown of SRm300 or SC35 exhibited dispersed ASF/SF2. (fig. 5C). This supports that ASF/SF2 is a more tentative and less stable component of the speckle domain, not required for this structure. In addition, it supports the idea that the splicing co-activator SRm300 may also be a more structural component of the speckle, similar to SC35.
Results thus far define conditions that will be helpful in further studies on the “Structural Dissection of a Speckle”. While these approaches can be applied to many more questions, results thus far are consistent with the hypothesis that the SC35 protein is an integral structural protein of the “speckle”, whose localization to the speckle may be required for localization of other protein components. Ultimately the respective roles of RNAs and structural proteins in domain formation will be investigated using a similar approach to tease apart domain structure while probing the molecular cytology of specific genes and RNAs.
MATERIALS AND METHODS
More details of our methods can be found in the references cited above.
Cell Culture, Drug Treatment and Cell Fixation
The TIG-1 cell line (AG06173) is a normal diploid female fibroblast line obtained from Coriell Cell Repositories. These cells were maintained in Minimum Essential Media (Gibco BRL) and 10% fetal calf serum (FCS). The phosphatase inhibitors okadaic acid (PP2A inhibitor), tautomycin (PP1 inhibitor) and cantharidin (PP2A inhibitor), were diluted in DMSO at stock concentrations of 500uM, 100uM, and 10uM, respectively. Diluted stocks were used before 6 months. Working concentration ranges used for these drugs were: okadaic acid: 0.3-1uM, tautomycin: 0.5-3uM, and cantharidin: 5-15uM. Cells were treated on glass cover slips for 4-6 hours in regular growth media at 37C, 5% CO2 before fixation. Our standard cell fixation protocol, in which cells are 0.5% triton extracted prior to fixation in 4% paraformaldehyde, has been previously described (Johnson et al., 1991; Tam et al., 2002).
Fluorescence In Situ Hybridization and Immunofluorescence
Human collagen 1A1 DNA clone, cosmid CG103, contains the entire 18-kb human gene (Barsh et al., 1984). The COL1A1 intron 26-specific probe was generated by PCR. These probes were nick translated with digoxigenin-11-dUTP or biotin-16-dUTP (Roche). Poly(A) RNA was detected using a biotin-labeled poly dT oligo. SC35 domains were detected with an antibody to the spliceosome assembly factor SC35 (Fu and Maniatis, 1990) (Sigma) or to an antibody to the splicing coactivator SRm300 (Blencowe et al., 1998) (B. Blencowe, U. Toronto). The SR protein ASF/SF2 was detected using a Splicing Factor 2 antibody (SF2/ASF) (Zymed). Our protocols for FISH and immunofluorescence, as well as combined in situ RNA and antibody detection, have been described previously in detail (Johnson et al., 1991; Tam et al., 2002), with antibody detection normally carried out prior to hybridization. The only alteration to our standard protocol for combined RNA/antibody detection is the substitution of VRC with 1unit/ul RNasin (Promega), as well as adding RNasin during antibody detection, and in the hybridization buffer.
Microscopy and Analysis
Images presented were acquired using an Axiovert 200 microscope or a Zeiss Axiophot microscope (Carl Zeiss, Thornwood, NY), equipped with a 100X PlanApo objective (NA 1.4) using Axiovision (Zeiss) imaging software. The Chroma (Brattleboro, VT) 83000 multiband pass dichroic and emission filter sets were used with excitation filters set up in a wheel to prevent optical shift. Images were captured on an Orca-ER camera (Hamamatsu, Bridgewater, NJ) or a 200 series CCD (Photometrics, Inc).
Conclusions presented here are based on quantitative analysis of large numbers of cells by multiple investigators, except where otherwise indicated.
