Abstract
Transcription is the first step in gene expression, and its regulation underlies multicellular development and the response to environmental changes. Most studies of transcriptional regulation have focused on the recruitment of RNA polymerase to promoters. However, recent work has shown that, for many promoters, post-recruitment steps in transcriptional initiation are likely to be rate-limiting. The rate at which RNA polymerase transitions from transcriptional initiation to elongation varies dramatically between promoters and between organisms, and is the target of multiple regulatory proteins that can function to both repress and activate transcription.
Introduction
Transcription is the first step in gene expression and is the major target of regulation. Transcription can be divided into three distinct phases: (i) initiation, (ii) elongation, and (iii) termination. It has been widely assumed that recruitment of RNA polymerase (RNAP) during transcription initiation is usually the rate-limiting step in transcription. Indeed, artificial recruitment of the transcriptional machinery is often sufficient for productive transcription in E. coli, yeast, and human cells [1–3]. However, recent genome-wide studies indicate that, for many promoters in both prokaryotes and eukaryotes, the rate-limiting step in transcription initiation is likely to occur after recruitment of RNAP. Furthermore, the transition from initiation to elongation is an important target of regulation in both prokaryotes and eukaryotes. In this review we discuss the mechanisms of transition from transcriptional initiation to elongation, how this transition varies between promoters and between species, and how it is regulated by proteins and small molecules.
The transition from transcription initiation to elongation in bacteria
In eubacterial species, transcription of all genes is mediated by a core RNAP complex, typically a 5-subunit (α2ββ'ω) enzyme. However, in order to recognize promoter DNA sequences, this core enzyme must associate with a σ factor to form RNAP holoenzyme [4]. Initiation occurs at a site that is a fixed distance from the σ recognition sequences. Eubacterial species typically contain multiple σ factors that form distinct classes of RNAP holoenzymes that recognize different promoter sequences and regulate distinct classes of genes [4]. σ does not usually associate with elongating RNAP in vivo, although this can occur at a minority of genes under certain environmental conditions [5,6].
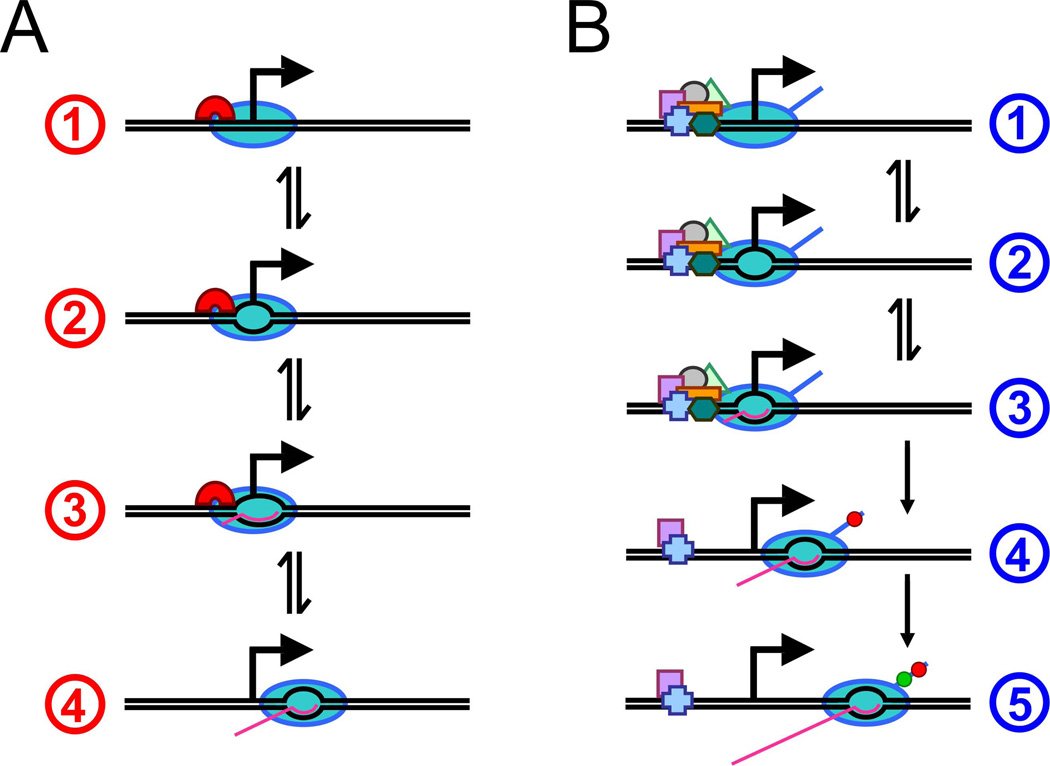
Transcription initiation by RNAP holoenzyme involves three biochemically defined steps (Figure 1A). RNAP holoenzyme binds to promoter DNA to form the closed “preinitiation” complex, melts the DNA around the transcription start site to form the open complex, and then transitions from initiation to elongation in a process known as promoter escape. Promoter escape typically includes multiple cycles of abortive initiation where RNAP synthesizes short RNAs of 2–15 nt [7]. Two recent studies have shown that promoter escape involves “scrunching” of the DNA immediately downstream of the transcription start site [8,9]. The upstream face of RNAP remains stationary relative to DNA during abortive initiation while the downstream DNA is drawn into RNAP. This scrunching creates a stressed intermediate state during transcription initiation due to the unwinding and compaction of DNA. It has been proposed that this stressed intermediate provides the driving force for either abortive initiation or promoter escape [8,9].
