Abstract
A new algorithm is described for label-free quantitation of relative protein abundances across multiple complex proteomic samples. Q-MEND is based on the denoising and peak picking algorithm, MEND, previously developed in our laboratory. Q-MEND takes advantage of the high resolution and mass accuracy of the hybrid LTQFT MS mass spectrometer (or other high resolution mass spectrometers, such as a Q-TOF MS). The strategy, termed “cross-assignment”, is introduced to increase substantially the number of quantitated proteins. In this approach, all MS/MS identifications for the set of analyzed samples are combined into a master ID list, and then each LC/MS run is searched for the features that can be assigned to a specific identification from that master list. The reliability of quantitation is enhanced by quantitating separately all peptide charge states, along with a scoring procedure to filter out less reliable peptide abundance measurements. The effectiveness of Q-MEND is illustrated in the relative quantitative analysis of E.coli samples spiked with known amounts of non-E.coli protein digests. A mean quantitation accuracy of 7% and mean precision of 15% is demonstrated. Q-MEND can perform relative quantitation of a set of LC/MS datasets without manual intervention and can generate files compatible with the Guidelines for Proteomic Data Publication.
Introduction
Many biological advances are expected as a result of the emerging quantitative methodologies in mass spectrometry-based proteomics that can provide fundamental understandings of biological systems as well as diagnostic biomarkers [1]. A number of methods for determining the relative abundances of proteins have been introduced, using either isotopic labeling [2-8] or label-free approaches [9-25]. Although quantitation by means of chemical or metabolic labeling with stable isotopes generally provides higher accuracy and precision, label-free quantitation has the advantages of (i) applicability to any samples, including human tissues, (ii) ability to quantitate and compare multiple samples, (iii) lower sample complexity, leading to a higher number of identified and quantitated peptides [23], and (iv) simpler and less expensive sample preparation.
The basis of label-free LC/MS quantitation was established in several papers [9-13], which demonstrated the linearity (with median CV of roughly 20%) of LC/ESI-MS ion chromatographic peak areas with protein concentration in model proteomic samples. To compensate for LC gradient changes from run to run and possible variations of sample loading and instrument sensitivity, methods were introduced for retention time alignment and normalization of peak intensities by the median peak intensity of components whose concentrations were assumed to remain constant in a series of samples [10, 13].
The complexity of proteomic samples results in a special challenge for the label-free quantitation methods. The presence of numerous coeluting peptides can not only cause ion suppression, thus compromising reproducibility of peak intensities, but can also make ambiguous the assignment of LC/MS peaks (features) to MS/MS based identifications. Several algorithms for high throughput label-free quantitation of complex proteomic samples have been introduced. These vary in how they (i) combine LC/MS and MS/MS data, (ii) perform retention time alignment, and (iii) normalize peptide abundances [10-24]. These algorithms often use common features for both retention time alignment and relative quantitation, with only differentially abundant features submitted for MS/MS-based identification [10, 17, 18, 24]. The obvious advantages of this approach are high throughput and reduction in data dependent bias, due to the minimization of the number of MS/MS acquisitions. However, retention time alignment and normalization based solely on LC/MS features may not be completely reliable for complex proteomic samples. The approach can best be used for comparison of similar samples where the majority of the features are conserved. Other algorithms employ both LC/MS features and MS/MS identifications for alignment and normalization (i) during the same LC/MS run [11, 20, 23] or (ii) from subsequent runs with settings optimized for identifications (high number of MS/MS acquisitions) and for quantitation (low number of MS/MS acquisitions, high number of LC/MS replicates) [21, 22], or even using different MS instruments for the identification and quantitation runs [12, 15, 16]. The most important challenge in the label-free quantitation of complex proteomic samples is to establish unambiguous assignments between the identified peptides and LC/MS features.
Similar to labeled quantitation, the label-free approach benefits from the high mass resolution data, such as provided by modern TOF, FTICR, or Orbitrap mass spectrometers, because high resolution greatly reduces the likelihood of incorrect peak assignment. Specifically, with high resolution and high mass accuracy (i) MS peak overlap is reduced, (ii) the charge state of the ion can be readily determined, and (iii) the probability of correct identification is increased, as discussed in [8]. An alternative, semi-quantitative method for determining protein abundance ratios was introduced in [26, 27]. This method, called “spectral counting”, compares the number of precursors submitted for MS/MS analysis (“sequencing events”) for each protein in a set of two or more samples. Spectral counting can be used both for labeled and label-free quantitation. For more comparison of quantitative and semi-quantitative methods, see [8].
The goal of the present paper is to introduce a new algorithm (Q-MEND) for label-free quantitation of complex proteomic samples using the high mass resolution and high mass accuracy of the hybrid LTQ-FT mass spectrometer. Similar to [21, 22], we use identified peptides as landmarks for reliable retention time alignment of LC/MS runs. We do not use specific “identification” and “quantitation” runs [15], but acquire both LC/MS and MS-MS data in the same run. This approach (i) minimizes the number of LC/MS runs, which can be important when the amount of sample is limited, and (ii) maximizes the number of identified, and quantitated, peptides (when sample amount is not limited) by combining identifications from several replicate runs.
In order to maximize the number of quantifiable peptides, a strategy termed “cross-assignment” has been implemented. Here, all identifications for the group of analyzed samples are combined into a master ID list, and then each LC/MS run is searched for the features that can reliably be assigned to a specific identification from that master list. Additionally, in order to maximize the number of independent measurements of peptide abundances and thus improve the precision of protein quantitation, m/z values of all likely peptide charge states for each identified peptide are calculated and added to the master ID list. Furthermore, since not all peptide abundance measurements are equally reliable, a scoring procedure, similar to that of our 15N/14N labeled quantitation algorithm QN [8], has been introduced in order to exclude less reliable MS intensity data.
The performance of Q-MEND is demonstrated in this paper in the analysis of relative protein abundance of E.coli samples that differ by the overexpression of the Hip A gene. Reproducibility, precision and accuracy of quantitation are evaluated and compared with that reported in other quantitative proteomic studies [10, 20]. The value of cross-assignment in significantly increasing the number of quantitated proteins and in improving the results of quantitation is demonstrated.