Acknowledgements
The authors thank John McNeil for help with figures. This work was made possible by NIH Public Health Service grant (R01 GM-68138) to J.B.L. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Grant Sponsor NIH grant number: R01 GM-68138 (to J.B.L)
Literature Cited
- Bangs P, Burke B, Powers C, Craig R, Purohit A, Doxsey S. Functional analysis of Tpr: identification of nuclear pore complex association and nuclear localization domains and a role in mRNA export. J Cell Biol. 1998;143:1801–1812. doi: 10.1083/jcb.143.7.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsh GS, Roush CL, Gelinas RE. DNA and Chromatin Structure of the Human alpha-1 (I) Collagen Gene. J.Biol.Chem. 1984;259(23):14906–14913. [PubMed] [Google Scholar]
- Bisotto S, Lauriault P, Duval M, Vincent M. Colocalization of a high molecular mass phosphoprotein of the nuclear matrix (p255) with spliceosomes. J Cell Sci. 1995;108(Pt 5):1873–1882. doi: 10.1242/jcs.108.5.1873. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ, Bauren G, Eldridge AG, Issner R, Nickerson JA, Rosonina E, Sharp PA. The SRm160/300 splicing coactivator subunits. Rna. 2000;6:111–120. doi: 10.1017/s1355838200991982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ, Issner R, Nickerson JA, Sharp PA. A coactivator of pre-mRNA splicing. Genes Dev. 1998;12:996–1009. doi: 10.1101/gad.12.7.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ, Nickerson JA, Issner R, Penman S, Sharp PA. Association of nuclear matrix antigens with exon-containing splicing complexes. J.Cell Biol. 1994;127(3):593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois CF, Lejeune F, Stevenin J. Broad specificity of SR (serine/arginine) proteins in the regulation of alternative splicing of pre-messenger RNA. Prog Nucleic Acid Res Mol Biol. 2004;78:37–88. doi: 10.1016/S0079-6603(04)78002-2. [DOI] [PubMed] [Google Scholar]
- Brede G, Solheim J, Prydz H. PSKH1, a novel splice factor compartment-associated serine kinase. Nucleic Acids Res. 2002;30:5301–5309. doi: 10.1093/nar/gkf648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman DB, Du L, van der Zee s, Warren SL. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J.Cell Biol. 1995;129(2):287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter KC, Bowman D, Carrington W, Fogarty K, McNeil JA, Fay FS, Lawrence JB. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 1993;259:1330–1335. doi: 10.1126/science.8446902. [DOI] [PubMed] [Google Scholar]
- Carter KC, Taneja KL, Lawrence JB. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J.Cell Biol. 1991;115:1191–1202. doi: 10.1083/jcb.115.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, Sementchenko V, Piccolboni A, Bekiranov S, Bailey DK, Ganesh M, Ghosh S, Bell I, Gerhard DS, Gingeras TR. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J.Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. Embo J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- Degot S, Le Hir H, Alpy F, Kedinger V, Stoll I, Wendling C, Seraphin B, Rio MC, Tomasetto C. Association of the breast cancer protein MLN51 with the exon junction complex via its speckle localizer and RNA binding module. J Biol Chem. 2004;279:33702–33715. doi: 10.1074/jbc.M402754200. [DOI] [PubMed] [Google Scholar]
- Dostie J, Lejbkowicz F, Sonenberg N. Nuclear eukaryotic initiation factor 4E (eIF4E) colocalizes with splicing factors in speckles. J Cell Biol. 2000;148:239–247. doi: 10.1083/jcb.148.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enukashvily N, Donev R, Sheer D, Podgornaya O. Satellite DNA binding and cellular localisation of RNA helicase P68. J Cell Sci. 2005;118:611–622. doi: 10.1242/jcs.01605. [DOI] [PubMed] [Google Scholar]
