Abstract
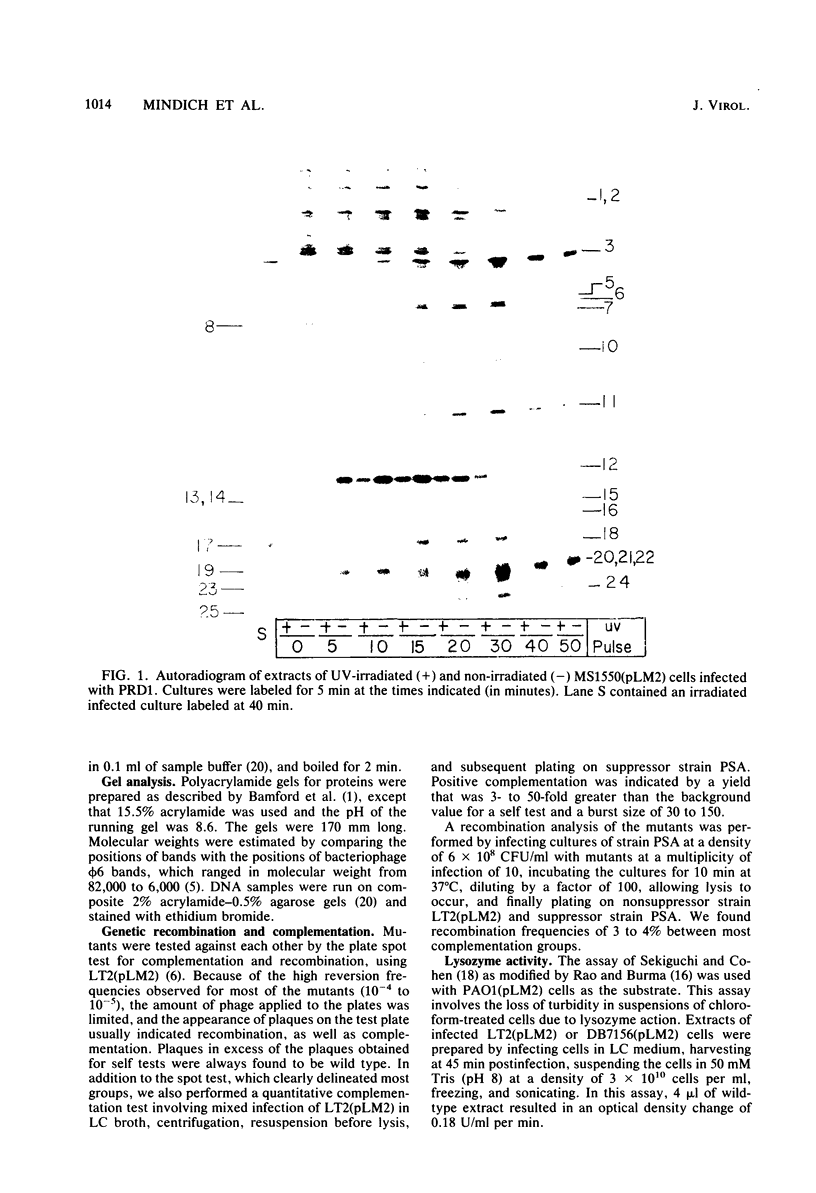
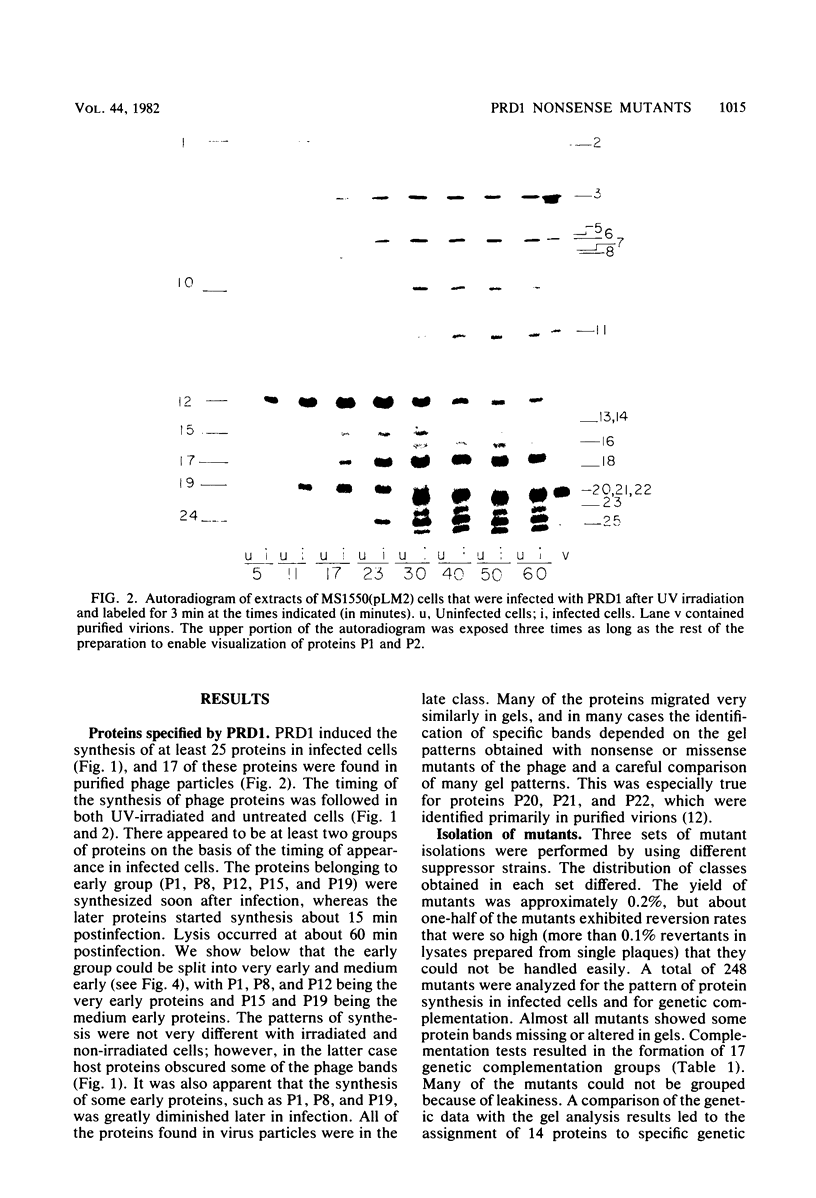
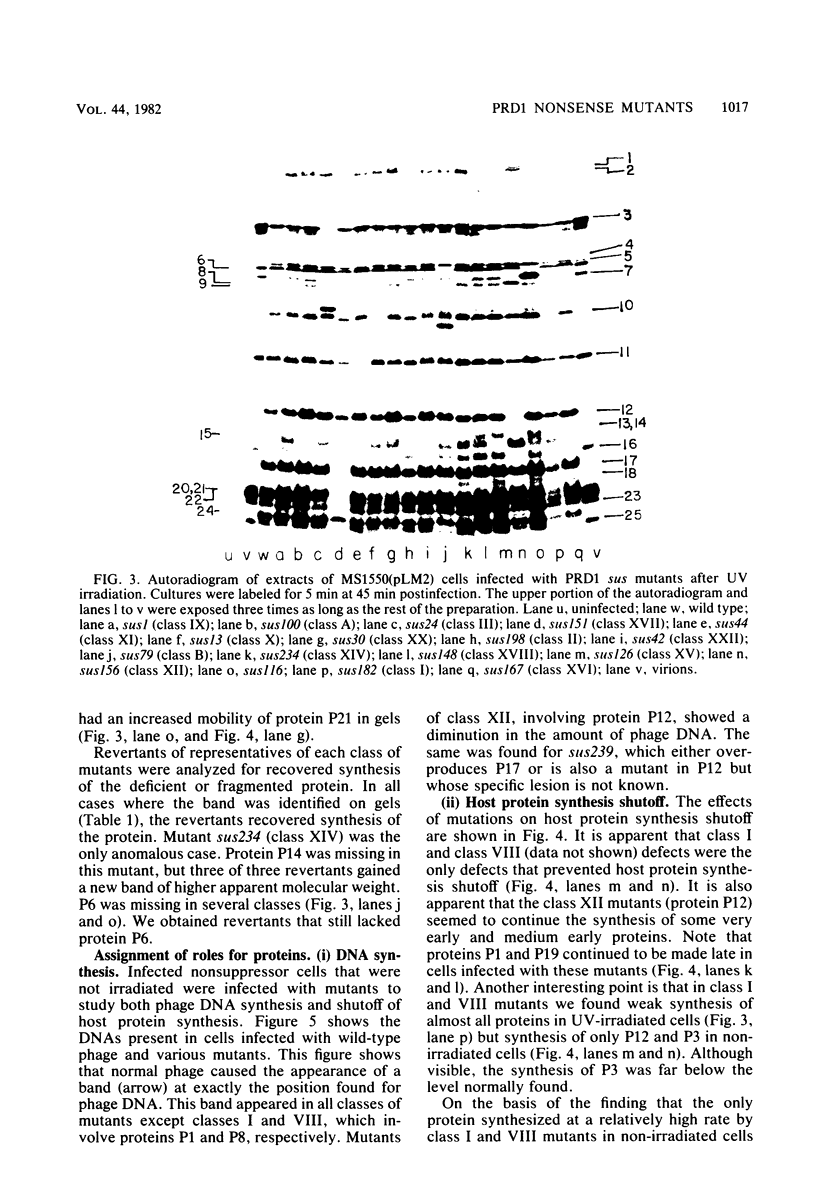
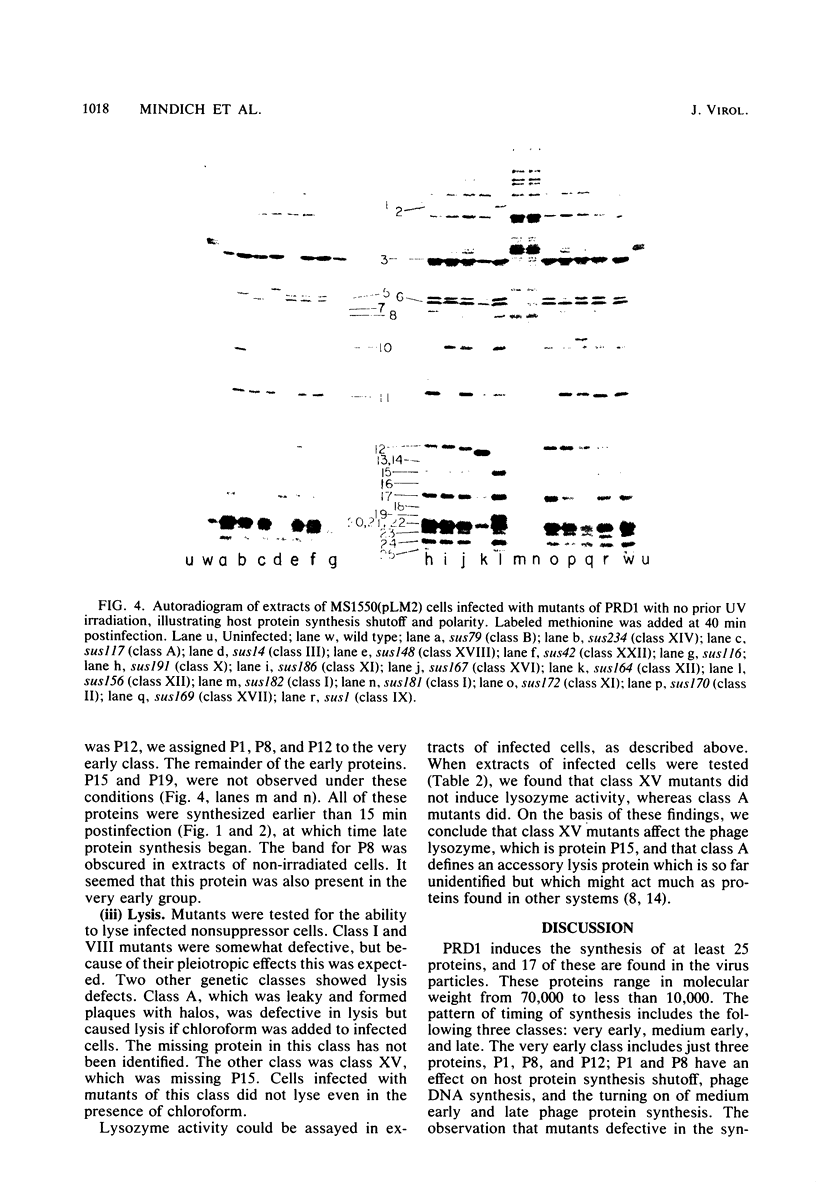
We isolated nonsense mutants of bacteriophage PRD1, a lipid-containing polyhedral virus capable of infecting many genera of gram-negative bacteria. These mutants were grouped into 19 classes on the basis of genetic complementation and sodium dodecyl sulfate-polyacrylamide gel electrophoretic analysis. PRD1 infection led to the synthesis of at least 25 viral proteins, 17 of which were components of mature virions. The synthesis of proteins fell into the following three classes: very early, middle early, and late. Two of the very early proteins, P1 and P8, had an effect on DNA synthesis, host protein synthesis shutoff, and the turning on of middle and late protein synthesis. Another very early protein, P12, was involved in the shutoff of early protein synthesis. Two genes were identified as affecting lysis of the host. One appeared to be a lysin, whereas the other was an accessory lytic factor.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bamford D. H., Rouhiainen L., Takkinen K., Söderlund H. Comparison of the lipid-containing bacteriophages PRD1, PR3, PR4, PR5 and L17. J Gen Virol. 1981 Dec;57(Pt 2):365–373. doi: 10.1099/0022-1317-57-2-365. [DOI] [PubMed] [Google Scholar]