Figure 1. Steps in transcription initiation.
- Preinitiation closed complex formation at the promoter by RNAP holoenzyme (containing a σ factor).
- DNA is unwound around the transcription start site to form an open complex.
- Abortive synthesis of 2–15 nt RNAs requiring DNA “scrunching”.
- Promoter escape is typically associated with loss of σ factor.
- Preinitiation complex formation at the promoter with Pol II and general transcription factors.
- DNA is unwound around the transcription start site to form an open complex.
- Abortive synthesis of 2–3 nt RNAs.
- Promoter escape is associated with release of most general transcription factors and with phosphorylation at Serine 5 of the C-terminal domain of the largest Pol II subunit (red circle). In some eukaryotes Pol II pauses after synthesis of 20–50 nt RNAs.
- Escape from promoter-proximal pauses is associated with phosphorylation at Serine 2 of the C-terminal domain of the largest Pol II subunit (green circle) by pTEFb.
The transition from transcription initiation to elongation in eukaryotes
Eukaryotic cells contain 3 nuclear RNA polymerases, with RNA polymerase II (Pol II) responsible for transcribing all mRNAs and numerous non-coding RNAs. Pol II, a 12-subunit enzyme with many similarities to bacterial RNAP, does not recognize promoter DNA by itself, but rather as part of the basal Pol II machinery that includes general transcription factors (TFIIA, B, D, E, F, H). Like σ factors, these general transcription factors do not associate with elongating Pol II, and hence rapidly dissociate from Pol II during the transition between initiation and elongation [10]. Numerous factors (e.g. FACT, Spt4, Paf1 and TREX complexes, Spt6, Swi/Snf) travel with elongating Pol II throughout the coding region [11]. Importantly, eukaryotic Pol II must contend with nucleosomes that inhibit both initiation and elongation, unlike the E. coli RNAP which interacts with a genome that is permissive for binding transcription factors [12].
The C-terminal domain (CTD) of the largest Pol II subunit contains multiple copies of a heptad repeat that is phosphorylated at serines 2 and 5 by different kinases [11,13]. After initial association of the unphosphorylated form of Pol II into the preinitiation complex, serine 5 is phosphorylated by TFIIH at the promoter, and then serine 2 is phosphorylated by P-TEFb/CTK1 as Pol II elongates through the mRNA coding region. CTD phosphorylation appears to be relatively unimportant for transcriptional initiation or elongation per se, but rather plays a critical role in coupling Pol II elongation to post-transcriptional steps such as mRNA capping, splicing, polyadenylation, export, and chromatin modifications such as histone methylation.
The steps of Pol II association into a closed complex, open complex formation, and promoter escape are analogous to, but mechanistically distinct from, those of bacterial RNAP (Figure 1B). Of particular significance, open complex formation requires the helicase activity of TFIIH, and promoter escape does not coincide with abortive initiation [14]. In addition, unlike most species in which initiation occurs at a fixed position downstream from the TATA element, initiation in Saccharomyces cerevisiae occurs at larger and more variable distances, suggesting that there are species-specific differences in how transcription is initiated in eukaryotes. Lastly, as will be discussed in more detail below, there can be an additional Pol II pausing step in the transition between initiation and elongation [15,16].
