Abstract
To identify new loci that are involved in the assembly and targeting of dynein complexes, we have screened a collection of motility mutants that were generated by insertional mutagenesis. One such mutant, 5B10, lacks the inner arm isoform known as the I1 complex. This isoform is located proximal to the first radial spoke in each 96-nm axoneme repeat and is an important target for the regulation of flagellar motility. Complementation tests reveal that 5B10 represents a new I1 locus, IDA7. Biochemical analyses confirm that ida7 axonemes lack at least five I1 complex subunits. Southern blots probed with a clone containing the gene encoding the 140-kDa intermediate chain (IC) indicate that the ida7 mutation is the result of plasmid insertion into the IC140 gene. Transformation with a wild-type copy of the IC140 gene completely rescues the mutant defects. Surprisingly, transformation with a construct of the IC140 gene lacking the first four exons of the coding sequence also rescues the mutant phenotype. These studies indicate that IC140 is essential for assembly of the I1 complex, but unlike other dynein ICs, the N-terminal region is not critical for its activity.
INTRODUCTION
The dyneins are a family of motor enzymes that produce force by converting the energy derived from binding and hydrolyzing ATP into minus end-directed movement along the surface of microtubules. These multisubunit ATPases are involved in many essential cellular processes including the intracellular trafficking of organelles, chromosome movement, axonal transport, and the sliding of doublet microtubules during ciliary and flagellar motility (reviewed by Holzbaur and Vallee, 1994; Mitchell, 1994; and Porter, 1996). The diverse functions that the dyneins perform within the cell require that these motors be targeted to specific cellular locations, that they bind to the appropriate cargoes, and that they are activated at the proper time.
The unicellular biflagellate alga Chlamydomonas is an excellent model system for studying the functional diversity of the axonemal dyneins, as well as the mechanisms by which the specific targeting of dynein isoforms occurs. For instance, both the sequence analysis of dynein subunits and the phenotypes of related flagellar mutations have shown that the axonemal dyneins can be separated into two distinct classes: the outer dynein arms and inner dynein arms. The outer dynein arms add power to the flagellar beat, whereas inner dynein arms are required to generate flagellar waveforms (Mitchell and Rosenbaum, 1985; Brokaw and Kamiya, 1987). The outer dynein arm is a three-headed isoform that is composed of three dynein heavy chains (DHCs)1 along with several intermediate chains (ICs) and light chains (LCs) and repeats every 24 nm along the length of the axoneme (reviewed in Mitchell, 1994; Witman et al., 1994). The inner dynein arms are considerably more complex than the outer dynein arms and, as a result, they have been less well characterized. Seven distinct inner arm subspecies have been identified by a combination of ion exchange chromatography and electron microscopy (Goodenough et al., 1987; Kagami and Kamiya, 1992): six single-headed isoforms that can be separated into two classes (I2 and I3) based on their association with specific LCs (Kagami and Kamiya, 1992; LeDizet and Piperno, 1995) and one two-headed isoform known as the I1 complex, which is composed of two DHCs, three ICs, and three LCs (Piperno et al., 1990; Smith and Sale, 1991; Porter et al., 1992; Harrison et al., 1998). Structural studies have shown that each axonemal dynein isoform occupies a unique site within the 96-nm axoneme repeat and therefore, like the cytoplasmic dyneins, they must be targeted to the appropriate locations (Mastronarde et al., 1992; Smith and Sale, 1992; Gardner et al., 1994). One approach to address how the specific targeting of dynein isoforms occurs is to determine the contribution of each dynein subunit in the assembly and function of the various motor complexes.
All dynein isoforms contain at least 1–3 DHCs (400–550 kDa), one or more ICs (43–140 kDa), and variable numbers of LCs (8–28 kDa). The conserved C-terminal two-thirds of the DHC includes the globular head, known as the motor domain, which contains the nucleotide binding site and interacts transiently with microtubules (reviewed by Gibbons, 1995). The more divergent N-terminal third of the DHC forms a stem domain that is complexed with other subunits and binds to cellular cargo (Sakikibara et al., 1993; reviewed by Holzbaur and Vallee, 1994; Mitchell, 1994; Porter, 1996). The ICs appear to play a central role in the assembly and targeting of the dynein complexes. For instance, both outer arm and cytoplasmic dynein ICs have been localized to the stem domain, close to the site of cargo binding (King and Witman, 1990; Steffen et al., 1996). Sequence analyses have indicated that several of these ICs contain WD repeat motifs (Wilkerson et al., 1995; Ogawa et al., 1995). WD repeats have previously been proposed to be involved in protein-protein interactions within multisubunit complexes (Neer et al., 1994). Not surprisingly, null mutations in two outer arm ICs result in the failure to properly assemble this dynein isoform (Mitchell and Kang, 1991; Wilkerson et al., 1995). No definitive role has been determined for the LCs of the complex, although sequence homologies of one of the outer arm LCs to calmodulin and of two others to thioredoxins have led to the suggestion that the LCs may serve a regulatory role (King and Patel-King, 1995a, 1995b).
To identify genes and gene products that are required for assembly of the inner dynein arms, we have screened a new collection of motility mutants that were generated by insertional mutagenesis. Although many mutations that affect the assembly of inner dynein arms have been previously isolated (Huang et al., 1979; Brokaw and Kamiya, 1987; Kamiya et al., 1991; Porter et al., 1992; Kato et al., 1993), in only three cases have the corresponding gene products been identified (LeDizet and Piperno, 1995; Myster et al., 1997; Kato-Minoura et al., 1997). In this report, we characterize a new motility mutant and demonstrate that the observed defects are due to a mutation in a novel I1 locus IDA7, which results in the failure to assemble the I1 inner arm complex. The I1 complex has previously been implicated as an important target for the regulatory signals that control flagellar motility (Porter et al., 1992; Habermacher and Sale, 1996, 1997; King and Dutcher, 1997). The I1 complex is also the axonemal dynein isoform that appears to be most closely related to cytoplasmic dynein; both subspecies are two-headed complexes that contain two identical LC subunits (8 kDa and 14 kDa) (Harrison et al., 1998). By Southern blot analysis, we have shown that the defects in ida7 are due to the insertion of a selectable marker into the gene that encodes the 140-kDa IC (IC140) of the I1 complex. The IC140 is a WD repeat containing polypeptide with significant homology in its C terminus to both the outer arm and cytoplasmic dynein IC subunits (Yang and Sale, 1998). Transformation with a wild-type copy of the IC140 gene rescues the motility, biochemical, and structural defects seen in ida7. These results demonstrate that IC140 is essential for the proper assembly of the I1 complex. Transformation with additional constructs of the IC140 gene has also revealed regions of the gene that appear to be required for the regulated expression of the IC140 transcript. Finally, we have recovered a novel transformant that expresses a truncated polypeptide lacking the N-terminal 283 amino acids of IC140. The truncated polypeptide retains the WD repeat sequences and can reassemble with other I1 subunits into the flagellar axoneme. In addition, the motility phenotype of this strain is indistinguishable from wild-type. These results indicate that the N-terminal region of the IC140 is not essential for its function and further demonstrate that the domains involved in both complex assembly and targeting are located elsewhere in the IC140 sequence.
MATERIALS AND METHODS
Cell Culture
The strains used in this study are listed in Table 1 or described below. All cells were grown on Tris-acetate phosphate (TAP) medium (Gorman and Levine, 1965) or rich (R) medium containing sodium acetate, as described by Sager and Granick (1953) and modified by Holmes and Dutcher (1989), unless otherwise noted. Solid media contained Bacto-agar (Difco Laboratories, Detroit, MI) that had been washed four times with Milli-Q purified water and then air dried before use. All arg- strains were grown on TAP media supplemented with 0.6 mg of l-arginine per ml of media.
Table 1.
Strains used in this study
| Strain name | Dynein defect | Motility | Swimming velocity (μm/s) | Reference |
|---|---|---|---|---|
| 137c | None | Wild type | 144.2 ± 17.1a | Harris, 1989 |
| ida7 (5B10) | I1 complex | Slow swimming | 81.5 ± 14.0a | This study |
| 27B3 | I1 complex | Slow swimming | 77.6 ± 15.4a | Our unpublished results |
| Rescued ida7 (AH1) | None | Wild type | 166.1 ± 27.3a | This study |
| Rescued ida7 (5A) | Truncated I1 IC | Wild type | 155.3 ± 20.3a | This study |
| ida2 | I1 complex | Slow swimming | 77.7 ± 15.2b | Kamiya et al., 1991 |
| ida3 | I1 complex | Slow swimming | 77.3 ± 11.0b | Kamiya et al., 1991 |
| ida4 | I2 inner arms | Slow swimming | 102.2 ± 10.7b | Kamiya et al., 1991 |
| Kagami and Kamiya, 1992 | ||||
| pf3 | DRC components, | Slow swimming | 105.6 ± 11.4c | Huang et al., 1982; |
| I2/I3 reduced | Piperno et al., 1992 | |||
| pf9-2 | I1 complex | Slow swimming | ND | Porter et al., 1992 |
| pf9-3 | I1 complex | Slow swimming | ND | Myster et al., 1997 |
| pf28 | Outer-arm complex | Slow swimming | ND | Mitchell and Rosenbaum, 1985 |
| arg2 | None | Wild type | ND | Harris, 1989 |
| arg7 | None | Wild type | ND | Harris, 1989 |
| L5 | None | Wild type | ND | Tam, personal communication |
Velocity determined in this study; n = 60, except from strain 137c, where n = 40.
Velocity determined by Kamiya et al., 1991.
Velocity determined by Gardner et al., 1994.