- Fakan S. Perichromatin fibrils are in situ forms of nascent transcripts. Trends in Cell Biology. 1994;4:86–90. doi: 10.1016/0962-8924(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Fakan S, Bernhard W. Localisation of rapidly and slowly labelled nuclear RNA as visualized by high resolution autoradiography. Exp.Cell Res. 1971;67:129–141. doi: 10.1016/0014-4827(71)90628-8. [DOI] [PubMed] [Google Scholar]
- Fakan S, Puvion E. The ultrastructural visualization of nuclear and extranucleolar RNA synthesis and distribution. Int.Rev.Cytol. 1980;65:255–299. doi: 10.1016/s0074-7696(08)61962-2. [DOI] [PubMed] [Google Scholar]
- Favre B, Turowski P, Hemmings BA. Differential inhibition and posttranslational modification of protein phosphatase 1 and 2A in MCF7 cells treated with calyculin-A, okadaic acid, and tautomycin. J Biol Chem. 1997;272:13856–13863. doi: 10.1074/jbc.272.21.13856. [DOI] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. Paraspeckles: a novel nuclear domain. Curr Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- Fu XD. The superfamily of arginine/serine-rich splicing factors. Rna. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- Fu X-D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–444. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Gama-Carvalho M, Krauss RD, Chiang L, Valcarcel J, Green MR, Carmo-Fonseca M. Targeting of U2AF65 to sites of active splicing in the nucleus. J Cell Biol. 1997;137:975–987. doi: 10.1083/jcb.137.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W, Guthrie C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell. 2004;13:201–212. doi: 10.1016/s1097-2765(04)00030-9. [DOI] [PubMed] [Google Scholar]
- Gui J-F, Lane WS, Fu X-D. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- Hall LL, Byron M, Sakai K, Carrel L, Willard HF, Lawrence JB. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc Natl Acad Sci USA. 2002;99:8677–8682. doi: 10.1073/pnas.132468999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock R, Wilde F, Scheer U, Bustin M. Dynamic relocation of chromosomal protein HMG-17 in the nucleus is dependent on transcriptional activity. Embo J. 1998;17:6992–7001. doi: 10.1093/emboj/17.23.6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Deerinck TJ, Ellisman MH, Spector DL. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J.Cell Biol. 1994;126(4):877–899. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Steitz JA. SRprises along a messenger’s journey. Mol Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Huang Y, Yario TA, Steitz JA. A molecular link between SR protein dephosphorylation and mRNA export. Proc Natl Acad Sci USA. 2004;101:9666–9670. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagatheesan G, Thanumalayan S, Muralikrishna B, Rangaraj N, Karande AA, Parnaik VK. Colocalization of intranuclear lamin foci with RNA splicing factors. J Cell Sci. 1999;112:4651–4661. doi: 10.1242/jcs.112.24.4651. [DOI] [PubMed] [Google Scholar]
- Johnson CV, Primorac D, McKinstry M, McNeil JA, Rowe D, Lawrence JB. Tracking COL1A1 RNA in Osteogenesis Imperfecta: Splice-defective transcripts initiate transport from the gene but are retained within the SC-35 domain. J. Cell Biol. 2000;150:417–431. doi: 10.1083/jcb.150.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CV, Singer RH, Lawrence JB. Fluorescent detection of nuclear RNA and DNA: Implication for genome organization. Methods Cell Biol. 1991;35:73–99. [PubMed] [Google Scholar]
- Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell. 2000;6:673–682. doi: 10.1016/s1097-2765(00)00065-4. [DOI] [PubMed] [Google Scholar]