Rate-limiting steps in transcription initiation vary among genes and organisms
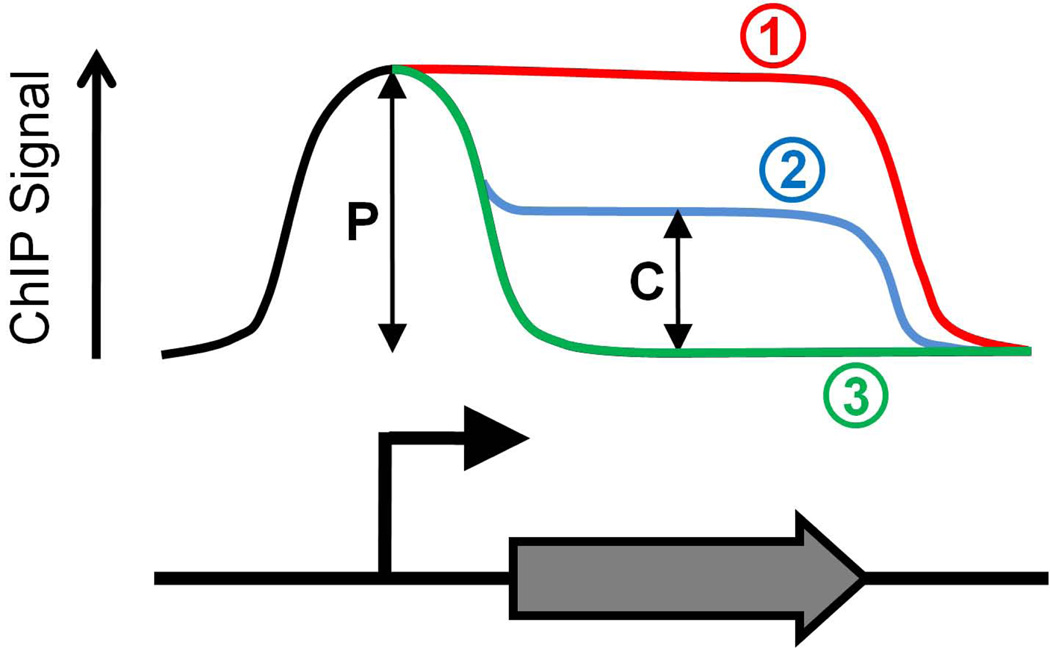
The transition between initiation and elongation in vivo can be investigated by using chromatin immunoprecipitation (ChIP) to determine RNAP association at promoters and transcribed regions. Recently, this transition has been analyzed on a genome-wide scale in several organisms using tiled, high-density microarrays (ChIP-chip). If the transition from initiation to elongation is rapid, the level of RNAP at a promoter will be roughly equivalent to the level in the corresponding coding sequence. If the transition is slow, RNAP association will be much greater at the promoter than in the corresponding coding sequence (Figure 2). Thus, the ratio of promoter association to coding sequence association is a simple measure of the rate of transition from initiation to elongation. We refer to this ratio as the “Traveling Ratio” (TR) [6].
Figure 2. Possible profiles of ChIP signal for RNAP.
The graph shows 3 possible ChIP profiles for RNAP across a gene. In all cases RNAP associates with promoter DNA sequences at an equivalent level. In case (1) the ChIP signal is constant throughout the promoter and coding sequence, indicating rapid transition of RNAP from initiation to elongation. In case (2) the ChIP signal is reduced within the coding sequence as compared to the promoter, although ChIP signal in the coding sequence is above background. This indicates that some or all RNAP complexes transition slowly from initiation to elongation. As ChIP measures a population of cells it is impossible to determine whether all RNAP complexes transition at the same rate. In case (3) the ChIP signal is only present at the promoter. This indicates “poised” RNAP at the promoter, i.e. RNAP that is unable to make the transition from initiation to elongation. If the peak of RNAP ChIP signal is downstream of the transcription start site this indicates that RNAP is paused in early elongation. In all cases a Traveling Ratio (TR) can be calculated as the ratio of coding sequence signal (C) to promoter signal (P). Hence, TR can be used as an measure of the rate of transition from initiation to elongation at a given promoter.
In rapidly growing E. coli, the TR values for different transcribed regions are highly variable, ranging from 0 to 1 [6]. In most cases, TR values are < 1, and the median value is 0.43, suggesting (i) that the transition from initiation to elongation is limiting at most transcribed regions, and (ii) that RNAP spends about 1–3 seconds at a promoter, which is ~50-fold more time than at a given position within the coding region. Strikingly, for 23% of transcribed regions where RNAP association is observed, there is no detectable transcript, indicating that RNAP at these promoters is unable to transition from initiation to elongation and hence is “poised”. It should be noted this genome-wide pattern of RNAP reflects the σ70-containing form which is responsible for transcription of the vast majority of E. coli genes. In contrast, σ54–containing RNAP is typically poised at the promoter, and transcription requires an activator protein and ATP hydrolysis [17].