Origin of Motility Mutations
Thirty-two of the motility mutants that were screened in this study were generated as described in Myster et al. (1997) based on the procedure of Tam and Lefebvre (1993). Twelve additional strains were generated by transformation of a nit1–305 strain with a pMN24 plasmid, which carries a wild-type copy of the nitrate reductase (NIT1) gene contained within a 14.5-kilobase (kb) fragment of Chlamydomonas genomic DNA (Fernandez et al., 1989; Kindle et al., 1989). These strains were generously provided by D. Mitchell (State University of New York Medical Center, Syracruse, NY).
Analysis of Motility Phenotypes
Motility phenotypes were assessed via phase contrast microscopy on a Zeiss Axioskop microscope (Carl Zeiss, Thornwood, NY) at a magnification of 200× or 400×. Measurements of swimming velocity were made from video recordings of live cells using a C2400 Newvicon camera and an Argus 10 video processor (Hamamatsu Photonic Systems, Bridgewater, NJ) that had been calibrated with a stage micrometer (Porter et al., 1992). The ability of strains to undergo phototaxis was monitored using a simple assay previously developed by King and Dutcher (1997).
Genetic Analysis
Genetic analysis was performed using standard techniques (Levine and Ebersold, 1960; Harris, 1989). To determine whether the motility phenotypes were linked to the selectable marker used in transformation, individual transformants were backcrossed to the nit1- strain L5 (nit1, apm1–19, mt+) provided by L.W. Tam, (University of Minnesota, St. Paul, MN). Because the viability of tetrad progeny from this cross was extremely low, we used the following method to isolate random progeny. After 24 h in the light, mating mixtures were plated on zygote plates containing 4% agar and kept in complete darkness for at least 4 d. Plates were then scraped with a razor blade to remove any nonmated, vegetative cells and exposed to light for 12–24 h to allow the zygotes to germinate. Examination of the zygote plates via a dissecting scope revealed that approximately 90% of the zygotes had germinated into colonies containing four to eight tetrad progeny. The zygote colonies were collected and resuspended in 2 ml of liquid media, and then 0.1 and 0.2 ml of cells were then plated at a low density on TAP medium and allowed to grow for 7 d. Single-colony isolates were then picked into liquid media and scored for motility as described above. The progeny were also scored for the presence of a functional nitrate reductase gene (NIT1) by plating on selective media (R-NO3), in which NH4NO3 is replaced by KNO3. Strains with a nit1 genotype are unable to grow on medium that contains nitrate as the sole nitrogen source (Fernandez et al., 1989).
Complementation tests were performed by constructing stable diploid cell lines (Ebersold, 1967). The 5B10 and 27B3 strains were first crossed into an arginine-requiring background (either arg2 or arg7) and then mated to one another and to other I1 mutants in the appropriate arg− background. Stable diploids were selected on R or TAP medium lacking arginine and then analyzed by phase contrast microscopy. At least six independently isolated diploid cell lines were scored for each complementation test, and all diploids were demonstrated to be mating type minus.
Southern Blot Analysis
Genomic DNA was prepared by cesium chloride density gradient purification as described by Porter et al. (1996). For Southern blots, 4 μg of genomic DNA were digested with the appropriate restriction enzyme and size fractionated on 0.8–1.0% agarose gels. The DNA was then transferred to either a Zetabind Nylon membrane (CUNO, Meridian, CT) or a Magnagraph membrane (Micron Separations, Westboro, MA) following manufacturer’s instructions. DNA probes for hybridization were purified in low melting point agarose gels (Gibco Life Technologies, Gaithersburg, MD) and radiolabeled with random hexamers and [32P] dCTP using the Rediprime Labeling Kit (Amersham, Uppsala, Sweden). Prehybridization and hybridization conditions were carried out as described by Myster et al. (1997).
Origin and Mapping of the IC140 clone
A phage clone containing the gene encoding the 140-kDa IC was obtained from large insert genomic library as described in the accompanying article (Yang and Sale, 1998). An 11.5-kb XbaI fragment was subcloned into the XbaI site of pBluescript KS II + (Stratagene, La Jolla, CA) in both orientations (pCP1 and RpCP1). Additional constructs of the IC140 gene used in transformation experiments were derived from pCP1 and RpCP1 by digesting with the appropriate restriction enzymes.
To place the IC140 gene on the genetic map, the 11.5-kb XbaI genomic fragment containing the IC140 gene was used to probe a series of mapping filters previously described by Porter et al. (1996). To identify a restriction length fragment polymorphism (RFLP) that could be used as a molecular marker, the 11.5-kb genomic fragment was first used as a hybridization probe on Southern blots of genomic DNA isolated from two polymorphic Chlamydomonas reinhardtii strains, 137c and S1-D2. RFLPs between 137c and S1-D2 were easily observed with genomic DNA that was digested with either EcoRI/XhoI or PvuII (Perrone and Porter, unpublished results). The 11.5-kb genomic fragment was then hybridized to a series of mapping filters containing EcoRI/XhoI digested genomic DNA that had been isolated from tetrad progeny derived from crosses between multiply marked C. reinhardtii strains and S1-D2. The mapping filters and the associated genetic and molecular markers are described in detail by Porter et al. (1996).
Northern Blot Analysis
Cells for RNA preparation were grown in 250 ml of liquid TAP media, and then total RNA was isolated both before and 45 min after deflagellation induced by pH shock (Wilkerson et al., 1994). Total RNA (20 μg per lane) was electrophoresed on 1.0% agarose-formaldehyde-denaturing gels, transferred overnight to a Magnagraph membrane, and then baked at 80°C under vacuum for 2 h. The RNA was further immobilized by irradiating with UV light using a Stratalinker II (Stratagene) at a 20,000 μJ setting. Prehybridization and hybridization conditions were the same as those used for Southern blots. Northern blots were probed with a 1.6-kb IC140 partial cDNA clone (pC1, see Yang and Sale, 1998) and with a clone for the CRY1 gene (Nelson et al., 1994).
Transformation and Screening for Rescue of Motility Defect
To identify regions of the IC140 gene that are required for rescue of the ida7 motility defects, the ida7 strain was crossed into an arginine-requiring background (ida7 arg7) and then cotransformed using a second selectable marker, pARG7.8, and the appropriate IC140 gene constructs. Plasmid pARG7.8 contains a wild-type copy of the arginino-succinate lyase gene (Debuchy et al., 1989). High-efficiency transformations were performed using the glass bead-mediated method of Kindle (1990) as described by Nelson and Lefebvre (1995). Briefly, the ida7 arg7 cells were grown under light for 4 d in liquid TAP medium supplemented with arginine. Cells were then centrifuged and concentrated to a density of 1 × 108 cells/ml, split into 0.6-ml aliquots, and incubated with 1 ml of autolysin at room temperature for 45 min to remove cell walls. The cells were collected in a clinical centrifuge to concentrate the cells and remove the autolysin, and then fresh TAP media were added to a final volume of 0.6 ml. Cells (0.3 ml) were then combined with 0.3 g of sterile glass beads, 0.1 ml filter-sterilized PEG 8000, and either 2 μg of BamHI-linearized pARG7.8 plasmid alone or both the pARG7.8 plasmid and 2 μg of linearized plasmid containing different portions of the IC140 genomic region. The transformation mixtures were vortexed at high speed for 45 s, and then immediately diluted with 10 ml of TAP media and transferred to a fresh tube, leaving the glass beads behind. Cells were reconcentrated to a final volume of 0.5 ml and plated on solid TAP media to select for arg+ transformants. Only cells that have received a functional copy of the pARG7.8 plasmid can grow under these conditions. After growth for 5–7 d, positive transformants were picked into 96-well plates containing TAP media and scored under a dissecting scope within 20–60 min for swimming phenotypes. Cells with apparent wild-type motility were grown in TAP media and rescored by phase contrast light microscopy to confirm the swimming phenotype.
Protein Purification and Sucrose Density Gradient Centrifugation
Large-scale cultures (20–40 l) of vegetative cells for protein purification were grown in rich medium containing sodium acetate and additional potassium phosphate as described by Witman (1986) and King et al. (1986), as modified by Gardner et al., (1994) and Myster et al. (1997). Dynein extracts were further purified by sucrose density gradient centrifugation as described by Porter et al. (1992) and Myster et al. (1997).
SDS-PAGE
The multiple DHCs were separated by SDS-PAGE using 3–5% polyacrylamide, 3–8 M urea gradient gels (Kamiya et al., 1991) and the Laemmli (1970) buffer system. To resolve intermediate and LCs, 5–15% polyacrylamide, 0–0.25 M glycerol gradient gels were used. Gels were stained with silver as described by Wray et al. (1981).
Antibody Characterization and Analysis
To test for the presence or absence of specific components of the I1 complex, whole axonemes or sucrose gradient fractions were electrophoresed on 5% or 7% polyacrylamide gels and then electroblotted at 800 mA for 90 min using a Genie electroblotter (Idea Scientific, Minneapolis, MN) to either PVDF or nitrocellulose membranes. The blots were then blocked for 2 h at room temperature in a buffer composed of PBS with 0.2% I-Block (Tropix, Bedford, MA) and 0.1% Tween-20 before incubating with primary antibody as described below.
To test for the presence or absence of the IC140 polypeptide, a rabbit polyclonal antiserum generated against an IC140 fusion protein (see Yang and Sale, 1998) was added to the blocking buffer at a dilution of 1:3000 and incubated with the blots at 4°C overnight. The blots were washed three times for 5 min in blocking buffer and then incubated with an alkaline phosphatase-conjugated secondary antibody (Sigma Chemical, St. Louis. MO) at a 1:1000 dilution for 2 h at room temperature. Subsequent detection of the secondary antibody with 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium were carried out per manufacturer’s instructions (Sigma Chemical).