Experimental
Test Sample Preparation
The Q-MEND algorithm and software were tested in the analysis of cell lysates of E.coli. Sample A was from wild type E.coli, and sample B was from E.coli with the Hip A gene product over-expressed, cultured in the same media [28] (Dr. Kim Lewis Northeastern University). The amount of extracted protein was measured with the Bradford assay (Bio-Rad, Hercules, CA). SDS-PAGE separation of the E.coli proteins was performed on a NuPAGE electrophoresis system (Invitrogen, Carlsbad, CA) with a Novex 4−12% Bis-Tris gel and MOPS SDS running buffer. Approximately 70 μg of the E.coli proteins were loaded on the gel, followed by a 30-min electrophoretic separation at 200 V. In-gel digestion was performed following a standard protocol [29]. Each lane was cut into seven fractions, each of which contained roughly the same total amount of protein as estimated by visual inspection. Only one pair of the gel bands from a given fraction (40−60 kDa) (Hip A sample and wild type) were used in the experiments.
A set of 3 model samples (A1, A2, and A3) was also prepared by spiking the 40−60 kDa gel band of sample A (wild type E.coli) with a tryptic digest of 3 non-E.coli proteins (bovine serum albumin, lactoperoxidase (bovine); myoglobin (equine), all from Sigma-Aldrich, St.Louis, MO). Table 1 lists the amounts of these digests added to each of these 3 samples.
Table 1.
Amounts of non-E.coli protein digests spiked into the wild type E.coli digest, sample A.
| Sample | Protein ratios (BSA: LPO: MYO) | Protein amounts |
|---|---|---|
| A1 | 1:1:1 | 100 fmol of each |
| A2 | 1:2:5 | 100: 200: 500 fmol |
| A3 | 0.5:1:5 | 50: 100: 500 fmol |
BSA: bovine serum albumin; LPO: lactoperoxidase (bovine); MYO: myoglobin (equus).
Nano-LC/MS
Digested samples were analyzed using a nano-LC system coupled to a hybrid linear ion trap-FTICR MS (LTQ-FT MS, Thermo Electron, San Jose, CA). Approximately 4 μL of the concentrated peptide extract was loaded onto a 75 μm × 15 cm column packed with 3 μm Magic C18 particles (Michrom BioResources, Auburn, CA), followed by a 75 min linear gradient from 2% to 35% ACN (v/v) in 0.1% formic acid using a 250 nL/min flow rate. Three LC/MS runs were performed for each digested sample. In order to reduce carry-over, each LC/MS run was followed by a blank injection of buffer and run with the same gradient. MS data were acquired in a repeating 4-second cycle: a high resolution FT MS scan (accumulation of 2 × 106 ions) followed by up to 10 MS/MS spectra in the linear ion trap (accumulation of 3 × 104 ions). Dynamic exclusion was set to 1 minute.
Peptide and Protein Identification
The data acquired from nano-LC/MS were searched using Sequest [30] against the NCBI database of E.coli (downloaded in June 2005). Sequences of non- E.coli proteins spiked into the model samples A1, A2, and A3 were added to the database for these samples. The precursor ion mass tolerance was set at ±1.4 Da, and trypsin was designated as the proteolytic enzyme with up to 2 missed cleavages. In order to minimize the false positive rate, peptide identifications were accepted only if all three of the following criteria were met: (i) Sequest Xcorr value was greater than 1.8, 2.2, and 3.0 (for +1, +2, +3 ions, respectively); (ii) the protein that the peptide was derived from had a probability of correct identification of greater than 0.9 (ProteinProphet [31]); and (iii) the observed precursor mass was within 15 ppm of the theoretical mass of the identified peptide. After tabulating a list of all peptides identified in at least one sample, the relative abundances of the parent proteins across all analyzed samples were determined as described in the Results and Discussion Section.
Results and Discussion
Algorithm Description
The Q-MEND algorithm was written to determine the relative abundances of proteins across a set of label-free samples analyzed by LC/ESI-MS using the LTQ-FT MS. It is important to note that the approach can also be used with other high resolution hybrid mass spectrometers, e.g. Q-TOF, LTQ-Orbitrap. The algorithm compares an arbitrary number of samples (N), pairwise comparison (N=2) being a special case. Q-MEND keeps track of samples prefractionated into a number (K) of gel bands, SCX fractions, etc., each of which may be analyzed in R replicate LC/MS runs (typically 2 or 3). Thus, quantitation is performed by analysis of N×K×R LC/MS data sets. The input data are: (i) the raw LC/MS data file (collected in the full profile mode) and (ii) a list of identified peptides presented either as an INTERACT [5] file resulting from an MS/MS database search with Sequest, verified by using PeptideProphet (Institute for Systems Biology, Seattle, WA), or as a Bioworks 3.2 (Thermo Electron, San Jose, CA) excel file.
Q-MEND consists of 6 modules (flowchart shown in Fig 1) which will be discussed in detail in the following sections. In summary, the first module reads a raw LC/MS data file and applies the denoising and peak picking MEND algorithm (matched filtration with experimental noise determination) that minimizes both chemical and electronic noise and then determines extracted ion chromatographic peak maxima and areas [32]. The module next outputs a peak list with the m/z, retention time and area of each extracted ion chromatographic peak. In parallel, the second module reads the list of identified peptides (INTERACT or BioWorks file) and calculates a list of m/z values for all potential peptide charge states (PCSs) within the m/z range of the MS instrument. The two modules process all N×K×R data sets one by one. The output files from the first two modules are sorted into K groups, corresponding to prefractionation sets. The third module then performs retention time alignment across multiple (N×R) data sets (applying alignment to both PCS lists from the second module and MEND peak lists from the first module) and generates a master list of all potential PCSs for each fraction. The fourth module assigns MS peaks from the MEND peak list to the PCSs from the master list, then quantitates the abundance of each PCS as the sum of extracted ion chromatographic peak areas of the three highest intensity isotopes in the isotopic cluster of the MS peak assigned to the PCS. The fifth module combines into one table the PCS abundances determined in all N×K×R runs. The abundances are normalized at this point in a manner described later. To account for cases where the same protein appears in adjacent bands or fractions, the normalized abundances of the PCSs present in the neighboring fractions are added. The resulting abundances for each PCS are then averaged across the R replicate runs. The sixth module quantitates at the protein level by averaging the relative abundances of individual PCSs that identify the given protein. The first module is written in C++ and the other modules in MATLAB. Q-MEND is available from the authors upon request. Below, we describe each of the modules in more detail.