In the budding yeast S. cerevisiae, genome-wide [18–20] analyses indicate that the level of Pol II association at promoters is roughly equivalent to that within the corresponding coding sequence in almost all cases; average TR values = ~0.9 [6,18]. This observation indicates a rapid transition from initiation to elongation, which is consistent with the very strong correlation between the level of promoter-bound TBP (a general transcription factor) and the level of transcription [21,22]. There are, however, isolated examples of transcriptionally inactive genes whose promoters bind the transcriptional machinery, probably with Pol II in a closed complex [21,23]. In addition, Pol II associates with many transcriptionally inactive promoters during stationary phase [19], particularly for genes whose transcription is induced within 3 minutes of exiting stationary phase, suggesting that Pol II is poised for rapid activation. Intriguingly, the Rpb1 CTD is hypophosphorylated in yeast cells during stationary phase, suggesting that global regulation of transcription could be brought about by controlling CTD phosphorylation [19].
The distribution of Pol II in human and Drosophila cells is very different from that in growing yeast cells, and in fact is more similar to the RNAP pattern in E. coli. For most transcribed regions, the level of Pol II association at the promoter is substantially higher than that in the corresponding coding sequence [24–30], indicating that post-recruitment steps in transcription initiation are generally slow. Furthermore, 20–50% of Pol II-bound promoters correspond to transcriptionally inactive genes [27,29–31]. Several lines of evidence suggest that Pol II at most of these transcriptionally inactive genes is “paused” in early elongation 20–50 bp beyond the initiation site, in a manner similar to that described two decades ago for Drosophila heat shock genes [14]. First, this class of genes contains chromatin with tri-methylated H3–K4, a histone mark that is generated after transcriptional initiation [27]. Second, the average position of Pol II at these genes is 50 bp downstream from the initiation site [30]. Third, in all cases tested, RNA can be detected at the extreme 5’ end of the gene, but not further downstream [27]. Fourth, in all cases tested, permanganate mapping reveals an open transcription bubble around the pause site [29,30]. As is the case for E. coli promoters with “poised” RNAP containing σ70, TFIID and presumably other general transcription factors are associated with promoters containing paused Pol II [14,25,28,31]. However, the paused Pol II observed at human and Drosophila promoters is transcriptionally engaged [14], and in this respect may differ from “poised” E. coli RNAP at promoters of inactive genes.
The prevalence of paused Pol II indicates that this is a major rate-limiting step in transcription in flies, human, and presumably most eukaryotic species. Strikingly, paused Pol II in Drosophila appears to be preferentially localized at genes involved in development, suggesting that Pol II pausing may have evolved to allow regulation of specific cellular processes [29,30].
Why does Pol II pause at many eukaryotic genes in vivo?
Three potential mechanisms, not mutually exclusive, might be considered for paused Pol II in vivo, given that such pauses do not occur in vitro with the minimal core Pol II machinery [14]. First, negative elongation factors (e.g. NELF and DSIF) associating with the preinitiation or early initiation complex might block the transition to full elongation. Second, positive factors might be required to dissociate Pol II from general initiation factors that are localized to the promoter, thereby limiting the distance Pol II can travel downstream from the initiation site. Third, nucleosomes near the initiation site might inhibit elongation, in which case variability in the position of paused Pol II among different genes might be explained by variations in position of the promoter-proximal nucleosome with respect to the initiation site. For all of these mechanisms, elongation beyond the pause requires the recruitment of positive elongation factors that remove the negative factors and/or mobilize or alter nucleosomes. Examples of such positive elongation factors are P-TEFb, which phosphorylates serine 2 of the CTD, and chromatin-modifying factors such as JIL-1 kinase (phosphorylates histone H3 at serine 10)[32], FACT (an H2A-H2B chaperone), Paf1 complex (required for H3 methylation at lysines 4, 36, and 79), and Spt6 (an H3–H4 chaperone). TFIIS, the transcript cleavage factor, also plays a role in releasing paused Pol II, and DSIF remains associated with elongating Pol II after release from the pause and may act as a positive elongation factor [14].