To test for the presence or absence of the 1α Dhc, rabbit polyclonal antibodies generated against a specific peptide in the 1α Dhc were used (Myster et al., 1997). To affinity purify antibodies against the 1α Dhc, dynein extracts from pf28 axonemes were run on a 5% polyacrylamide gel and electroblotted to a PVDF membrane. The Dhc region of the blot was visualized by staining with Ponceau S, excised, incubated with blocking buffer for 30 min at room temperature, and then incubated in a 1:100 dilution of antiserum in PBS, 0.2% I-Block, 0.05% Tween-20, and 0.05% sodium azide at room temperature overnight. After the diluted antiserum was removed, the PVDF strips were washed three times for 5 min in TBS, 0.05% Tween 20. Bound antibodies were eluted with a glycine elution buffer (pH 2.8, 0.1 M glycine, 0.5 M NaCl, 0.05% Tween 20) two times for 3 min and neutralized as described by Tang (1993). The affinity-purified antibody was used at a 1:10 dilution in blocking buffer. All other steps were carried out as described above, with the exception that the secondary antibody was diluted 1:5000.
Electron Microscopy and Image Analysis
Axonemes were prepared for electron microscopy as described by Porter et al., (1992). The methods used for digitization, averaging, and comparison of longitudinal sections were as described by Mastronarde et al. (1992) and by O’Toole et al. (1995).
RESULTS
Recovery of New Motility Mutations
To identify new mutations in genes that affect inner dynein arm function, we screened a collection of 41 motility mutants generated by insertional mutagenesis with a selectable marker. The transformants were examined by phase contrast light microscopy and sorted into five phenotypic classes. The first class contained 9 strains that swam forward with a slow, shaky phenotype characteristic of outer dynein arm mutations. The second class contained 6 strains that jiggled or twitched in place and made little forward progress. The third class contained 13 strains that appeared to have subtle differences in motility when compared with a wild-type strain, but were determined to be too difficult to differentiate easily from wild-type motility in a backcross. The fourth class included 2 strains that were either aflagellate or had no motility. The last class contained 12 strains that swam forward with the slow, smooth motility characteristic of mutants with inner dynein arm defects. In particular, the swimming behavior of two transformants, strains 5B10 and 27B3, appeared strikingly similar to that of inner arm mutants with defects in the assembly of the I1 complex. Measurements of swimming velocities confirmed that these 2 strains swim at speeds similar to those of other I1 mutants, but were slower than mutants with defects in the assembly of other inner dynein arm isoforms (see Table 1 and Kamiya et al., 1991; Gardner et al., 1994). In addition, other assays revealed that neither strain could phototax toward a directional light source, consistent with the phenotype of mutants that have I1 defects (King and Dutcher, 1997). This article will address the characterization of one of these strains, the mutant 5B10.
Strain 5B10 Represents a Mutation at a New I1 Locus, IDA7
To determine whether the 5B10 mutant represents a new allele of a previously identified I1 locus or a mutation in a novel gene, we performed a series of complementation tests (Table 2). Earlier studies have identified only three I1 loci (PF9/IDA1, IDA2, IDA3), but all three are represented by multiple alleles (Kamiya et al., 1991, Porter et al., 1992). Recent work has shown the PF9/IDA1 locus corresponds to the gene that encodes the 1α DHC of the I1 complex (Myster et al., 1997), but the gene products of the other I1 loci are unknown. As shown in Table 2, when 5B10 was mated to either 27B3, pf9, ida2, or ida3, all of the resulting diploid strains swam with a wild-type motility phenotype. These results indicated that the 5B10 strain represents a mutation in a novel I1 locus. We therefore now refer to the 5B10 strain as ida7, in keeping with the revised nomenclature for Chlamydomonas mutations (Dutcher, 1995).
Table 2.
Complementation analysis of I1 loci
| Diploid genotype | Swimming phenotype | Conclusion |
|---|---|---|
| Wild type | 5B10 complements ida2 | |
| Wild type | 5B10 complements ida3 | |
| Wild type | 5B10 complements pf9-2 | |
| Wild type | 5B10 complements 27B3 | |
ida7 Axonemes Lack the I1 Complex
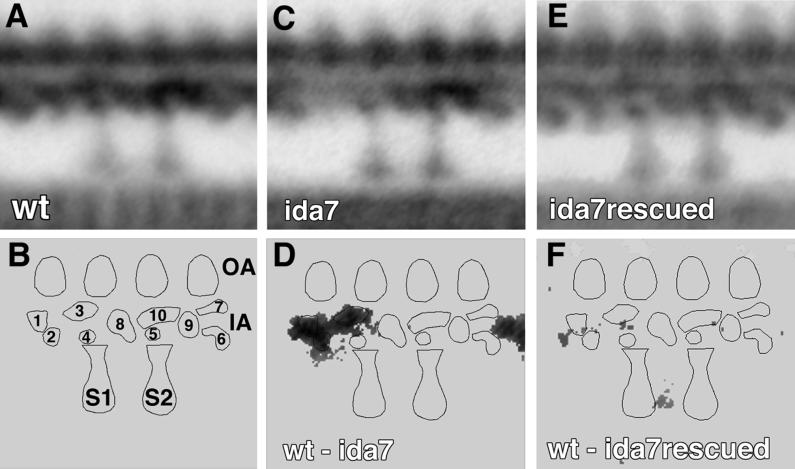
Previous work has demonstrated that the I1 complex occupies a specific position proximal to the first radial spoke within each 96-nm axoneme repeat (Piperno et al., 1990, Mastronarde et al., 1992). To determine whether the ida7 strain fails to assemble the I1 complex into the axoneme, we isolated axonemes from both ida7 and a wild-type strain, prepared them for electron microscopy, and analyzed longitudinal sections using image averaging procedures (Mastronarde et al., 1992). The average from several wild-type longitudinal sections is shown in Figure 1A, and the relative positions of the radial spokes, outer arms, and the inner dynein arms are indicated in the model in Figure 1B. The I1 complex is a trilobed density proximal to the first radial spoke (identified as lobes 1, 2, and 3 in Figure 1B). This complex appears to be missing in the average of ida7 axonemes shown in Figure 1C. A pixel-by-pixel analysis of variance between the wild-type and ida7 averages confirms that the only significant difference between the strains is in the region corresponding to the I1 complex, as shown in the difference plot in Figure 1D. These images demonstrate that the ida7 strain fails to assemble the I1 complex at the appropriate location within the axoneme.
Figure 1.
Analysis of longitudinal images of axonemes isolated from wild-type, ida7, and a rescued ida7 strain (AH1). Grand averages from wild-type (A), ida7 (C), and the rescued ida7 (E) are based on 8, 8, and 9 individual axonemes and 74, 65, and 84 axoneme repeats, respectively. (B) Model of axoneme structures within the 96-nm axoneme repeat, with the major lobes of density in the inner arm region labeled 1–10. The proximal and distal radial spokes are labeled S1 and S2, respectively; the outer arms (OA) are on top, and the inner arm (IA) region is below. (D) Difference plot between wild-type and ida7, and (F) difference plot between wild-type and the rescued ida7 strain (AH1). Differences not significant at 0.005 confidence level have been set to zero.
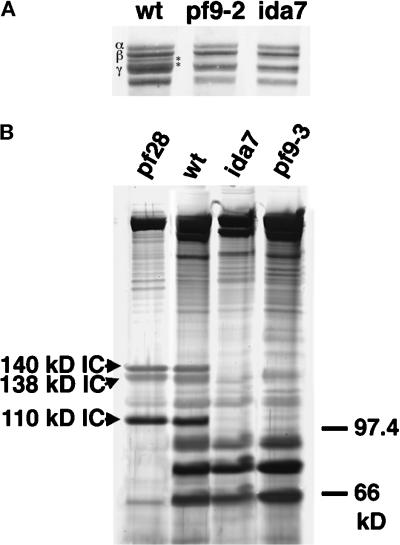
The I1 complex contains at least five subunits: two DHCs known as 1α and 1 β, and three ICs of 140, 138, and 110 kDa.2 As ida7 axonemes appear to lack the structures associated with the I1 complex, we wished to determine which I1 components were specifically missing. To examine the DHCs, we isolated whole axonemes from wild-type, ida7, and pf9–2, a previously characterized I1 mutant (Porter et al., 1992), and analyzed them on 3–5% polyacrylamide gradient gels. As shown in Figure 2A, the 1α and the 1β DHCs migrate as two faint bands between the β and γ DHCs of the outer arm in the wild-type sample, but these two bands appear to be missing in both the ida7 and pf9 samples. To analyze the ICs of the I1 complex, we obtained crude dynein extracts by high-salt extraction of isolated axonemes and then partially purified the I1 complexes by sucrose density gradient centrifugation. The wild-type I1 complex typically cosediments in the 18–19S region of the gradient, as a shoulder on the larger peak of outer arm components (Piperno et al., 1990; Porter et al., 1992). The 5–15% polyacrylamide gradient gels shown in Figure 2B illustrate the polypeptides present in this region in extracts prepared from wild-type and the mutants pf28, pf9, and ida7. The three ICs of 110, 138, and 140 kDa are clearly visible in both the wild-type sample and the pf28 sample, which lacks the outer arm components. However, these three polypeptides are missing in both the pf9 and ida7 samples. Analysis of the remaining gradient fractions on polyacrylamide gels confirmed that these I1 ICs could not be detected elsewhere in the sucrose gradient (Perrone and Porter, unpublished results). Therefore, ida7 mutant axonemes lack all of the intermediate and heavy-chain components of the I1 complex.
Figure 2.