- Kojima T, Zama T, Wada K, Onogi H, Hagiwara M. Cloning of human PRP4 reveals interaction with Clk1. J Biol Chem. 2001;276:32247–32256. doi: 10.1074/jbc.M103790200. [DOI] [PubMed] [Google Scholar]
- Krause S, Fakan S, Weis K, Wahle E. Immunodetection of poly(A) binding protein II in the cell nucleus. Exp Cell Res. 1994;214:75–82. doi: 10.1006/excr.1994.1235. [DOI] [PubMed] [Google Scholar]
- Kruhlak MJ, Lever MA, Fischle W, Verdin E, Bazett Jones DP, Hendzel MJ. Reduced mobility of the ASF splicing factor through the nucleoplasm and steady-state speckle compartments. J Cell Biol. 2000 doi: 10.1083/jcb.150.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Tarn WY. Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J Biol Chem. 2004;279:31745–31749. doi: 10.1074/jbc.C400173200. [DOI] [PubMed] [Google Scholar]
- Larsson SH, Charlieu JP, Miyagawa K, Engelkamp D, Rassoulzadegan M, Ross A, Cuzin F, van Heyningen V, Hastie ND. Subnuclear localization of WT1 in splicing or transcription factor domains is regulated by alternative splicing. Cell. 1995;81:391–401. doi: 10.1016/0092-8674(95)90392-5. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Carter KC, Xing X. Probing Functional Organization within the Nucleus: Is Genome Structure Integrated with RNA Metabolism? Cold Spring Harbor Symp.on Quant.Biol. 1993;LVIII:807–818. doi: 10.1101/sqb.1993.058.01.088. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Braun IC, Forler D, Izaurralde E. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep. 2001;2:1119–1124. doi: 10.1093/embo-reports/kve245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Li YM, Casida JE. Cantharidin-binding protein: identification as protein phosphatase 2A. Proc Natl Acad Sci USA. 1992;89:11867–11870. doi: 10.1073/pnas.89.24.11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Xiao R, Sun P, Xu X, Fu XD. Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Mol Cell. 2005;20:413–425. doi: 10.1016/j.molcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Ma X, He F. Advances in the study of SR protein family. Genomics Proteomics Bioinformatics. 2003;1:2–8. doi: 10.1016/S1672-0229(03)01002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maita H, Kitaura H, Keen TJ, Inglehearn CF, Ariga H, Iguchi Ariga SM. PAP-1, the mutated gene underlying the RP9 form of dominant retinitis pigmentosa, is a splicing factor. Exp Cell Res. 2004;300:283–296. doi: 10.1016/j.yexcr.2004.07.029. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Mattaj IW. Splicing in space. Nature. 1994;372:727–728. doi: 10.1038/372727a0. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Badolato J, Kobayashi R, Zhang MQ, Gardiner EM, Krainer AR. Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. Embo J. 1999;18:4560–4570. doi: 10.1093/emboj/18.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Lambermon M, Blencowe BJ. SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol Cell Biol. 2002;22:148–160. doi: 10.1128/MCB.22.1.148-160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz PJ, Patterson SD, Neuwald AF, Spahr CS, Spector DL. Purification and biochemical characterization of interchromatin granule clusters. Embo J. 1999;18:4308–4320. doi: 10.1093/emboj/18.15.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Cell biology of transcription and pre-mRNA splicing: nuclear architecture meets nuclear function. J Cell Sci. 2000;113:1841–1849. doi: 10.1242/jcs.113.11.1841. [DOI] [PubMed] [Google Scholar]
- Moen J, P. T, Smith KP, Lawrence JB. Compartmentalization of specific pre-mRNA metabolism: an emerging view. Hum. Mol. Genet. 1995;4:1779–1789. doi: 10.1093/hmg/4.suppl_1.1779. [DOI] [PubMed] [Google Scholar]