Why does paused Pol II not occur in S. cerevisiae, especially considering that almost all the relevant factors are present in this organism? Possible explanations include 1) the absence of NELF, 2) differences in histone variants and possibly stability or positioning of nucleosomes, or 3) the absence of activators that (directly or indirectly) recruit the preinitiation complex but not elongation factors. An intriguing possibility is that lack of paused Pol II is linked to the longer and more variable distance between the TATA element and initiation site in S. cerevisiae than in most eukaryotic organisms [33]. This difference in start-site selection occurs in vitro, and is due primarily to TFIIB and Pol II [34]. As the stereochemistry of TFIIB with respect to initiation site is conserved, we speculate that S. cerevisiae Pol II rapidly dissociates from the preinitiation complex due to a weak interaction with TFIIB, whereupon it travels down the gene for a variable distance prior to initiating transcription at a site defined by the initiator element. An inherently unstable preinitiation complex in yeast cells is supported by an unusually large open complex between the TATA element and initiation site [35], and the lack of Mediator at core promoters [36].
Controlling the rate of transition by DNA sequence
Gene-specific variation in the rate of transition from initiation to elongation can be due to differences in promoter DNA sequence. In E. coli, such differences can affect both the affinity of RNAP for the promoter and the rate of post-recruitment steps [37], such as the level of abortive initiation and promoter escape [38,39], the formation of unproductive “moribund” RNAP complexes [40,41], and σ-dependent pausing of RNAP in the initial transcribed region [42,43]. As initial transcription occurs through scrunching, an intriguing possibility is that the presence of promoter-proximal σ-dependent pause sites may cause RNAP to favor forward translocation and hence promoter escape, rather than repeated cycles of abortive initiation. In addition, relative differences in DNA melting temperature around promoters may influence the rate of transition from initiation to elongation [6].
In eukaryotes, sequence-dependent effects on the transition between initiation and elongation have yet to be described, although some of the above mechanisms may be involved. Sequence-dependent effects might also arise from the fact that core promoters have a great deal of structural and functional diversity, particularly with respect to the TATA, initiator, and downstream promoter elements [44]. In addition, Pol II initiation and elongation is strongly inhibited by nucleosomes, and DNA sequences in the vicinity of the promoter may differ significantly with respect to nucleosome positioning and stability.
Regulating the rate of transition by proteins and small molecules
In bacteria, the transition from initiation to elongation is regulated by a wide variety of proteins and mechanisms. Some DNA-binding repressors stabilize initiating RNAP:promoter DNA complexes, thereby trapping RNAP at the promoter [45]. The nucleoid-associated protein, H-NS, can trap RNAP at promoters by forming looping interactions between binding sites upstream and downstream of the promoter [46]. Intriguingly, loop formation, and hence repression, occur when RNAP is bound to σ70 but not to the alternative σ factor, σ38 [46]. The transition from initiation to elongation can also be accelerated by certain activator proteins that bind directly to both promoter DNA and RNAP and stimulate transcription at a post-recruitment step [47–50]. The transcription elongation factor GreA, a homologue of eukaryotic TFIIS, can also increase the rate of transition from initiation to elongation [51,52], presumably through its RNA cleavage activity that can rescue unproductive RNAP complexes [40,41].
The transition from initiation to elongation can also be regulated by small molecules. For example, RNAP is poised at the osmY promoter under conditions of low glutamate, but is released when the glutamate concentration increases, indicating a direct role for glutamate in controlling the conformation of RNAP [53]. Small molecules can function through transcription factors. ArgP either represses or activates transcription at the argO promoter in the presence or lysine or arginine respectively. In both cases RNAP binds promoter DNA and forms open complex but in the presence of lysine RNAP is unable to complete promoter escape [54]. ppGpp, a small molecule produced during nutrient starvation, binds directly to RNAP and down-regulates transcription at promoters that have intrinsically unstable open complexes [55]. ppGpp can also upregulate transcription of certain genes, and DksA has been implicated in modulating ppGpp function [55]. Promoters with unstable open complexes are also regulated by the concentration of the initiating nucleotide, which increases the half-life of open complexes by mass action [56].
In eukaryotes, the widespread existence of paused Pol II (except in S. cerevisiae) strongly suggests that this a major step at which the transition between initiation and elongation is regulated [14]. The classic example of such regulation is the induction of heat shock genes in Drosophila, in which the release of paused Pol II is mediated by HSF, a DNA-binding transcriptional activator protein. It is presumed that HSF recruits factors that release paused Pol II and permit it to traverse the gene, although it is unclear which factors are direct targets and exactly how Pol II release occurs. p-TEFb plays an important role, because artificial recruitment bypasses the pause [57], and chemical inhibition blocks release from the pause [58]. It is likely that other activators (e.g. the HIV Tat protein) will function in a manner analogous to HSF, although the precise details may differ [59]. Conversely, some repressors (e.g. PIE-1) [60] might function by inhibiting CTD-serine 2 phosphorylation and/or recruitment of elongation factors. P-TEFb is negatively regulated through its association with a complex containing 7SK RNA, HEXIM, and other proteins [59].