SDS-PAGE analysis of dynein polypeptides in wild-type and mutant axonemes. (A) Shown here is the high molecular weight region of a 3–5% polyacrylamide gradient loaded with whole axonemes from wild-type, pf9–2, and ida7. The outer arm DHCs (α, β, and γ) are indicated on the left in wild type. The 1α and 1β DHCs of the I1 complex migrate between the β and γ DHCs of the outer arm, as indicated by asterisks on the right. Note that both pf9–2 and ida7 axonemes lack the 1α and 1β DHCs. (B) Sucrose density gradient centrifugation of dynein extracts from pf28, wild-type, ida7, and pf9–3. Shown here is a 5–15% polyacrylamide gradient gel loaded with fractions from the 19S region. This region typically contains the peak of the I1 complex and the trailing edge of the outer arm components that peak at 21S. The three ICs associated with the I1 complex are indicated on the left in the pf28 sample, which lacks the outer arms (Mitchell and Rosenbaum, 1985). The I1 ICs are also present in the wild-type sample, but are missing in both ida7 and pf9–3.
The ida7 Mutation Is Linked to the Insertion of the NIT1 Plasmid
To confirm that the motility defect in ida7 was the direct result of a plasmid insertion event into the IDA7 locus, we backcrossed ida7 to a nit1 strain with a wild-type motility and checked that the slow-swimming phenotype cosegregated with the Nit+ phenotype. The viability of tetrad progeny from this cross was extremely low, so we analyzed cosegregation using random progeny as described in MATERIALS AND METHODS. Following growth on nonselective media, single colony isolates were analyzed by phase contrast microscopy to determine motility phenotypes. The progeny were then plated on both selective and nonselective media. Analysis of 118 colonies indicated that all 53 slow swimming progeny were able to grow on selective media and were therefore Nit+, whereas all 65 progeny with wild-type motility were unable to grow on selective media and were therefore nit−. These results indicated that the Nit+ phenotype was tightly linked to the mutant motility phenotype (<0.8 cM) and suggested that the mutation in ida7 was the direct result of the plasmid insertion into the IDA7 locus.
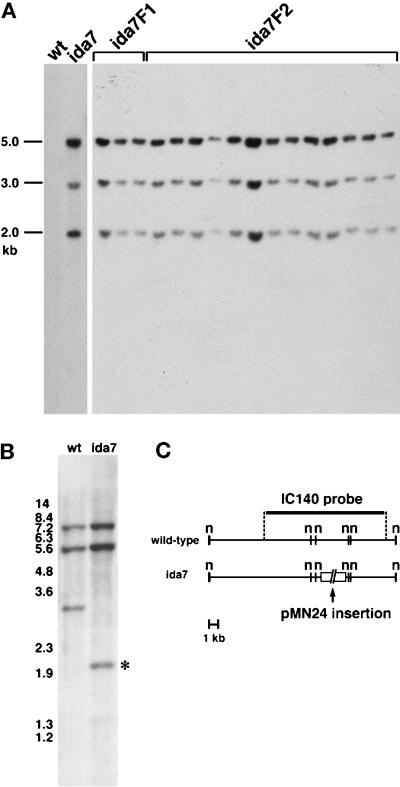
To determine whether the IDA7 locus might be amenable to cloning by plasmid rescue or other methods, genomic DNA was isolated from several ida7 progeny and analyzed on Southern blots probed with the vector portion of the pMN24 plasmid used for the initial transformation. As shown in Figure 3A, the original ida7 strain contains at least three copies of the pMN24 plasmid, and all three copies of the plasmid cosegregate with the ida7 progeny through multiple backcrosses. These results indicate that the ida7 mutation was caused by the insertion of three tandemly linked copies of the pMN24 plasmid into the IDA7 locus.
Figure 3.
Genomic Southern analyses of ida7 probed with plasmid sequences and the IC140 gene. (A) Cosegregation of the ida7 motility defects and the NIT1 plasmid. Shown here are Southern blots of PstI-digested genomic DNA isolated from wild-type, the original ida7 strain, and the slow swimming progeny from two successive generations (F1, F2) of ida7 x nit1 crosses. The blot was hybridized to a probe containing vector sequence from the pMN24 (NIT1) plasmid. No vector bands are detectable in the wild-type sample, but three bands are visible in ida7 and in all of its slow swimming progeny. (B) Disruption of the IC140 gene by plasmid insertion. A Southern blot of NotI-digested genomic DNA from wild-type and ida7 was hybridized with an 11.5-kb XbaI genomic fragment containing the IC140 gene. One of the restriction fragments (indicated by the asterisk to the right) is shifted in the ida7 lane. (C) Shown here is a NotI restriction map of the genomic region surrounding the IC140 gene in wild-type (top), and a diagram of the pMN24 plasmid insertions in ida7 (bottom).
The ida7 Mutation Is Associated with a Plasmid Insertion into the IC140 Gene
Plasmid insertions into the nuclear genome of Chlamydomonas are often accompanied by deletions or rearrangements of the host DNA. We therefore reasoned that if the ida7 mutation represented a plasmid insertion into a previously cloned I1 gene, we might be able to identify the affected gene by hybridizing Southern blots of ida7 and wild-type genomic DNA with gene-specific probes and looking for RFLPs between the two DNA samples. We have previously used this approach to identify a mutation in the Dhc1 gene, which encodes the 1α DHC of the I1 complex (Myster et al., 1997). We therefore screened blots of wild-type and ida7 DNA with probes corresponding to seven different DHC genes (Porter et al., 1996), but in each case we failed to detect an RFLP (Perrone and Porter, unpublished results). Given the crucial role of ICs in the proper assembly of outer arm dynein complexes (Mitchell and Kang, 1991; Wilkerson et al., 1995), we reasoned that a mutation in an I1 IC gene might also result in the failure to assemble the I1 complex. We therefore screened the ida7 DNA with an 11.5-kb genomic fragment containing the gene predicted to encode the 140-kDa IC of the I1 complex (Yang and Sale, 1998). As shown in the blot in Figure 3B, DNA corresponding to the IC140 genomic region could be easily detected in the ida7 mutant, but a small RFLP was observed in a 3.0-kb NotI fragment. Analysis of this and other Southern blots indicated that the ida7 mutation was caused by the insertion of the pMN24 plasmids into a 3.0-kb NotI fragment located in the middle of the IC140 gene (Figure 3C).
Mapping of the IC140 Gene
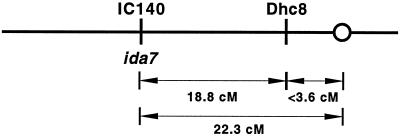
To determine the location of the IC140 gene on the genetic map of Chlamydomonas, we used the IC140 gene as a molecular marker to identify a RFLP between polymorphic strains of C. reinhardtii (see MATERIALS AND METHODS). We then analyzed the segregation of the IC140 RFLP with respect to the segregation of 42 other genetic and molecular markers. As shown in Figure 4, linkage of the IC140 gene could be detected with only one other molecular marker, the Dhc8 gene. Because both Dhc8 and the IC140 gene have not yet been linked to any other markers tested thus far, they may represent a new linkage group in Chlamydomonas.
Figure 4.
Genetic map location of the IC140 gene/IDA7 locus. The IC140 gene maps to a new linkage group, approximately 18.8 cM from the DHC gene Dhc8 and 22.3 cM from the centromere. The ratio of parental ditype:nonparental ditype:tetratype tetrads (PD:NPD:TT) was 15:0:9 for IC140 vs. Dhc8 and 6:11:14 for IC140 vs. the centromere-linked marker ac17.
Transformation of ida7 with the IC140 Gene Rescues the Motility Defect
To determine whether the motility defect in the ida7 mutant could be rescued by supplying this strain with a functional copy of the IC140 gene, the ida7 strain was crossed into an arg7 background and then cotransformed with one plasmid containing the selectable marker ARG7 and a second plasmid containing 11.5 kb of the genomic region surrounding the IC140 gene (see the pCP1 construct (XbaI to XbaI) in Figure 5). In Chlamydomonas, 10–70% of cells that are transformed with a selectable marker also receive the second, unselected gene (Diener et al., 1990). Positive (Arg+) transformants were first obtained by plating on selective media. The resulting colonies were then picked into liquid media in 96-well microtiter plates and scored for motility phenotype on a dissecting scope; 22% of the Arg+ colonies appeared to have a wild-type swimming phenotype. As expected, all of the Arg+ colonies that were obtained from control transformations with the ARG7 plasmid alone had a slow-swimming phenotype identical to the original ida7 strain. These results demonstrate that the 11.5-kb fragment contains a functional copy of the IC140 gene capable of rescuing the ida7 motility defect.
Figure 5.
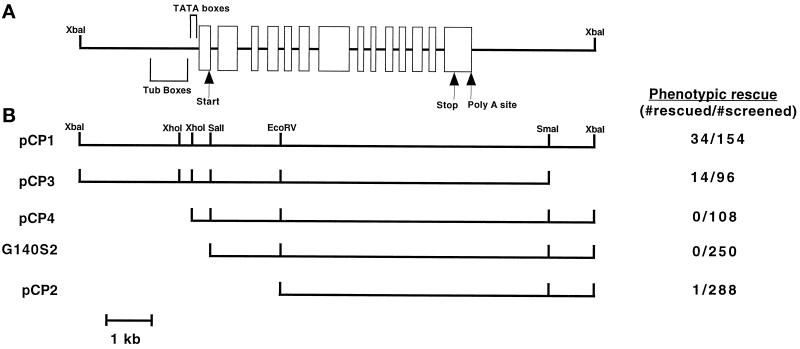
Rescue of the ida7 motility defects by transformation with constructs of the IC140 genomic region. (A) Diagram of the 11.5-kb wild-type genomic fragment containing the IC140 gene. Areas of interest based on the sequence analysis of Yang and Sale (1998) are shown here. The open rectangles indicate the exons of the IC140 gene, and the solid lines indicate the introns. The predicted translation start and stop sites are indicated, as well as the probable polyadenylation site. The regions containing TATA boxes and “tub” boxes are denoted by brackets. (B) Diagram of the IC140 gene constructs used to transform the ida7 mutant. The constructs are drawn with respect to the diagram of the IC140 gene above. The name of each plasmid construct is listed on the left, and the number of rescued transformants relative to the total number of transformants screened is presented as a ratio on the right.