- Moen PT, Jr., Johnson CV, Byron M, Shopland LS, de la Serna IL, Imbalzano AN, Lawrence JB. Repositioning of muscle-specific genes relative to the periphery of SC-35 domains during skeletal myogenesis. Mol Biol Cell. 2004;15:197–206. doi: 10.1091/mbc.E03-06-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar C, Abdulle A, Gena A, Tanke HJ, Dirks RW. Poly(A)+ RNAs roam the cell nucleus and pass through speckle domains in transcriptionally active and inactive cells. J Cell Biol. 2004;165:191–202. doi: 10.1083/jcb.200310139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyman U, Hallman H, Hadlaczky G, Pettersson I, Sharp G, Ringertz NR. Intranuclear localization of snRNP antigens. J.Cell Biol. 1986;102:137–144. doi: 10.1083/jcb.102.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendergrast PS, Wang C, Hernandez N, Huang S. FBI-1 can stimulate HIV-1 Tat activity and is targeted to a novel subnuclear domain that includes the Tat-PTEFb-containing nuclear speckles. Mol Biol Cell. 2002;13:915–929. doi: 10.1091/mbc.01-08-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salditt-Georgieff M, Harpold MM, Wilson MC, Darnell JE., Jr. Large heterogeneous nuclear ribonucleic acid has three times as many 5′ caps as polyadenylic acid segments, and most caps do not enter polyribosomes. Mol.Cell.Biol. 1981;1(2):179–187. doi: 10.1128/mcb.1.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schul W, de Jong L, van Driel R. Nuclear neighbours: the spatial and functional organization of genes and nuclear domains. J Cell Biochem. 1998;70:159–171. doi: 10.1002/(sici)1097-4644(19980801)70:2<159::aid-jcb2>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Shibata S, Matsuoka Y, Yoneda Y. Nucleocytoplasmic transport of proteins and poly(A)+ RNA in reconstituted Tpr-less nuclei in living mammalian cells. Genes Cells. 2002;7:421–434. doi: 10.1046/j.1365-2443.2002.00525.x. [DOI] [PubMed] [Google Scholar]
- Shopland LS, Johnson CV, Byron M, McNeil J, Lawrence JB. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J Cell Biol. 2003;162:981–990. doi: 10.1083/jcb.200303131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland LS, Johnson CV, Lawrence JB. Evidence that all SC-35 domains contain mRNAs and that transcripts can be structurally constrained within these domains. J Struct Biol. 2002;140:131–139. doi: 10.1016/s1047-8477(02)00507-5. [DOI] [PubMed] [Google Scholar]
- Shopland LS, Lawrence JB. Seeking common ground in nuclear complexity [comment] J Cell Biol. 2000;150:F1–4. doi: 10.1083/jcb.150.1.f1. [DOI] [PubMed] [Google Scholar]
- Skalsky YM, Ajuh PM, Parker C, Lamond AI, Goodwin G, Cooper CS. PRCC, the commonest TFE3 fusion partner in papillary renal carcinoma is associated with pre-mRNA splicing factors. Oncogene. 2001;20:178–187. doi: 10.1038/sj.onc.1204056. [DOI] [PubMed] [Google Scholar]
- Skare P, Kreivi JP, Bergstrom A, Karlsson R. Profilin I colocalizes with speckles and Cajal bodies: a possible role in pre-mRNA splicing. Exp Cell Res. 2003;286:12–21. doi: 10.1016/s0014-4827(03)00102-2. [DOI] [PubMed] [Google Scholar]
- Smith KP, Moen PT, Wydner KL, Coleman JR, Lawrence JB. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J Cell Biol. 1999;144:617–629. doi: 10.1083/jcb.144.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somanathan S, Stachowiak EK, Siegel AJ, Stachowiak MK, Berezney R. Nuclear matrix bound fibroblast growth factor receptor is associated with splicing factor rich and transcriptionally active nuclear speckles. J Cell Biochem. 2003;90:856–869. doi: 10.1002/jcb.10672. [DOI] [PubMed] [Google Scholar]
- Spector DL. Macromolecular Domains Within the Cell Nucleus. Ann.Rev.Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Spector DL, Schrier WH, Busch H. Immunoelectron microscopic localization of SnRNPs. Biol.Cell. 1983;49:1–10. doi: 10.1111/j.1768-322x.1984.tb00215.x. [DOI] [PubMed] [Google Scholar]