The transition between initiation and elongation can be regulated in other ways. For example, the interaction between phosphorylated ELK1 and the Med23 subunit of Mediator is not only important for recruitment of Mediator to the Egr1 promoter, but also for a step after preinitiation complex assembly that permits Pol II to escape the promoter [61]. However, the salt-sensitivity of Pol II in the uninduced state indicates that regulation does not involved paused Pol II. It is also worth noting that transcription of ribosomal RNA by Pol I is also regulated after recruitment to the promoter, with UBF1 and nuclear actin being implicated in promoter clearance in human cells [62,63].
In vivo, the basic Pol II machinery is unable to access the chromatin template unless it is recruited (directly or indirectly) by DNA-binding activator proteins [64]. As a consequence, and given the existence of paused Pol II, there must be a class of “activators” that can recruit the core machinery to the promoter, but is unable to recruit elongation factors and hence stimulate Pol II transcription. Based on the Drosophila heat shock genes, the GAGA factor is likely to be such an “activator”. Such “activators” are analogous to, although mechanistically distinct from, bacterial activators that require regulatory signals to stimulate post-recruitment steps. It is possible that S. cerevisiae lacks activator proteins of this type.
Conclusions
Post-recruitment steps in transcription initiation are rate-limiting at many promoters in species ranging from E. coli to humans. There is great variability within the rate of these steps both within and between species, and under different growth conditions. Since most studies of transcriptional regulation have focused on recruitment of RNAP to promoters there is still much to learn about the transition from initiation to elongation, in particular with regard to the proteins and small molecules that regulate this process.
ACKNOWLEDGEMENTS
We thank John Lis for thoughtful discussion. This work was supported by a Charles A. King Trust Postdoctoral Fellowship, Bank of America, Co-Trustee (Boston, MA) to J.T.W. and a research grant from the NIH (GM30186) to K.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference Comments
**Reppas et al., 2006
The authors show that the transition from initiation to elongation is slow at most promoters and that nearly a quarter of promoters that bind RNAP are transcriptionally silent.
**Kapanidis et al., 2006; **Revyakin et al., 2006
The authors show that DNA in the initial transcribed region must be “scrunched” during promoter escape, suggesting the formation of a stressed intermediate during the transition from initiation to elongation.
*Radonjic et al., 2005
The authors show that many promoters bind poised Pol II in S. cerevisiae during stationary phase, and that transcription of the corresponding genes is induced rapidly upon the transition to exponential growth.
**Guenther et al., 2007
The authors show that most promoters in human stem cells, including many that are transcriptionally inactive, bind Pol II and have histone modifications associated with transcriptionally active regions.
**Zeitlinger et al., 2007; **Muse et al., 2007
The authors show that promoter-proximal pausing of Pol II is widespread in Drosophila embryos. Intriguingly, genes binding paused Pol II are enriched for those involved in development.
*Ivaldi et al., 2007
The authors identify a histone modification that is associated with release of Pol II from promoter-proximal pauses in Drosophila.
*Laishram and Gowrishankar, 2007
The authors show that ArgP, a transcription factor in E. coli, has different effects on promoter escape depending on whether it is bound to arginine or lysine.