To better define the position of the IC140 transcription unit and identify the minimal region required for expression of a functional gene, we cotransformed the ida7 arg7 strain with the ARG7 plasmid and a series of IC140 gene constructs shown in Figure 5. Truncation of the 11.5-kb genomic fragment near the 3′ end of the IC140 transcription unit (pCP3 construct) did not significantly alter the cotransformation frequency. However, two different truncations near the 5′ end of the IC140 gene (pCP4 and G140S2) completely abolished the ability of these constructs to rescue the ida7 motility defects (see Figure 5). These results indicated the presence of sequence elements near the 5′ end of the transcription unit that appear to be required for efficient expression of the IC140 transgene.
Subsequent sequence analysis of the 11.5-kb fragment containing the IC140 gene has shown that this construct (pCP1) contains 2779 base pairs (bp) upstream of the proposed translation start site (Yang and Sale, 1998). Within this region, it is possible to identify at least three TATA box sequences that might be required for transcription initiation and several “tub” box sequences (Brunke et al., 1984; Davies and Grossman, 1994), which have been previously implicated as important sequence elements for the expression of flagellar transcripts (see Figure 5A and Yang and Sale, 1998). The truncation of the IC140 gene in the pCP4 construct would therefore eliminate all of the “tub” box sequences as well as the three TATA box sequences. Similarly, the G140S2 construct begins 29 bp downstream of the predicted translation start site and therefore lacks all of the upstream regulatory sequences. Given our cotransformation results with the IC140 gene constructs, it appears that at least some of these upstream sequence elements are required for efficient expression of the IC140 gene and rescue of the ida7 motility defects. We were therefore very surprised when we analyzed a group of 288 Arg+ transformants that had been cotransformed with a smaller IC140 gene construct (pCP2) and identified a single strain with wild-type motility. As shown in Figure 5, the pCP2 construct removes all of the 5′ regulatory regions as well as exons 1–4 of the IC140 coding sequence. Any resulting polypeptide would therefore be missing the first 283 amino acids out of a total of 1024 residues of the IC140 protein. Nonetheless, the rescued strain 5A swims at a velocity of 155.3 ± 20.3 μm/s, which is comparable to the swimming velocity measured for wild-type cells (144.2 ± 17.1 μm/s; see Table 1). In addition, the 5A transformant has recovered the ability to phototax toward a directional light source. The 5A strain, therefore, has a motility phenotype indistinguishable from wild-type.
Expression of IC140 Transcripts in Wild-Type and Mutant Strains
To analyze the IC140 transcripts in both wild-type and mutant strains, we isolated total RNA from the wild-type strain 137c, the original ida7 strain, the transformant strain AH1 (which was rescued with the 11.5-kb genomic fragment contained in pCP1), and the strain 5A (which was rescued with the smaller pCP2 construct). Because the transcription of flagellar genes is typically up-regulated after deflagellation in Chlamydomonas, total RNA was isolated both before and after deflagellation. As shown in Figure 6, hybridization with a 1.6-kb partial cDNA clone of the IC140 gene identifies a single 3.7-kb transcript in both wild-type and AH1 that is up-regulated in response to deflagellation. No transcript is detected in the ida7 sample, although a faint band can be observed at ∼7.5 kb. These observations suggest that the insertion of the NIT1 plasmids into the IC140 transcription unit resulted in the formation of a hybrid transcript that is relatively unstable as compared with wild-type. Interestingly, however, the 5A strain produces a smaller IC140 transcript that is expressed at the same level both before and after deflagellation (see Figure 6). Thus, although the IC140 transcript is not appropriately regulated in the 5A strain, it appears that a truncated transcript is now constitutively expressed at levels sufficient for the restoration of wild-type motility.
Figure 6.
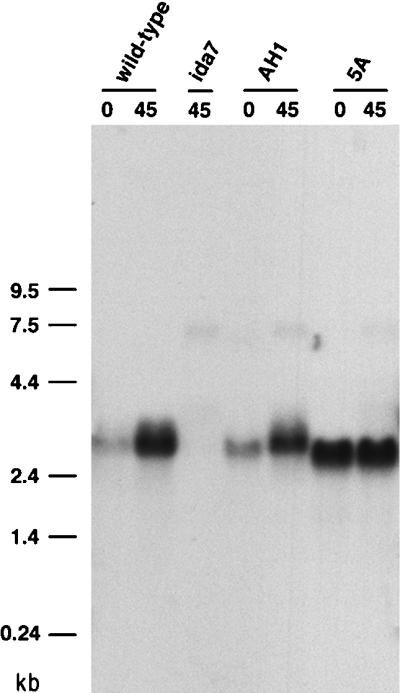
Expression of IC140 transcripts in wild-type, ida7, and rescued strains. Shown here are Northern blots of total RNA isolated before (0) or after (45) deflagellation. RNA (20 μg per lane) was separated on a 1% formaldehyde agarose gel, transferred to a Magnagraph membrane, and hybridized to a 1.6-kb cDNA fragment of the IC140 gene. A single 3.7-kb transcript is present in both the wild-type and AH1, and the expression of this transcript is up-regulated by deflagellation. The 3.7-kb transcript is completely missing in ida7. A smaller transcript is expressed at high levels both before and after deflagellation in the 5A strain. Hybridizing the blot with a control probe for the CRY1 gene, which encodes the S14 ribosomal protein subunit, confirmed that equal amounts of RNA were loaded in each lane (Perrone and Porter, unpublished results).
Reassembly of the I1 Complex in Transformed Strains
To confirm that the structures of the I1 complex were assembled at the appropriate axonemal location, we prepared isolated axonemes from the AH1 strain for electron microscopy and analyzed longitudinal sections using image averaging procedures. As shown in Figure 1E, the structures of the I1 complex (lobes 1, 2, and 3 in the model) are present in the AH1 strain. The difference plot between wild-type and AH1 shown in Figure 1F illustrates that no significant differences are detected between the two strains. Therefore we can conclude that the presence of a wild-type IC140 gene has resulted in the reassembly and targeting of I1 structures to the appropriate location.
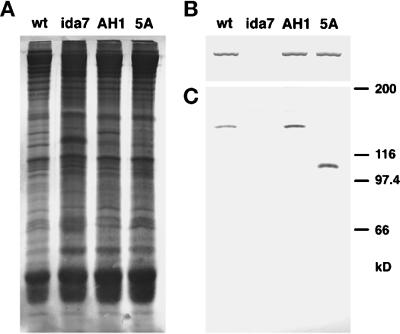
To determine which polypeptides of the I1 complex reassembled into the axoneme in the transformants, we isolated whole axonemes from wild-type, ida7, and two of the transformants, AH1 and 5A. Duplicate samples of the axonemes were electrophoresed on 5–15% PAGE gels and blotted to two membranes. Figure 7A shows the first blot stained with a colloidal gold stain, which demonstrates the whole axoneme samples were loaded at approximately equal concentrations. Figure 7B shows the high molecular weight region of the second blot that was probed with an antibody generated against the 1α DHC of the I1 complex. A single immunoreactive band is present in the wild-type, AH1, and 5A lanes, but not in the ida7 lane. These data indicate that the 1α DHC has been restored to both the AH1 and 5A axonemes. Figure 7C shows the bottom portion of the same blot probed with an antibody to the IC140. An immunoreactive band of ∼140 kDa is present in both wild-type and AH1, and, as expected, no band is present in the ida7 lane. These data demonstrate that the IC140 polypeptide has been reassembled into the AH1 axonemes, as predicted for a full-length rescue. To examine whether other components of the I1 complex are also reassembled in AH1 axonemes, we fractionated dynein extracts on sucrose density gradients and analyzed the polypeptides present in each fraction by SDS-PAGE. Analysis of the 19S region indicated that the 110- and 138-kDa ICs are also restored in AH1 axonemes (Perrone and Porter, unpublished results). These results indicate that the presence of a wild-type IC140 is sufficient to rescue all of the motility, structural, and biochemical defects observed in the ida7 mutant.
Figure 7.
Assembly of I1 complex polypeptides in wild-type and mutant axonemes. Whole axonemes were isolated from wild-type, the ida7 mutant, and two of the rescued transformants, strain AH1 and strain 5A, and then loaded in duplicate onto a 7% polyacrylamide gel (25 μg/lane), and blotted to two membranes. (A) The first blot was stained with colloidal gold to confirm that equal amounts of total protein were loaded in all the lanes. (B) The high molecular weight region of the second blot was incubated with an antibody specific for the 1α DHC of the I1 complex. A single, high molecular weight band was seen in the wild-type, AH1, and 5A lanes, but not in the ida7 lane. (C) The bottom of the blot was incubated with an antibody specific for the C-terminal half of the IC140 (Yang and Sale, 1998). No band was detected in the ida7 lane, whereas both wild-type and AH1 contained a single immunoreactive band at approximately 140 kDa. Strain 5A, which was recovered after transformation with a fragment of the IC140 gene, has an immunoreactive band at ∼108 kDa.