Fig. 1.
Flow chart of the Q-MEND algorithm.
Module 1. Processing of LC/MS Data by MEND
Raw LC/MS data files are first denoised using the MEND algorithm, as described in detail in [32]. Briefly, denoising is performed in the chromatographic time domain by matched filtration with the assumption of Gaussian chromatographic peak shapes and with experimentally determined noise profiles. Denoising is followed by peak picking based on the scoring of candidates according to the similarity of the observed and expected chromatographic and MS peak shapes. Importantly, this procedure excludes chemical noise from the peak list since the former does not have a Gaussian-like shape in the chromatographic time domain. The final peak list generated by MEND includes the m/z value of the centroid of the MS peak, the retention time of the extracted ion chromatographic peak maximum (scan number), as well as the intensity and area of the peak.
An option is provided in the algorithm to reduce the time for computation by splitting the raw LC/MS data files into M m/z regions (M = 8 in this study), processing the regions separately by MEND, and generating M peak lists. This step enables MEND processing to be performed in parallel on M nodes of a computer cluster, thus reducing the time of computation M-fold.
Module 2. Generating the List of Potential Peptide Charge States
The second module reads a list of identified peptides (INTERACT or Bioworks files) for each of the N×K×R runs, calculates their theoretical molecular weights based on the amino acid sequences of the peptides, and then generates a list of all potential PCSs, i.e. the m/z values of all possible charge states within the m/z range of the MS instrument (400<m/z<2000, for the LTQ-FT MS). Quantitation of all charge states increases the number of independent quantitative measurements of each identified peptide, thus leading to improved accuracy of quantitation. This procedure is especially beneficial in the analysis of experiments with large peptides, which can have charge states well in excess of 3 [33]. For each PCS, the theoretical m/z and expected relative intensities of the three highest intensity isotopes are calculated, for use in the fourth module (assignment of PCSs to the experimentally observed MS peaks).
Module 3. Retention Time Alignment
The third module performs retention time alignment across multiple runs, both on the potential PCS lists (from module 2) and on the MEND peak lists (from module 1). First, one of the data sets (i.e. an LC/MS run) is selected by the user as a template for alignment; typically, it is a run that has numerous identified peptides common with other runs. Then, PCS lists for each of the other (N×R −1) runs are compared with the template list, and a number of common identified peptides are selected as landmarks for alignment. Generally, roughly 50 landmark peptides are selected, distributed as uniformly as possible across the chromatogram. Retention time alignment of (N×R-1) data sets to the template is then performed by linear compression or extension of the intervals between the landmarks. This recalculation of retention time is performed both for PCS lists (a master PCS list is composed) and MEND peak lists for each run. Thus, a “universal” retention time is established for correlating PCSs and MS peaks across the entire N×R data sets. The same procedure is performed for each of the K fractions.
Module 4. Assignment of MS Peaks to PCSs
The high mass accuracy of the LTQ-FT MS, or other high resolution MS instruments, together with the alignment of extracted ion chromatographic retention time across multiple samples enables cross-assignment, i.e. MS peaks from each run may be assigned not only to PCSs identified in the same run, but in other runs as well. Cross-assignment is important in LC/MS analysis of complex proteomic samples since there is competition between potential precursors for the limited number of MS/MS analyses that can be selected by data dependent scanning. Fig. 2 illustrates how cross-assignment increases the number of common quantitated peptides in the case of two runs of different samples from a given sample set. If cross-assignment is not employed, only peptides identified in both samples, A and B, are used for quantitation. When cross-assignment is implemented, the number of quantitated peptides is increased by adding those peptides identified by MS/MS in only one of the samples, while the peptides are detected and quantitated in the MS mode of both samples. More specific examples will be provided in the Algorithm Performance Evaluation Section demonstrating that cross-assignment nearly doubles the number of quantitated proteins relative to quantitation without cross assignment. This result occurs even though three replicate runs of each sample were performed.
Fig. 2. Illustration of the increase in the number of quantitated peptides due to cross-assignment.

Quantitation without cross-assignment. Only peptides identified in both samples A and B are used for calculation of relative abundances. Quantitation with cross-assignment. Peptides identified in only one of the samples and visible in the LC/MS of both samples are used for calculation of relative abundances.
The main function of the fourth module is to assign MS peaks, tabulated in the MEND peak list for each of N×R runs, to identified peptides from the master PCS list. This assignment is performed by comparing the retention time, m/z and isotopic cluster structure (ratio of isotopic peaks and Δm/z between the peaks in the cluster) for each PCS with the experimentally observed features from the MEND peak lists. When more than one candidate is within the m/z and retention time tolerance windows, the candidate with the smallest deviation from the theoretical predictions is selected. A fuzzy logic approach is then applied taking into account the m/z and retention time deviations as well as deviations in the isotopic cluster structure. The exact equality of one of the parameters is less important than the approximate equality of multiple parameters. The abundance of each PCS is calculated as the sum of the extracted ion chromatographic peak areas of the three highest intensity isotopic peaks of the selected isotopic cluster.
To reduce the time of computation, it is beneficial to run module 4 by distributed processing on a computer cluster. Similar to parallelizing the LC/MS raw data in module 1, the PCS master list is split into M m/z regions. Assignment is performed in parallel on a separate node for each m/z region, and the results of assignment are then combined into one file.