REFERENCES
- 1.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 2.Nevado J, Gaudreau L, Adam M, Ptashne M. Transcriptional activation by artificial recruitment in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2674–2677. doi: 10.1073/pnas.96.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorris DR, Struhl K. Artificial recruitment of TFIID, but not RNA polymerase II holoenzyme, activates transcription in mammalian cells. Mol. Cell. Biol. 2000;20:4350–4358. doi: 10.1128/mcb.20.12.4350-4358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gross CA, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harbor Symp. Quant. Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- 5.Mooney RA, Darst SA, Landick R. Sigma and RNA polymerase: an on-again, off-again relationship? Mol. Cell. 2005;20:335–345. doi: 10.1016/j.molcel.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 6.Reppas NB, Wade JT, Church G, Struhl K. The transition between transcriptional initiation and elongation in E. coli is often rate-limiting, variable, and associated with rapid release of σ70. Mol. Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 7.McClure W. Mechanism and control of transcription initiation in prokaryotes. Annu. Rev. Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 8.Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 11.Sims RJ, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes & Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 12.Wade JT, Reppas NB, Church GM, Struhl K. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes & Dev. 2005;19:2619–2630. doi: 10.1101/gad.1355605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komarnitsky P, Cho E-J, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes & Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell. Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 15.Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5' end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 16.Krumm A, Hickey LB, Groudine M. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes & Dev. 1995;9:559–572. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- 17.Buck M, Gallegos MT, Studholme DJ, Guo Y, Gralla JD. The bacterial enhancer-dependent sigma(54)(sigmaN) transcription factor. J. Bacteriol. 2000;182:4129–4136. doi: 10.1128/jb.182.15.4129-4136.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radonjic M, Andrau JC, Lijnzaad P, Kemmeren P, Kockelkorn TT, van Leenen D, van Berkum NL, Holstege FC. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol. Cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Z AA, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 21.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–612. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 22.Li X-Y, Virbasius A, Zhu X, Green MR. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 23.Martens C, Krett B, Laybourn PJ. RNA polymerase II and TBP occupy the repressed CYC1 promoter. Mol. Microbiol. 2001;40:1009–1019. doi: 10.1046/j.1365-2958.2001.02445.x. [DOI] [PubMed] [Google Scholar]
- 24.Cheng C, Sharp PA. RNA polymerase II accumulation in the promoter-proximal region of the dihydrofolate reductase and gamma-actin genes. Mol. Cell. Biol. 2003;23:1961–1967. doi: 10.1128/MCB.23.6.1961-1967.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ENCODE project consortium. The ENCODE (ENCyclopedia of DNA Elements) project. Science. 2004;304:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 26.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 27.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 29.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeitlinger J, Stark A, Kellis M, Hong J-W, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ivaldi MS, Karam CS, Corces VG. Phosphorylation of histone H3 at Ser10 facilitates RNA polymerase II release from promoter-proximal pausing in Drosophila. Genes Dev. 2007;21:2818–2831. doi: 10.1101/gad.1604007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Struhl K. Yeast transcriptional regulatory mechanisms. Ann. Rev. Genet. 1995;29:651–674. doi: 10.1146/annurev.ge.29.120195.003251. [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Flanagan PM, Tschochner H, Kornberg RD. RNA polymerase II initiation factor interactions and transcription start site selection. Science. 1994;263:805–807. doi: 10.1126/science.8303296. [DOI] [PubMed] [Google Scholar]
- 35.Giardina C, Lis JT. Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol. Cell. Biol. 1995;15:2737–1744. doi: 10.1128/mcb.15.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan X, Chou D, Struhl K. Activator-specific recruitment of Mediator in vivo. Nat. Struct. Mol. Biol. 2006;13:117–120. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- 37.Xu J, McCabe BC, Koudelka GB. Function-based selection and characterization of base-pair polymorphisms in a promoter of Escherichia coli RNA polymerasesigma(70) J. Bacteriol. 2001;183:2866–2873. doi: 10.1128/JB.183.9.2866-2873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu LM, Vo NV, Kane CM, Chamberlin MJ. In vitro studies of transcript initiation by Escherchia coli RNA polymerase. 1. RNA chain initiation, abortive initiation, and promoter escape at three bacteriophage promoters. Biochemistry. 2003;42:3777–3786. doi: 10.1021/bi026954e. [DOI] [PubMed] [Google Scholar]
- 39.Vo NV, Hsu LM, Kane CM, Chamberlin MJ. In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 3. Influences of individual DNA elements within the promoter recognition region on abortive initiation and promoter escape. Biochemistry. 2003;42:3798–3811. doi: 10.1021/bi026962v. [DOI] [PubMed] [Google Scholar]