Given the apparent requirement for a functional IC140 gene, we were especially interested in examining axonemes from the 5A strain, which appears to have wild-type motility (Table 1) but only expresses a truncated IC140 transcript (Figure 6). Western blots probed with the IC140 antibody (Figure 7C) revealed that 5A axonemes contain an immunoreactive band that migrates at ∼108 kDa. Together with the Northern blots described above, these observations indicate that transformation of ida7 with a construct predicted to encode only 73% of the IC140 polypeptide results in the expression of a truncated IC140 that is capable of assembling with other I1 components into the axoneme and restoring motility.
DISCUSSION
Isolation of a New I1 Locus
To identify the genes and gene products that are required for the assembly of the multiple inner dynein arm subspecies, we have screened a new collection of motility mutants that were generated by insertional mutagenesis (Myster et al., 1997 and present study). Analysis of the motility phenotypes along with structural studies have indicated that several of these strains have defects in the assembly of inner arm components (Perrone, O’Toole, and Porter, unpublished data). In particular, two strains are defective in the assembly of the I1 complex (Figures 1 and 2 and Perrone and Porter, unpublished results). Previous work has shown that the I1 complex is composed of two DHCs, three ICs, and three LCs (Piperno et al., 1990; Porter et al., 1992; Harrison et al., 1998). Mutations in any one of these subunits might be expected to disrupt the assembly of the I1 complex, but thus far only three distinct I1 loci have been identified: PF9/IDA1, which encodes the 1α DHC, IDA2, and IDA3 (Kamiya et al., 1991; Porter et al., 1992; Myster et al., 1997). Complementation tests with different I1 mutations have now revealed that one of the insertional mutants, 5B10, represents a mutation in a new I1 locus, IDA7 (Table 2). Southern blot analysis has further demonstrated that the IDA7 locus is tagged by the insertion of three tandemly linked copies of the pMN24 plasmid used as the selectable marker (Figure 3). These results indicated that it would be possible to identify the IDA7 gene product, either by recovery of genomic DNA flanking the site of plasmid insertion or by screening for RFLPs with cloned I1 genes (see below). This strategy should also be useful for the identification of other inner arm loci (Myster et al., 1997; Perrone, Bower, and Porter, unpublished data).
The ida7 Mutation Is the Result of Plasmid Insertions into the IC140 Gene
Because the ida7 mutation was tagged, we screened genomic Southern blots of ida7 and wild-type DNA with gene-specific probes corresponding to specific I1 subunits and thereby determined that the ida7 mutation is the result of plasmid insertion into the gene that encodes the IC140 (Figure 3). Northern blot analysis further demonstrated that the ida7 strain completely lacks the 3.7-kb IC140 transcript and therefore represents a true null allele (Figure 6). Transformation of the slow swimming ida7 mutant with a wild-type copy of the IC140 gene rescued all of the mutant defects (Figure 5). The rescued strains contain an IC140 transcript that is appropriately up-regulated after deflagellation (Figure 6), reassemble the I1 complex polypeptides at the correct location (Figure 1F and 7), and swim with wild-type motility (Table 1). These results demonstrate that a functional IC is required for the assembly of the I1 complex.
Defining Essential and Nonessential Regions of the IC140 Gene by Transformation of ida7
To further define regions within the IC140 gene required for expression and assembly, we generated a series of constructs that deleted different regions of the IC140 gene sequence and tested these constructs for their ability to rescue the ida7 motility defects (Figure 5). These experiments suggested that regulatory sequences located within the 5′ region, such as “tub” boxes and TATA boxes, are apparently essential for the expression of the IC140 transcript and subsequent rescue of the ida7 motility defects (Figures 5 and 6). However, the pCP2 construct lacks all of the upstream 5′ regions thought to be important for regulated expression, as well as the predicted translation start site and the first four exons of the IC140 coding sequence. Nonetheless, we were able to identify one transformant, 5A, that swims with a wild-type phenotype and contains a truncated IC140 transcript. These observations suggest that the pCP2 construct probably integrated into the regulatory region of another gene to form a chimeric transcript that is constitutively expressed. Such an integration event should be relatively rare, and consistent with this hypothesis, the frequency of rescue with the pCP2 construct (0.35%) was much lower than that observed with the full-length gene (22%) (see Figure 5).
Analysis of the nucleotide sequence (see Yang and Sale, 1998) indicates that any polypeptide expressed by the pCP2 construct would lack the first 283 amino acids of the IC140 sequence. Interestingly, there is a potential start codon located 27 bp downstream of the 5′ end of the pCP2 construct (see Figure 3, nucleotide 1873, Yang and Sale, 1998); if this codon is used as the translation start site in the chimeric gene, 18 additional amino acids would be added to the N terminus of exon 5, and the resulting protein product would be approximately 27% smaller than the original IC140. Blots of 5A axonemes probed with polyclonal antibodies against IC140 have revealed the presence of an immunoreactive band migrating at ∼108 kDa (Figure 7); this decrease in size is consistent with the smaller protein product that would be encoded by the pCP2 construct. These results demonstrate that the truncated IC140 is capable of assembly into the axoneme.
We have shown that the 1α DHC is also present in 5A axonemes, which suggests that other components of the I1 complex are reassembled as well (Figure 7B). Although we have not directly demonstrated the presence of the 1β DHC, we predict that both I1 DHCs are present, because strains that lack either the 1α Dhc or the 1β Dhc motor domains have a reduced swimming speed as compared with wild-type (Myster, Perrone, and Porter, unpublished results), whereas the 5A strain swims at a wild-type velocity (Table 1). Analysis of dynein extracts on sucrose density gradients has also indicated that the 110-kDa IC is present in 5A axonemes, but we have not yet conclusively demonstrated that the 138-kDa IC is restored (Perrone and Porter, unpublished results). However, given that the 5A strain undergoes wild-type phototaxis, it is highly likely that the IC138 is also present, as previous work has shown that defects in the IC138 result in the failure to phototax (King and Dutcher, 1997). Finally, recent studies by Harrison et al. (1998) have shown that the I1 complex also contains three LC components of 8, 12, and 14 kDa. Immunoblots probed with specific antibodies have confirmed that 14-kDa LC is present in 5A axonemes (Perrone and Porter, unpublished results). Thus, all of the available evidence indicates that several subunits of the I1 complex have reassembled in the 5A axonemes.
Functional Domains within the IC140
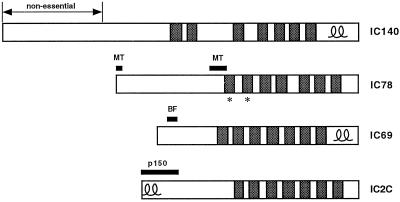
Sequence comparisons have shown that the IC140 shares significant regions of homology with other dynein ICs, including IC69 and IC78 of the Chlamydomonas outer arm, IC2 and IC3 from the sea urchin outer arm, and IC74 of cytoplasmic dynein (Yang and Sale, 1998). This homology is greatest in the C-terminal one-half to one-third of the ICs, particularly in regions that correspond to the WD repeat motifs (Ogawa et al., 1995; Wilkerson et al., 1995; Yang and Sale, 1998). WD repeats have been previously implicated as participating in protein–protein interactions in multisubunit complexes (Neer et al., 1994). These observations have led to the proposal that the WD repeats play similar roles in each dynein IC, such as in binding to other dynein components (King et al., 1995). Based on our transformation results (Figure 5) and by analogy to the other dynein ICs, we predict that the WD repeats of the IC140 play an important structural role in the assembly of the I1 complex and may serve to link subunits of the I1 complex together. Interestingly, chemical cross-linking of purified I1 complexes has suggested that the IC140 is closely associated with the IC110 subunit (see Yang and Sale, 1998). The ability to introduce modified constructs of the IC140 gene into the ida7 strain will allow us to determine the role of the WD repeats in mediating specific interactions with individual subunits.
The more divergent N-terminal domains of the dynein ICs are thought to perform isoform-specific roles, such as the targeting of dynein complexes to appropriate locations, or in regulating the activity of the complex (see Figure 8). For example, in the outer arm IC78, the N-terminal region appears to contain several highly charged domains that are essential for binding microtubules in vitro (King et al., 1995). The N-terminal region of the IC74 of cytoplasmic dynein also appears be necessary for targeting dynein to vesicles, as treatment of cell extracts with antibodies directed against this region blocks the dynein-dependent formation of membranous networks in vitro (Steffen et al., 1997). Furthermore, N-terminal regions of the IC74 have been shown to be required for interaction with the p150 subunit of the dynactin complex (Vaughan and Vallee, 1995). The dynactin complex has been implicated as a cofactor that stimulates the interaction between cytoplasmic dynein and vesicles in the cytoplasm (Gill et al., 1989; Schroer and Sheetz, 1991). In the case of the outer arm IC69, a 28-amino acid portion contained in the N-terminal third of the polypeptide appears to modulate flagellar beat frequency, as sequence alterations in this region reduce outer arm activity without altering outer arm assembly (Mitchell and Kang, 1993). It is therefore surprising that a construct that lacks the N-terminal 27% of the IC140 is able to support the assembly of several I1 components onto the axoneme. These results imply that the N-terminal 283 amino acids of the IC140 play no significant role in targeting the I1 complex to the axoneme. In addition, as the motility phenotype of the 5A strain is indistinguishable from wild-type (Table 1), it appears that this N-terminal region is not required to modulate I1 activity.
Figure 8.
Functional domains in dynein ICs. The IC140 is shown diagramatically in comparison to other dynein ICs. The shaded boxes indicate WD repeats; the double loops indicate predicted coiled-coil regions, and the asterisks denote WD repeats that have been implicated in subunit interactions (redrawn following Yang and Sale, 1998). IC78 is the outer arm IC that has been implicated in binding to the axoneme (see MT regions indicated, King et al., 1995). IC69 is another IC required for outer arm assembly (Mitchell and Kang, 1991). Mutations in the N-terminal region can alter flagellar beat frequency (see BF, Mitchell and Kang, 1993). IC2C (IC74) is a cytoplasmic dynein IC that appears to mediate interaction with the p150glued subunit of the dynactin complex (see p150, Vaughan and Vallee, 1995; Steffen et al., 1997).