Module 5. Generation of Combined Table of PCSs Abundances
The procedures described above are repeated for each fraction (gel band, SCX, etc.), and then a combined table of PCS abundances measured in the N×K×R runs is formed. PCS abundances for each run are normalized by the median ratio of abundances of PCSs present both in the given run and the template run. As an alternative, the normalization coefficient can be calculated as the median relative abundance for a user-defined group of peptides (from housekeeping proteins or internal standards). If a peptide is present in two or more fractions, it is excluded from the list of PCSs used for calculation of normalization coefficient. After normalization, the relative abundances of PCSs in the neighboring fractions are added together, and the resulting values are then averaged across the R replicates. Thus, the (N×K×R+1) column table is transformed into an N+1 column table presenting abundances of PCSs in N samples.
Module 6. Protein Quantitation
Obviously, not all measurements of PCS abundances are equally reliable. Errors in PCS quantitation can result from ion suppression, overlapping of LC/MS peaks, electrospray variations or other factors. Analogous to the QN algorithm for 15N/14N quantitation [8], a reliability score is introduced based on the assumption that high intensity peaks are measured with higher accuracy and precision, and therefore such peaks should be preferentially selected for quantitation of the parent protein. Additionally, PCSs with a normalized abundance that are consistent across several replicates is considered to be more representative than PCSs with greater deviations of abundance. Finally, PCSs detected and quantitated in several samples are taken to be more reliable for protein quantitation than PCSs present in only two samples. These assumptions are incorporated in a score that is calculated for each PCS by summing across all N samples:
| (1) |
where Ii,j is the mean abundance of the ith PCS in the jth sample across R replicate runs and CVi,j is the coefficient of variation. If a PCS identifying a protein has a score lower than 1/100 of the top score for a given protein, it is not included in the protein average, with the exception that PCSs with scores above 3 times the median score always remain on the list. (The above exception is used to avoid the situation where one very high abundance PCS forces the exclusion of other reliably quantitated PCSs.)
Relative abundances of the proteins are calculated from the corresponding PCSs by the following procedure. The abundances of PCSs that remain on the list are normalized along the rows (across samples) by dividing by the maximum PCS abundance in the given row. Next, the relative abundances of all PCSs identifying the same protein are evaluated by a Dean-Dixon test (90% confidence level) [34], removing outliers, and finally, the mean of the relative abundances together with the coefficient of variation are calculated and included in the output file listing for all quantitated proteins. It is important to emphasize that, as in our QN algorithm [8], the Dean-Dixon test alone was not sufficient to remove outliers. If a protein were represented by two PCSs or two groups of PCSs with significantly different relative abundances, the Dean-Dixon test would fail to determine which of the two PCSs or two groups of PCSs were correct. In contrast, using scoring measures such as eq.1, the less reliable PCSs are eliminated. The scoring approach differs from the weighted average [6], by taking into account not only the CV, but also peak intensity and the number of PCS that appear in the replicate runs. These additional considerations were found to be important in cases of a low number of measurements for a given PCS where the CV was not representative.
Finally, to comply with the Guidelines for Proteomic Data Publication [35], another output file is generated that includes all information on the MS peaks used in quantitation: raw file name (sample, fraction, replicate), scan number, molecular weight of the peptide, charge state, peptide sequence, protein name, centroids m/z values, intensities and areas of chromatographic peaks, PeptideProphet probability (or p-value from BioWorks) , and the Xcorr and ΔCn parameters from the Sequest database search.
Performance of the Q-MEND Algorithm
The performance of Q-MEND was examined in the determination of the relative protein abundances in lysates of a wild type E.coli (sample A) and a Hip A over-expressor E.coli (sample B). Specifically, a single (40−60 kDa) SDS PAGE gel band from each of the samples was analyzed in triplicate by LC/MS/MS, followed by Sequest search, resulting in a total of 2773 peptide charge states (PCSs) identified in 6 runs and associated with 351 proteins.
Retention time alignment
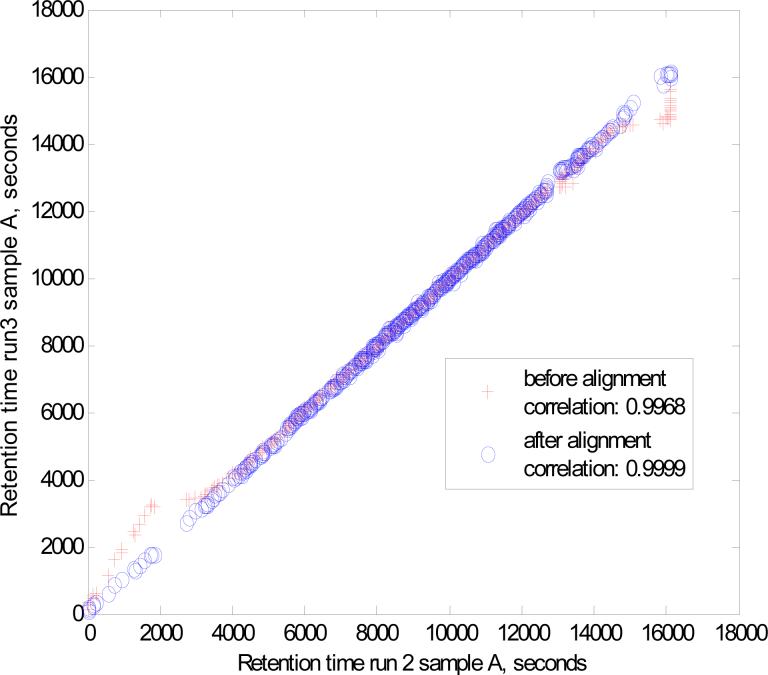
The first run of sample A was used as the template for retention time alignment. Thirty five peptides with relatively high intensity precursor ions, identified in all the runs, were selected as landmarks for alignment, performed by linear compression or extension of the intervals between the landmarks. Fig. 3 illustrates the effectiveness of alignment by comparing corrected and uncorrected retention times of the 1112 peptides detected in both runs 2 and 3 of sample A. Alignment was found to be most important at the beginning and end of LC gradient. Alignment improved the Pearson's correlation coefficient from 0.9968 to 0.9999; reduced the maximum run-torun variation in retention time of peptides from 442 s to 198 s; and decreased the standard deviation of retention time variation from 63 s to 42 s. Similar performance was observed (data not shown) for runs 2 and 3 of sample B (Hip A E.coli) aligned to run 1 of sample A (correlation coefficients before and after alignment, 0.9977 and 0.9998, respectively). The efficiency and simplicity of the alignment algorithm is attributed to using as landmarks multiple peptides identified with high probabilities (p>0.99 in these experiments). Thus, it is not necessary to use complicated nonlinear alignment algorithms [36, 37] developed for applications where sample component identifications are not available at the alignment stage. In general, our alignment algorithm worked well for chromatograms having at least 10 −15 common identified peptides across the chomatogram to use as landmarks. In our experience, the choice of the template is not important for successful alignment. All that is required is to have at least 10 common peptides identified with more that 99% probability in all runs. Any run that has all the common identified peptides can be used as the template.