- Bradley D. E., Rutherford E. L. Basic characterization of a lipid-containing bacteriophage specific for plasmids of the P, N, and W compatibility groups. Can J Microbiol. 1975 Feb;21(2):152–163. doi: 10.1139/m75-023. [DOI] [PubMed] [Google Scholar]
- Cadden S. P., Sands J. A. Structural proteins of a lipid-containing bacteriophage which replicates in Escherichia coli: phage PR4. Can J Microbiol. 1977 Aug;23(8):1084–1087. doi: 10.1139/m77-163. [DOI] [PubMed] [Google Scholar]
- Das A., Court D., Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1959–1963. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day L. A., Mindich L. The molecular weight of bacteriophage phi 6 and its nucleocapsid. Virology. 1980 Jun;103(2):376–385. doi: 10.1016/0042-6822(80)90196-8. [DOI] [PubMed] [Google Scholar]
- EDGAR R. S., DENHARDT G. H., EPSTEIN R. H. A COMPARATIVE GENETIC STUDY OF CONDITIONAL LETHAL MUTATIONS OF BACTERIOPHAGE T4D. Genetics. 1964 Apr;49:635–648. doi: 10.1093/genetics/49.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassler J. S., Tessman I. Suppression of polarity in the gal operon by ultraviolet irradiation. J Bacteriol. 1980 Dec;144(3):1205–1207. doi: 10.1128/jb.144.3.1205-1207.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama M., Sano Y., Shinomiya T. Suppressor mutation in Pseudomonas aeruginosa. J Bacteriol. 