- Tam R, Shopland LS, Johnson CV, McNeil J, Lawrence JB. Applications of RNA FISH for visualizing gene expression and nuclear architecture. Oxford University Press; New York: 2002. [Google Scholar]
- Trinkle-Mulcahy L, Sleeman JE, Lamond AI. Dynamic targeting of protein phosphatase 1 within the nuclei of living mammalian cells. J Cell Sci. 2001;114:4219–4228. doi: 10.1242/jcs.114.23.4219. [DOI] [PubMed] [Google Scholar]
- Valdez BC, Perlaky L, Henning D. Expression, cellular localization, and enzymatic activities of RNA helicase II/Gu(beta) Exp Cell Res. 2002;276:249–263. doi: 10.1006/excr.2002.5538. [DOI] [PubMed] [Google Scholar]
- van den IP, Wheelock R, Prescott A, Russell P, Quinlan RA. Nuclear speckle localisation of the small heat shock protein alpha B-crystallin and its inhibition by the R120G cardiomyopathy-linked mutation. Exp Cell Res. 2003;287:249–261. doi: 10.1016/s0014-4827(03)00092-2. [DOI] [PubMed] [Google Scholar]
- Visa N, Puvion-Dutilleul F, Harper F, Bachellerie J-P, Puvion E. Intranuclear distribution of Poly(A) RNA determined by electron microscope in situ hybridization. Exp.Cell Res. 1993;208:19–34. doi: 10.1006/excr.1993.1218. [DOI] [PubMed] [Google Scholar]
- Wagner S, Chiosea S, Nickerson JA. The spatial targeting and nuclear matrix binding domains of SRm160. Proc Natl Acad Sci USA. 2003;100:3269–3274. doi: 10.1073/pnas.0438055100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Somanathan S, Samarabandu J, Berezney R. Three-dimensional visualization of transcription sites and their association with splicing factor-rich nuclear speckles. JCB. 1999;146:543–558. doi: 10.1083/jcb.146.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. Embo J. 1998;17:6359–6367. doi: 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Dobner PR, Lawrence JB. Higher level organization of individual gene transcription and RNA splicing. Science. 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Moen PT, McNeil JA, Lawrence JB. Nonrandom gene organization: Structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J. Cell Biol. 1995;131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Taneja KL, Singer RH, Green MR. Localization of pre-mRNA splicing in mammalian nuclei. Nature. 1994;372:809–812. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Luo MJ, Straesser K, Katahira J, Hurt E, Reed R. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature. 2000;407:401–405. doi: 10.1038/35030160. [DOI] [PubMed] [Google Scholar]
- Zhu J, Krainer AR. Pre-mRNA splicing in the absence of an SR protein RS domain. Genes Dev. 2000;14:3166–3178. doi: 10.1101/gad.189500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YQ, Lu Y, Tan XD. Monochloramine induces reorganization of nuclear speckles and phosphorylation of SRp30 in human colonic epithelial cells: role of protein kinase C. Am J Physiol Cell Physiol. 2003;285:C1294–1303. doi: 10.1152/ajpcell.00090.2003. [DOI] [PubMed] [Google Scholar]
- Zimowska G, Shi J, Munguba G, Jackson MR, Alpatov R, Simmons MN, Shi Y, Sugrue SP. Pinin/DRS/memA interacts with SRp75, SRm300 and SRrp130 in corneal epithelial cells. Invest Ophthalmol Vis Sci. 2003;44:4715–4723. doi: 10.1167/iovs.03-0240. [DOI] [PubMed] [Google Scholar]
- Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, Rudolph C, Alt F, Luther K, Braz C, Sadoni N, Rosenecker J, Schindelhauer D. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirbel RM, Mathieu UR, Kurz A, Cremer T, Lichter P. Evidence for a nuclear compartment of transcription and splicing located at chromosome domain boundaries. Chrom.Res. 1993;1:92. doi: 10.1007/BF00710032. [DOI] [PubMed] [Google Scholar]