The N-terminal region (amino acids 284–472) remaining in the truncated IC140 polypeptide will be an important region to target in future studies. Indeed, sequence alignment programs indicate that this domain shares limited homology with the N termini of some of the other ICs (Perrone and Porter, unpublished results; Yang and Sale, 1998). Transformation of ida7 with specific constructs containing mutations in this region may allow us to more precisely define domains within the IC140 polypeptide that are required for regulation or targeting of the I1 complex in vivo. In addition, the predicted coiled-coil domain located within the C terminus of the IC140 (see Yang and Sale, 1998), which may be analogous to the coiled-coil domains observed in both IC69 and IC74, is another region of high priority to analyze in future transformation experiments.
The Role of the IC140 in I1 Complex Assembly and Targeting
Our transformation experiments have demonstrated that the presence of a functional IC140 is critical for the assembly of the I1 complex, but it remains uncertain whether the IC140 is directly involved in binding the I1 complex to the axoneme. Fusion protein constructs containing the C-terminal region of the IC140 will bind selectively to isolated axonemes lacking the I1 complex in co-sedimentation experiments (Yang and Sale, 1998). These data are consistent with our transformation results with the 5A strain and further suggest that the IC140 could be directly involved in targeting. However, chemical cross-linking experiments have not yet identified a specific binding partner in the axoneme (Yang and Sale, 1998). Previous binding studies using purified I1 complex and repolymerized brain microtubules have demonstrated that the I1 dynein can bind and cross-link microtubules in the absence of ATP, but the complex can be released from the microtubules in the presence of ATP (Smith and Sale, 1991). These observations indicate that the I1 complex can bind to microtubules by means of its two DHC motor domains, but it does not bind directly to tubulin by an ATP-insensitive anchoring site. In addition, incubation of purified I1 complexes with wild-type and mutant axonemes further demonstrated that the I1 complex would not bind to wild-type axonemes, but it could rebind specifically to vacant sites in I1 mutant axonemes (Smith and Sale, 1992). These results suggest the presence of accessory proteins at specific sites within the 96-nm repeat that function as docking structures for the binding of the I1 complex, similar to those seen with the Chlamydomonas outer arm dyneins (Takada and Kamiya, 1994; Koutoulis et al., 1997). The isolation and characterization of additional I1 loci should allow us to identify these accessory polypeptides and to analyze their interactions with IC140 and other subunits of the I1 complex.
Note Added in Proof. We have recently found that the ida7 (5B10) mutation fails to complement another I1 mutation previously isolated by R. Kamiya (University of Tokyo).
ACKNOWLEDGMENTS
We wish to thank D. Mitchell (SUNY Health Sciences Center at Syracuse) for contributing several motility mutants, including strain 5B10. We also thank L.W. Tam (University of Minnesota) for providing the L5 strain. We are grateful to the members of the laboratories of C. Silflow and P. Lefebvre (University of Minnesota) and members of the Porter laboratory for their advice on this project. Parts of this work were completed by C.A.P. in partial fulfillment of the requirements for a Ph.D degree (University of Minnesota). This work was supported by grants from the March of Dimes (FY-1031), the National Science Foundation (MCB 9305217), and the National Institute of General Medical Sciences (GM 55667) to M.E.P, by a NIH postdoctoral fellowship (F32GM17666-00) to P.Y., and also by a NIH grant (GM51173) to W.S.S. C.A.P. was also supported in part by a research training grant from the National Science Foundation for Interdisciplinary Studies on the Cytoskeleton (DIR 91134444). E.O.T. was supported by a NIH Biotechnology Resource (RR00592) grant to J.R. McIntosh.
Abbreviations used:
- cM
centimorgan
- DHC
dynein heavy chain
- IC
intermediate chain
- LC
light chain
- RFLP
restriction fragment length polymorphism
Footnotes
The 110-kDa polypeptide was identified as an I1 subunit by Porter et al. (1992), based on an apparent molecular mass of 108 kDa. This polypeptide is the same as the 97-kDa subunit described by Smith and Sale (1991) and referred to by Yang and Sale (1998) as IC97. The discrepancy in molecular masses is apparently due to differences in electrophoretic conditions.
REFERENCES
- Brokaw CJ, Kamiya R. Bending patterns of Chlamydomonas flagella. IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil Cytoskel. 1987;8:68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- Brunke K, Anthony J, Sterberg E, Weeks D. Repeated consensus sequence and pseudopromoters in the four coordinately regulated tubulin genes of Chlamydomonas reinhardtii. Mol Cell Biol. 1984;4:1115–1124. doi: 10.1128/mcb.4.6.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Grossman AR. Sequences controlling transcription of the Chlamydomonas reinhardtii β2-tubulin gene after deflagellation and during the cell cycle. Mol Cell Biol. 1994;14:5165–5174. doi: 10.1128/mcb.14.8.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy R, Purton S, Rochaix J-D. The arginosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 1989;8:2803–2809. doi: 10.1002/j.1460-2075.1989.tb08426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener DR, Curry AM, Johnson KA, Williams BD, Lefebvre PA, Kindle KA. Rescue of a paralyzed-flagella mutant of Chlamydomonas by transformation. Proc Natl Acad Sci USA. 1990;87:5739–5743. doi: 10.1073/pnas.87.15.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S. Chlamydomonas reinhardtii. Trends Genet. 1995;12S:18–19. [PubMed] [Google Scholar]
- Ebersold WT. Chlamydomonas reinhardti: heterozygous diploid strains. Science. 1967;57:446–449. doi: 10.1126/science.157.3787.447. [DOI] [PubMed] [Google Scholar]
- Fernandez E, Schnell R, Ranum LPW, Hussey SC, Silflow CD, Lefebvre PA. Isolation and characterization of the nitrate reductase structural gene in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1989;86:6449–6453. doi: 10.1073/pnas.86.17.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner LC, O’Toole E, Perrone CA, Giddings T, Porter ME. Components of a “dynein regulatory complex” are located at the junction between the radial spokes and the dynein arms in Chlamydomonas flagella. J Cell Biol. 1994;127:1311–1325. doi: 10.1083/jcb.127.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons IR. Dynein family of motor proteins: present status and future questions. Cell Motil Cytoskel. 1995;32:136–144. doi: 10.1002/cm.970320214. [DOI] [PubMed] [Google Scholar]
- Gill S, Schroer T, Szilak I, Steur E, Sheetz M, Cleveland D. Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J Cell Biol. 1989;115:1639–1650. doi: 10.1083/jcb.115.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough UW, Gebhart B, Mermall V, Mitchell DR, Heuser JE. High pressure liquid chromatography fractionation of Chlamydomonas dynein extracts and characterization of inner arm dynein subunits. J Mol Biol. 1987;194:481–494. doi: 10.1016/0022-2836(87)90676-0. [DOI] [PubMed] [Google Scholar]
- Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermacher G, Sale W. Regulation of flagellar dynein by an axonemal type-1 phosphatase in Chlamydomonas. J Cell Sci. 1996;109:1899–1907. doi: 10.1242/jcs.109.7.1899. [DOI] [PubMed] [Google Scholar]
- Habermacher G, Sale W. Regulation of flagellar dynein by phosphorylation of a 138-kD inner arm dynein intermediate chain. J Cell Biol. 1997;136:167–176. doi: 10.1083/jcb.136.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E. The Chlamydomonas Sourcebook. San Diego, CA: Academic Press; 1989. [Google Scholar]
- Harrison A, Olds-Clarke P, King S. Identification of the t complex-encoded cytoplasmic dynein light chain Tctex1 in inner arm I1 supports the involvement of flagellar dyneins in meiotic drive. J Cell Biol. 1998;140:1137–1147. doi: 10.1083/jcb.140.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes J, Dutcher S. Cellular asymmetry in Chlamydomonas reinhardtii. J Cell Sci. 1989;94:273–285. doi: 10.1242/jcs.94.2.273. [DOI] [PubMed] [Google Scholar]
- Holzbaur ELF, Vallee RB. Dyneins: molecular structure and cellular function. In: Spudich JA, McKnight SL, Schekman R, editors. Annual Review of Cell Biology. Palo Alto, CA: Annual Reviews; 1994. pp. 339–372. [DOI] [PubMed] [Google Scholar]
- Huang B, Piperno G, Luck DJL. Paralyzed flagella mutants of Chlamydomonas reinhardtii defective for axonemal doublet microtubule arms. J Biol Chem. 1979;254:3091–3099. [PubMed] [Google Scholar]
- Huang B, Ramanis Z, Luck DJ L. Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for flagellar function. Cell. 1982;28:115–124. doi: 10.1016/0092-8674(82)90381-6. [DOI] [PubMed] [Google Scholar]
- Kagami O, Kamiya R. Translocation and rotation of microtubules caused by multiple species of Chlamydomonas inner-arm dynein. J Cell Sci. 1992;103:653–664. [Google Scholar]