1979 Jun;138(3):748–755. doi: 10.1128/jb.138.3.748-755.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundström K. H., Bamford D. H., Palva E. T., Lounatmaa K. Lipid-containing bacteriophage PR4: structure and life cycle. J Gen Virol. 1979 Jun;43(3):583–592. doi: 10.1099/0022-1317-43-3-583. [DOI] [PubMed] [Google Scholar]
- Mindich L., Bamford D., McGraw T., Mackenzie G. Assembly of bacteriophage PRD1: particle formation with wild-type and mutant viruses. J Virol. 1982 Dec;44(3):1021–1030. doi: 10.1128/jvi.44.3.1021-1030.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindich L., Cohen J., Weisburd M. Isolation of nonsense suppressor mutants in Pseudomonas. J Bacteriol. 1976 Apr;126(1):177–182. doi: 10.1128/jb.126.1.177-182.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., Siak J. S., Gray R. H. Characteristics of PRD1, a plasmid-dependent broad host range DNA bacteriophage. J Virol. 1974 Sep;14(3):689–699. doi: 10.1128/jvi.14.3.689-699.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao G. R., Burma D. P. Purification and properties of phage P22-induced lysozyme. J Biol Chem. 1971 Nov;246(21):6474–6479. [PubMed] [Google Scholar]