Fig. 3. Performance of retention time alignment algorithms.
Retention times of common peptides in replicate runs for sample A are compared before and after alignment.
Reproducibility of PCS abundance measurements
Ideally, the same PCSs should be detected and the ratios of PCS abundances should be equal to 1 in sample replicates. The two main reasons for non-ideality of replicate runs in the same column are the LC gradient and electrospray variations. As shown in the above section, LC gradient variation results in the retention time shifts that can be corrected by the alignment algorithm. Electrospray variations results not only in lower reproducibility of PCS abundance measurements, but also in non-detection of lower abundance PCSs in some of the replicate runs.
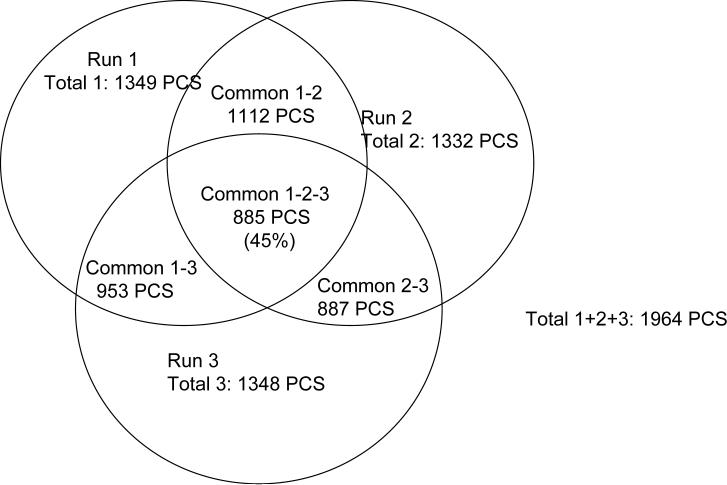
Fig. 4A illustrates the number of PCSs detected in each of three separate runs of sample A and the overlap between PCSs detected in different runs. Out of a total of 1964 PCSs combined from the triplicate runs, only 885 PCSs (45%) were detected in all 3 runs. PCSs common to all 3 runs have on average 12 fold higher abundance (median value of peak area 60.5·105 a.u.) than PCSs detected in only one of the runs (median peak area 4.8·105 a.u.). Less reproducible detection of low abundance peptides most probably results from noise or ion suppression by the coeluting higher abundance peptides.
Fig. 4. Run-to-run reproducibility.
A. Venn diagram presenting the numbers of PCSs detected and quantitated in replicate runs of sample A (wild type E.coli). B. Reproducibility of PCSs abundances (chromatographic peak areas) in replicate runs of sample A
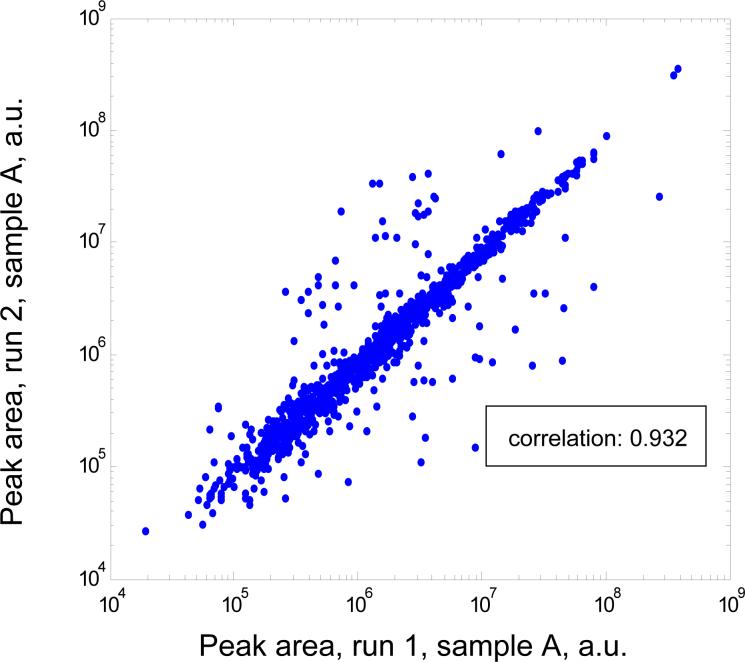
Fig. 4B compares peak areas of 1112 PCSs detected in both replicate runs 1 and 2 of sample A. The Pearson correlation coefficient was 0.932, within the range of the previously reported run-to-run abundance reproducibility in label-free quantitation using LC/LTQ-FT MS [19]. The median value of the ratio of peak areas between the two runs was 0.82. The value was most likely due to differences in the injected sample amount (compensated by the normalization coefficient in module 5). Roughly 70% of the peaks have a ratio within 20% of the median value. However, 7% of peaks have more than a two-fold deviation from the value of the median ratio. These outliers are filtered out when the data from the third run is included, as well as by statistical analysis at the protein quantitation level where the ratios from multiple PCSs from the same protein are averaged. As expected, abundance ratio reproducibility is significantly improved (correlation coefficient 0.96) for higher abundance PCSs detected in all 3 runs (data not shown).