- Kamiya R, Kurimoto E, Muto E. Two types of Chlamydomonas flagellar mutants missing different components of inner-arm dynein. J Cell Biol. 1991;112:441–447. doi: 10.1083/jcb.112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Kagami O, Yagi T, Kamiya R. Isolation of two species of Chlamydomonas reinhardtii flagellar mutants, ida5 and ida6, that lack a newly identified heavy chain of the inner dynein arm. Cell Struct Funct. 1993;18:371–377. doi: 10.1247/csf.18.371. [DOI] [PubMed] [Google Scholar]
- Kato-Minoura T, Hirono M, Kamiya R. Chlamydomonas inner-arm dynein mutant, ida5, has a mutation in an actin-encoding gene. J Cell Biol. 1997;137:649–656. doi: 10.1083/jcb.137.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL, Schnell RA, Fernandez E, Lefebvre PA. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J Cell Biol. 1989;109:2589–2601. doi: 10.1083/jcb.109.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S, Dutcher S. Phosphoregulation of an inner dynein arm complex in Chlamydomonas reinhardtii is altered in phototactic mutant strains. J Cell Biol. 1997;136:177–191. doi: 10.1083/jcb.136.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S, Patel-King R. Identification of a Ca++ binding light chain within Chlamydomonas outer arm dynein. J Cell Sci. 1995a;108:3757–3764. doi: 10.1242/jcs.108.12.3757. [DOI] [PubMed] [Google Scholar]
- King S, Patel-King R. The Mr = 8000 and 11,000 outer arm dynein light chains from Chlamydomonas flagella have cytoplasmic homologues. J Biol Chem. 1995b;265:11445–11452. doi: 10.1074/jbc.270.19.11445. [DOI] [PubMed] [Google Scholar]
- King S, Patel-King R, Wilkerson C, Witman G. The 78, 000-Mr intermediate chain of Chlamydomonas outer arm dynein is a microtubule binding protein. J Cell Biol, 1995;131:399–409. doi: 10.1083/jcb.131.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S, Witman G. Localization of an intermediate chain of outer arm dynein by immunoelectron microscopy. J Biol Chem. 1990;265:19807–19811. [PubMed] [Google Scholar]
- King SM, Otter T, Witman GB. Purification and characterization of Chlamydomonas flagellar dyneins. Methods Enzymol. 1986;134:291–306. doi: 10.1016/0076-6879(86)34097-7. [DOI] [PubMed] [Google Scholar]
- Koutoulis A, Pazour GJ, Wilkerson CG, Inaba K, Sheng H, Takada S, Witman GB. The Chlamydomonas reinhardtii ODA3 gene encodes a protein of the outer dynein arm docking complex. J Cell Biol. 1997;137:1069–1080. doi: 10.1083/jcb.137.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- LeDizet M, Piperno G. The light chain p28 associates with a subset of inner dynein arm heavy chains in Chlamydomonas axonemes. Mol Biol Cell. 1995;6:697–711. doi: 10.1091/mbc.6.6.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RP, Ebersold WT. The genetics and cytology of Chlamydomonas. Annu Rev Microbiol. 1960;14:197–216. doi: 10.1146/annurev.mi.14.100160.001213. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN, O’Toole ET, McDonald KL, McIntosh JR, Porter ME. Arrangement of inner dynein arms in wild-type and mutant flagella of Chlamydomonas. J Cell Biol. 1992;118:1145–1162. doi: 10.1083/jcb.118.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR. Cell and molecular biology of flagellar dyneins. In: Jeon KW, Jarvik J, editors. International Review of Cytology. San Diego, CA: Academic Press; 1994. pp. 141–175. [DOI] [PubMed] [Google Scholar]
- Mitchell DR, Kang Y. Identification of oda6 as a Chlamydomonas dynein mutant by rescue with the wild-type gene. J Cell Biol. 1991;113:835–842. doi: 10.1083/jcb.113.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR, Kang Y. Reversion analysis of dynein intermediate chain function. J Cell Sci. 1993;105:1069–1078. doi: 10.1242/jcs.105.4.1069. [DOI] [PubMed] [Google Scholar]
- Mitchell DR, Rosenbaum JL. A motile Chlamydomonas flagellar mutant that lacks outer dynein arms. J Cell Biol. 1985;100:1228–1234. doi: 10.1083/jcb.100.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myster S, Knott J, O’Toole E, Porter M. The Chlamydomonas Dhc1 gene encodes a dynein heavy chains subunit required for assembly of the I1 inner arm complex. Mol Biol Cell. 1997;8:607–620. doi: 10.1091/mbc.8.4.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- Nelson J, Lefebvre P. Transformation of Chlamydomonas reinhardtii. Methods Cell Biol. 1995;47:513–516. doi: 10.1016/s0091-679x(08)60854-7. [DOI] [PubMed] [Google Scholar]
- Nelson JAE, Savereide PB, Lefebvre PA. The CRY1 gene in Chlamydomonas reinhardtii, structure and use as a dominant selectable marker for nuclear transformation. Mol Cell Biol. 1994;14:4011–4019. doi: 10.1128/mcb.14.6.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole E, Mastronarde D, McIntosh JR, Porter ME. Computer-assisted image analysis of flagellar mutants. Methods Cell Biol. 1995;47:183–191. doi: 10.1016/s0091-679x(08)60808-0. [DOI] [PubMed] [Google Scholar]
- Ogawa K, Kamiya R, Wilkerson CG, Witman GB. Interspecies conservation of outer arm dynein intermediate chain sequences defines two intermediate chain subclasses. Mol Biol Cell. 1995;6:685–696. doi: 10.1091/mbc.6.6.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Mead K, Shestak W. The inner dynein arms I2 interact with a “dynein regulatory complex” in Chlamydomonas flagella. J Cell Biol. 1992;118:1455–1463. doi: 10.1083/jcb.118.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Ramanis Z. The proximal portion of Chlamydomonas flagella contains a distinct set of inner dynein arms. J Cell Biol. 1991;112:701–709. doi: 10.1083/jcb.112.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME. Axonemal dyneins: assembly, organization, and regulation. Curr Opin Cell Biol. 1996;8:10–17. doi: 10.1016/s0955-0674(96)80042-1. [DOI] [PubMed] [Google Scholar]
- Porter ME, Knott JA, Myster SH, Farlow SJ. Characterization of the dynein gene family in Chlamydomonas. Genetics. 1996;144:569–585. doi: 10.1093/genetics/144.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Power J, Dutcher SK. Extragenic suppressors of paralyzed flagellar mutations in Chlamydomonas reinhardtii identify loci that alter the inner dynein arms. J Cell Biol. 1992;118:1163–1176. doi: 10.1083/jcb.118.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R, Granick S. Nutritional studies in Chlamydomonas reinhardtii. Ann NY Acad Sci. 1953;56:831–838. doi: 10.1111/j.1749-6632.1953.tb30261.x. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Takada S, King SM, Witman GB, Kamiya R. A Chlamydomonas outer arm dynein mutant with a truncated beta heavy chain. J Cell Biol. 1993;122:653–661. doi: 10.1083/jcb.122.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer T, Sheetz M. Two activators of microtubule-based vesicle transport. J Cell Biol. 1991;115:1309–1318. doi: 10.1083/jcb.115.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EF, Sale WS. Microtubule binding and translocation by inner dynein arm subtype I1. Cell Motil Cytoskel. 1991;18:258–268. doi: 10.1002/cm.970180403. [DOI] [PubMed] [Google Scholar]
- Smith EF, Sale WS. Structural and functional reconstitution of inner dynein arms in Chlamydomonas flagellar axonemes. J Cell Biol. 1992;117:573–581. doi: 10.1083/jcb.117.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen W, Hodgkinson J, Wiche G. Immunogold localization of the intermediate chain within the protein complex of cytoplasmic dynein. J Struct Biol. 1996;117:227–235. doi: 10.1006/jsbi.1996.0087. [DOI] [PubMed] [Google Scholar]
- Steffen W, Karki S, Vaughan K, Vallee R, Holzbaur E, Weiss D, Kuznetsov S. The involvement of the intermediate chain of cytoplasmic dynein in binding the motor complex to membranous organelles of Xenopus oocytes. Mol Biol Cell. 1997;8:2077–2088. doi: 10.1091/mbc.8.10.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Kamiya R. Functional reconstitution of Chlamydomonas outer dynein arms from α-β and γ subunits: requirement of a third factor. J Cell Biol. 1994;131:399–409. doi: 10.1083/jcb.126.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam LW, Lefebvre PA. Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics. 1993;135:375–384. doi: 10.1093/genetics/135.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W-J. Blot-affinity purification of antibodies. Methods Cell Biol. 1993;37:95–104. doi: 10.1016/s0091-679x(08)60245-9. [DOI] [PubMed] [Google Scholar]
- Vaughan KT, Vallee RB. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150glued. J Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson CG, King SM, Koutoulis A, Pazour GJ, Witman GB. The 78, 000 Mr intermediate chain of Chlamydomonas outer arm dynein is a WD-repeat protein required for arm assembly. J Cell Biol. 1995;129:169–178. doi: 10.1083/jcb.129.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson CG, King SM, Witman GB. Molecular analysis of the γ heavy chain of Chlamydomonas flagellar outer arm dynein. J Cell Sci. 1994;107:497–506. doi: 10.1242/jcs.107.3.497. [DOI] [PubMed] [Google Scholar]
- Witman G, Wilkerson C, King S. The biochemistry, genetics, and molecular biology of flagellar dynein. In: Hyams HS, editor. Microtubules. C.W. Lloyd, New York: Wiley-Liss; 1994. pp. 229–249. [Google Scholar]
- Witman GB. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 1986;134:280–290. doi: 10.1016/0076-6879(86)34096-5. [DOI] [PubMed] [Google Scholar]
- Wray W, Boulikas T, Wray VP, Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981;118:197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- Yang P, Sale W. Mol Biol. Cell 9:3335–3349. 1998. The Mr 140,000 intermediate chain of Chlamydomonas flagellar inner arm dynein is a WD-repeat protein implicated in dynein arm anchoring. [DOI] [PMC free article] [PubMed] [Google Scholar]