- SEKIGUCHI M., COHEN S. S. THE SYNTHESIS OF MESSENGER RNA WITHOUT PROTEIN SYNTHESIS. II. SYNTHESIS OF PHAGE-INDUCED RNA AND SEQUENTIAL ENZYME PRODUCTION. J Mol Biol. 1964 May;8:638–659. doi: 10.1016/s0022-2836(64)80114-5. [DOI] [PubMed] [Google Scholar]
- Sands J. A., Cadden S. P. Phospholipids in an Escherichia coli bacteriophage. FEBS Lett. 1975 Oct 15;58(1):43–46. doi: 10.1016/0014-5793(75)80221-3. [DOI] [PubMed] [Google Scholar]
- Sinclair J. F., Tzagoloff A., Levine D., Mindich L. Proteins of bacteriophage phi6. J Virol. 1975 Sep;16(3):685–695. doi: 10.1128/jvi.16.3.685-695.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. Analysis of bacteriophage T7 early RNAs and proteins on slab gels. J Mol Biol. 1973 Sep 15;79(2):237–248. doi: 10.1016/0022-2836(73)90003-x. [DOI] [PubMed] [Google Scholar]
- Winston F., Botstein D., Miller J. H. Characterization of amber and ochre suppressors in Salmonella typhimurium. J Bacteriol. 1979 Jan;137(1):433–439. doi: 10.1128/jb.137.1.433-439.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong F. H., Bryan L. E. Characteristics of PR5, a lipid-containing plasmid-dependent phage. Can J Microbiol. 1978 Jul;24(7):875–882. doi: 10.1139/m78-145. [DOI] [PubMed] [Google Scholar]
- Youderian P., Susskind M. M. Identification of the products of bacteriophage P22 genes, including a new late gene. Virology. 1980 Nov;107(1):258–269. doi: 10.1016/0042-6822(80)90291-3. [DOI] [PubMed] [Google Scholar]