- 40.Susa M, Sen R, Shimamoto N. Generality of the branched pathway in transcription initiation by Escherichia coli RNA polymerase. J. Biol. Chem. 2002;277:15407–15412. doi: 10.1074/jbc.M112481200. [DOI] [PubMed] [Google Scholar]
- 41.Vo NV, Hsu LM, Kane CM, Chamberlin MJ. In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 2. Formation and characterization of two distinct classes of initial transcribing complexes. Biochemistry. 2003;42:3787–3797. doi: 10.1021/bi0269613. [DOI] [PubMed] [Google Scholar]
- 42.Brodolin K, Zenkin N, Mustaev A, Mameava D, Heumann H. The sigma 70 subunit of RNA polymerase induces lacUV5 promoter-proximal pausing of transcription. Nat. Struct. Mol. Biol. 2004;11:551–557. doi: 10.1038/nsmb768. [DOI] [PubMed] [Google Scholar]
- 43.Nickels BE, Mukhopadhyay J, Garrity SJ, Ebright RH, Hochschild A. The sigma(70) subunit of RNA polymerase mediates a promoter-proximal pause at the lac promoter. Nat. Struct. Mol. Biol. 2004;11:544–550. doi: 10.1038/nsmb757. [DOI] [PubMed] [Google Scholar]
- 44.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 45.Choy HE, Hanger RR, Aki T, Mahoney M, Murakami K, Ishihama A, Adhya S. Repression and activation of promoter-bound RNA polymerase activity by Gal repressor. J. Mol. Biol. 1997;272:293–300. doi: 10.1006/jmbi.1997.1221. [DOI] [PubMed] [Google Scholar]
- 46.Shin M, Song M, Rhee JH, Hong Y, Kim YJ, Seok YJ, Ha KS, Jung SH, Choy HE. DNA looping-mediated repression by histone-like protein H-NS: specific requirement of Esigma70 as a cofactor for looping. Genes Dev. 2005;19:2388–2398. doi: 10.1101/gad.1316305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eichenberger P, Dethiollaz S, Buc H, Geiselmann J. Structural kinetics of transcription activation at the malT promoter of Escherichia coli by UV laser footprinting. Proc. Natl. Acad. Sci. U.S.A. 1997;94:9022–9027. doi: 10.1073/pnas.94.17.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dove SL, Huang FW, Hochschild A. Mechanism for a transcriptional activator that works at the isomerization step. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13215–13220. doi: 10.1073/pnas.97.24.13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu M, Gupte G, Roy S, Bandwar RP, Patel SS, Garges S. Kinetics of transcription initiation at lacP1. Multiple roles of cyclic AMP receptor protein. J. Biol. Chem. 2003;278:39755–39761. doi: 10.1074/jbc.M305995200. [DOI] [PubMed] [Google Scholar]
- 50.Roy S, Lim HM, Liu M, Adhya S. Asynchronous basepair openings in transcription initiation: CRP enhances the rate-limiting step. EMBO J. 2004;23:869–875. doi: 10.1038/sj.emboj.7600098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Susa M, Kubori T, Shimamoto N. A pathway branching in transcription initiation in Escherichia coli. Mol. Microbiol. 2006;59:1807–1817. doi: 10.1111/j.1365-2958.2006.05058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stepanova E, Lee J, Ozerova M, Semenova E, Datsenko K, Wanner BL, Severinov K, Borukhov S. Analysis of promoter targets for E. coli transcription elongation factor GreA in vivo and in vitro. J. Bacteriol. 2007;189:8772–8785. doi: 10.1128/JB.00911-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee SJ, Gralla JD. Osmo-regulation of bacterial transcription via poised RNA polymerase. Mol. Cell. 2004;14:153–162. doi: 10.1016/s1097-2765(04)00202-3. [DOI] [PubMed] [Google Scholar]
- 54.Laishram RS, Gowrishankar J. Environmental regulation operating at the promoter clearance step of bacterial transcription. Genes Dev. 2007;21:1258–1272. doi: 10.1101/gad.1520507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Magnusson LU, Farewell A, Nystrom T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 56.Schneider DA, Ross W, Gourse RL. Control of rRNA expression in Escherichia coli. Curr. Opin. Microbiol. 2003;6:151–156. doi: 10.1016/s1369-5274(03)00038-9. [DOI] [PubMed] [Google Scholar]
- 57.Lis J, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- 58.Ni Z, Saunders A, Fuda NJ, Yao J, Suarez JR, Webb WW, Lis JT. P-TEFb is critical for the maturation of RNA Polymerase II into productive elongation in vivo. Mol. Cell. Biol. 2008 doi: 10.1128/MCB.01859-07. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peterlin BM, Price DH. Controlling the elongation phase of transcription with PTEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 60.Zhang F, Barboric M, Blackwell TK, Peterlin BM. A model of repression: CTD analogs and PIE-1 inhibit transcriptional elongation by P-TEFb. Genes & Dev. 2003;17:748–758. doi: 10.1101/gad.1068203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mediator requirement for both recruitment and post-recruitment steps in transcription initiation. Mol. Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 62.Philimonenko VV, Zhao J, Iben S, Dingova H, Kysela K, Kahle M, Zentgraf H, Hofmann WA, de Lanerolle P, Hozak P, et al. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat. Cell Biol. 2004;6:1165–1172. doi: 10.1038/ncb1190. [DOI] [PubMed] [Google Scholar]
- 63.Panov KI, Friedrich JK, Russell J, Zomerdijk JC. UBF activates RNA polymerase I transcription by stimulating promoter escape. EMBO J. 2006;25:3310–3322. doi: 10.1038/sj.emboj.7601221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Struhl K. Fundamentally different logic of gene expression in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]