Prior to protein quantitation, PCS abundances for each run were normalized as described in the Algorithm Description Section. For the samples used in this work, the number of differentially abundant proteins (due to spiking and Hip A over expression) was expected to be limited. Consequently, the normalization coefficient was calculated as a median ratio of all PCSs in common between the run being processed and the template run (sample A run 1). In the cases where the number of differentially abundant proteins is substantial, abundances of housekeeping proteins or internal standards should be used for normalization.
Accuracy and Precision of quantitation for samples spiked with known amounts of protein digests
In order to determine the accuracy and precision of Q-MEND for label-free quantitation of complex proteomic samples, 3 model samples were produced by spiking known amounts of digests of non-E.coli proteins (see Table 1) into the wild type E.coli sample A. Analysis of raw data for sample A1 showed that abundances of non-E.coli PCSs in the model sample were comparable to the abundances of E.coli PCSs.
Table 2 compares the observed relative protein abundances to the spiked protein ratios. The maximum observed quantitation error was 23%, while the mean quantitation error was 9%, and the maximum and mean CV were 16% and 7%, respectively. These values are similar to the maximum error of 14%, mean error of 6% and mean CV of 15% reported for label-free quantitation of 5 equimolar exogeneous proteins spiked at various levels (5, 2, 1, 0.5, 0.25, and 0.1 pmol) in human plasma [10] and also to the 16% average error for quantitation of bovine serotransferrin spiked at the levels of 1000, 500, and 250 fmol in depleted plasma [20]. Thus, accuracy and precision of Q-MEND is comparable with other state of the art label-free quantitation algorithms.
Table 2.
Quantitation of relative abundance of spiked proteins in samples A1, A2, A3.
| Sample A2 versus sample A1 | ||||
|---|---|---|---|---|
| Protein | Expected ratio | Mean measured ratio | CV | Quantitation error |
| BSA | 1.0 | 1.09 | 16 % | 9 % |
| LPO | 2.0 | 2.46 | 2 % | 23 % |
| MYO | 5.0 | 4.96 | 3 % | 0.8 % |
| Sample A3 versus sample A1 | ||||
|---|---|---|---|---|
| Protein | Expected ratio | Mean measured ratio | CV | Quantitation error |
| BSA | 0.5 | 0.60 | 15 % | 20 % |
| LPO | 1.0 | 0.98 | 4 % | 2 % |
| MYO | 5.0 | 5.06 | 2 % | 1.2 % |
BSA: bovine serum albumin; LPO: lactoperoxidase (bovine); MYO: myoglobin (equus).
Cross-assignment in protein abundance quantitation
Use of cross-assignment, i.e. assignment and quantitation of MS peaks from each run, not only to PCSs identified in the same run, can significantly increase the number of quantitated PCSs. To illustrate this, we compared Q-MEND with and without cross-assignment for quantitation of relative protein abundances in samples A and B, see Table 3. With cross-assignment, the number of PCSs used for quantitation (3 replicate runs used for each sample) was found to be more than double (981 versus 483), and the number of quantitated proteins increased by 85% (351 versus 190). Importantly, the number of proteins quantitated with 2 or more PCSs was doubled as well (222 versus 114). Cross-assignment resulted in quantitation of 161 additional proteins (56 of which have 2 or more PCSs) and in improvement in the reliability of quantitation by an increase in the number of proteins quantitated with 2 or more PCS from 114 to 166 (out of 190 proteins quantitated by both approaches).
Table 3.
Quantitation with cross-assignment versus quantitation without cross-assignment (Hip A over-expressed versus normal E.coli, 40−60 kDa SDS-PAGE band)
| Without cross-assignment (Xb) | With cross-assignment (Yc) | |||
|---|---|---|---|---|
| Total | Proteins from Y common with Xd | Additional proteinse | ||
| No. PCS | 486 | 982 | 754 | 228 |
| No. Proteins | 190 | 351 | 190 | 161 |
| No. Proteins2a (No. PCS ≥ 2) | 114 | 222 | 166 | 56 |
| Mean no. PCS (for proteins2) | 3.6 | 3.8 | 4.4 | 2.2 |
| Mean no. peaks (per protein2) | 21.6 | 22.7 | 27.6 | 8.3 |
| Mean CV (per protein2) | 0.18 | 0.15 | 0.13 | 0.23 |
| Mean no. peaks per PCS | 6.00 | 5.97 | 6.27 | 3.77 |
Proteins2 - proteins quantitated with 2 or more PCSs.
X- protein quantitation without using cross-assignment
Y- protein quantitation with using cross-assignment
Proteins from Y common with X — proteins that were quantitated with and without cross-assignment.
Additional proteins — proteins that were quantitated only with cross-assignment
No. - number
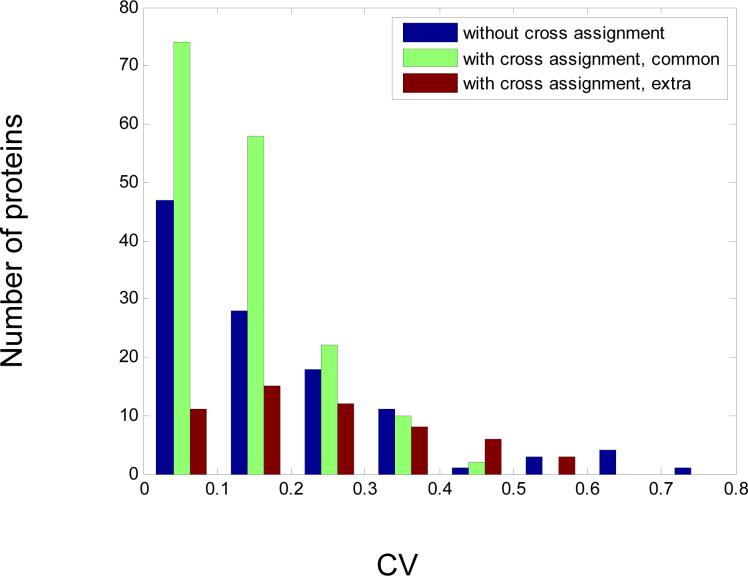
For the 190 proteins that were quantitated both with and without cross-assignment, the change in the values of the relative abundances due to cross-assignment was not significant, i.e. for 94% of the proteins, the difference being less than 10%. However, the precision of quantitation improved (mean CV = 0.13 for processing with cross-assignment versus mean CV=0.18 for processing without cross-assignment). Also, the number of proteins quantitated with CV values lower than 30% was increased by using cross-assignment from 93 (or 49% of common 190 proteins) to 154 (81%). Thus, and importantly, the additional PCSs used for quantitation from cross-assignment did not significantly change the values of relative protein abundance, but did improve the precision of quantitation. The histogram of coefficient of variation for proteins quantitated with and without cross-assignment is presented in Fig. 5.
Fig. 5. Histogram of coefficient of variation for proteins quantitated with and without cross-assignment.
Additional proteins – proteins that were quantitated due to cross-assignment and were not quantitated without cross-assignment.
Generally, quantitative proteomics studies seek to determine the differentially abundant proteins (e.g. two-fold or greater differences in abundance between the two states) for consideration as biomarker candidates or as indications of how metabolic or signaling pathways have been perturbed. Out of 190 proteins that were quantitated both with and without cross-assignment, 50 (or 26%) were differentially abundant, while for the additional 161 quantitated proteins, the number that were differentially abundant was higher, i.e. 65 (or 40%). Thus, implementation of the cross-assignment increased the number of differentially abundant proteins by a factor of 2.3 (115 versus 50).
Conclusion
In this paper, we have described a new algorithm, Q-MEND, for label-free quantitation of LC/ESI-MS data acquired using an LTQ-FT MS, or other high resolution hybrid mass spectrometer. Q-MEND improved the accuracy and precision of quantitation by taking advantage of the high mass accuracy and resolution of the LTQ-FT MS instrument and by using our LC/MS denoising and peak picking algorithm, MEND. The number of quantitated proteins was doubled by the strategy of cross-assignment, while the precision of quantitation was improved by including abundance measurements of all charge states of identified peptide, as well as by cross-assignment.
The performance of Q-MEND has been illustrated in the comparative analysis of two E.coli samples (wild type and a Hip A over-expresser). For one SDS PAGE band (40−60 kDa), a total of 222 proteins were reliably quantitated with two or more peptide charge states (PCSs) and a mean CV of 15% or less. The mean accuracy of quantitation was estimated to be roughly 7%, by spiking known amounts of non-E.coli protein digests into the E.coli samples. Cross-assignment nearly doubled the number of quantitated proteins relative to processing without cross-assignment. In other studies, Q-MEND has been successfully used for discovery of biomarkers for cervical dysplasia [38]. Q-MEND can be applied to perform quantitative analysis using other high resolution instruments, such as an LTQ-Orbitrap MS or Q-TOF MS. Q-MEND allows quantitation without manual intervention and generates files compatible with the Guidelines for Proteomic Data Publication [35]. It is available from the authors upon request.
Acknowledgement
The authors gratefully acknowledge the support of NIH GM 15847. The authors wish to thank Drs. Kim Lewis and Fred Correia for the E.coli samples, and Dr. Roger Kautz for helpful discussions. The authors thank one of the reviewers for suggesting the format of Fig. 2. This is contribution number 900 from the Barnett Institute.
References
- 1.Ong S-E, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 2.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope- coded affinity tags. Nat. Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 3.Washburn MP, Ulaszek R, Deciu C, Schieltz DM, Yates JR. Analysis of quantitative proteomic data generated via multidimensional protein identification technology. Anal. Chem. 2002;74:1650–1657. doi: 10.1021/ac015704l. [DOI] [PubMed] [Google Scholar]
- 4.Schulze WX, Mann M. A novel proteomic screen for peptide-protein interactions. J. Biol. Chem. 2003;279:10756–10764. doi: 10.1074/jbc.M309909200. [DOI] [PubMed] [Google Scholar]
- 5.Han DK, Eng J, Zhou H, Aebersold R. Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat. Biotechnol. 2001;19:946–951. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Zhang H, Ranish JR, Aebersold R. Automated statistical analysis of protein abundance ratios from data generated by stable-isotope dilution and tandem mass spectrometry. Anal.Chem. 2003;75:6648–6657. doi: 10.1021/ac034633i. [DOI] [PubMed] [Google Scholar]
- 7.Qian W-J, Monroe ME, Liu T, Jacobs JM, Anderson GA, Shen Y, Moore RJ, Zhang R, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Camp DG, Smith RD. Quantitaive proteome analysis of human plasma following in vivo lipopolysaccharide administration using 16O/18O labeling and the accurate mass and time tag approach. Mol. Cell. Proteomics. 2005;4:700–709. doi: 10.1074/mcp.M500045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreev VP, Li L, Rejtar T, Qingbo Li Q, Ferry JG, Karger BL. A New Algorithm for 15N/14N Quantitation with LC-ESI-MS Using an LTQ-FT Mass Spectrometer. J. Proteome Res. 2006;5:2039–2045. doi: 10.1021/pr060105m. [DOI] [PubMed] [Google Scholar]
- 9.Chelius D, Bondarenko PV. Quantitative Profiling of Proteins in Complex Mixtures Using Liquid Chromatography and Mass Spectrometry. J. Proteome Res. 2002;1:317–323. doi: 10.1021/pr025517j. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, Zhou H, Lin H, Roy S, Shaler TA, Hill LR, Norton S, Kumar P, Anderle M, Becker CH. Quantification of Proteins and Metabolites by Mass Spectrometry without Isotopic Labeling or Spiked Standards. Anal. Chem. 2003;75:4818–4826. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- 11.Silva JC, Denny R, Dorschel CA, Gerenstein M, Kass IJ, Li G-Z, McKenna T, Nold MJ, Richardson K, Young P, Geromanos S. Quantitative proteomic analysis by accurate mass retention time pairs. Anal. Chem. 2005;77:2187–2200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 12.Qian W-J, Camp DG, Smith RD. High-throughput proteomics using Fourier transform ion cyclotron resonance mass spectrometry. Expert Rev. Proteomics. 2004;1:87–95. doi: 10.1586/14789450.1.1.87. [DOI] [PubMed] [Google Scholar]
- 13.Becker CH, Hastings CA, Norton SM. Mass Spectrometric quantitation of chemical mixture components. US Patent: US 6,835,927 B2, 12/28/2004. [Google Scholar]
- 14.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Fang R, Elias DA, Monroe ME, Shen Y, Mcintosh M, Wang P, Goddard CD, Calister SJ, Moore RJ, Gorby YA, Adkins JN, Fredrickson JK, Lipton MS, Smith RD. Differential label-free quantitative proteomics analysis of shewanella oneidensis cultured under aerobic and sub-oxic conditions by accurate mass and time (AMT) tag approach. Mol. Cell. Proteomics. 2006;5:714–725. doi: 10.1074/mcp.M500301-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Yi EC, Li X, Mallick P, Kelly-Spratt KS, Masselon CD, Camp DG, Smith RD, Kemp CJ, Aebersold R. High throughput quantitative analysis of serum proteins using glycopeptide capture and liquid chromatography mass spectrometry. Mol. Cell. Proteomics. 2005;4:144–155. doi: 10.1074/mcp.M400090-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Yi EC, Kemp CJ, Zhang H, Aebersold R. A software suite for the generation and comparison of peptide arrays from sets of data collected by liquid chromatography – mass spectrometry. Mol. Cell. Proteomics. 2005;4:1328–1340. doi: 10.1074/mcp.M500141-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Ono M, Shitashige M, Honda K, Isobe T, Kuwabara H, Matsuzuki H, Hirohashi S, Yamada T. Label-free quantitative proteomics using large peptide data sets generated by nano-flow liquid chromatography and mass spectrometry. Mol. Cell. Proteomics. 2006;5:1338–1347. doi: 10.1074/mcp.T500039-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Higgs RE, Knierman MD, Gelfanova V, Butler JP, Hale JE. Comprehensive label- free method for the relative quantitation of proteins from biological samples. J. Proteome Res. 2005;4:1442–1450. doi: 10.1021/pr050109b. [DOI] [PubMed] [Google Scholar]
- 20.Wang G, Wu WW, Zeng W, Chou C, Shen R. Label-free protein quantitation using LC-coupled ion trap or FT mass spectrometry: reproducibility, linearity, and application with complex proteomes. J. Proteome Res. 2006;5:1214–1223. doi: 10.1021/pr050406g. [DOI] [PubMed] [Google Scholar]
- 21.Leptos KC, Sarracino DA, Jaffe JD, Krastins B, Church GM. MapQuant: open source software for large-scale protein quantitation. Proteomics. 2006;6:1770–1782. doi: 10.1002/pmic.200500201. [DOI] [PubMed] [Google Scholar]
- 22.Jaffe JD, Mani DR, Leptos KC, Church GM, Gillette MA, Carr SA. PEPPeR: A platform for experimental proteomic pattern recognition. Mol. Cell. Proteomics. 2006;5:1927–1941. doi: 10.1074/mcp.M600222-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreev VP, Li Zang L, Cao L, Rejtar T, Pillai S, Li L, Wang Y, Wu S-L, Karger BL. ASMS. San-Antonio, TX: 2005. A new Algorithm for Quantitation of LC-MS Proteomic Data. [Google Scholar]
- 24.Askenazi M, Sutton J, Sadygov R, Richmond T, Shi X, Gerszten R, Bonilla L, Zumvalt A. HUPO. Munich, Germany: 2005. The Biomarker SIEVE: A computational framework for label-free MS-based proteomic biomarker discovery. [Google Scholar]
- 25.Qian W-J, Jacobs JM, Liu T, Camp DG, Smith RD. Advances and Challenges in Liquid Chromatography-Mass Spectrometry Based Proteomic Profiling for Clinical Applications. Mol. Cell. Proteomics. 2006;5:1727–1744. doi: 10.1074/mcp.M600162-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H, Sadygov RG, Yates JR. A Model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 27.Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal. Chem. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 28.Correia FF, D'Onofrio A, Rejtar T, Li L, Karger BL, Makarova K, Koonin EV, Lewis K. Kinase Activity of Overexpressed HipA is Required for Growth Arrest and Multidrug Tolerance in E. coli. J. Bacteriol. 2006 doi: 10.1128/JB.01237-06. JB.01237-06v1 – published online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steen H, Kuster B, Fernandez M, Pandey A, Mann M. Detection of tyrosine phosphorylated peptides by precursor ion scanning quadrupole TOF mass spectrometry in positive ion mode. Anal. Chem. 2001;73:1440–1448. doi: 10.1021/ac001318c. [DOI] [PubMed] [Google Scholar]
- 30.Eng JK, Ashley L, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 31.Nevsvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 32.Andreev VP, Rejtar T, Chen H-S, Moskovets EV, Ivanov AR, Karger BL. A universal denoising and peak picking algorithm for LC-MS based on matched filtration in the chromatographic time domain. Anal Chem. 2003;75:6314–6326. doi: 10.1021/ac0301806. [DOI] [PubMed] [Google Scholar]
- 33.Wu S-L, Kim J, Hancock WS, Karger BL. Extended Range Proteomic Analysis (ERPA): A New and Sensitive LC-MS Platform for High Sequence Coverage of Complex Proteins with Extensive Post-translational Modifications-Comprehensive Analysis of Beta-Casein and Epidermal Growth Factor Receptor (EGFR). J. Proteome Res. 2005;4:1155–1170. doi: 10.1021/pr050113n. [DOI] [PubMed] [Google Scholar]
- 34.Rorabacher DB. Statistical treatment for rejection of deviant values: critical values of Dixon's “Q“parameter and related subrange ratios at the 95% confidence level. Anal. Chem. 1991;63:139–146. [Google Scholar]
- 35. http://www.mcponline.org.
- 36.Nederkassel AM, Daszykowski M, Eilers PHC, Heyden YV. A comparison of three algorithms for chromatograms alignment. J. Chromatogr A. 2006;1118:199–210. doi: 10.1016/j.chroma.2006.03.114. [DOI] [PubMed] [Google Scholar]
- 37.Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching and identification. Anal.Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 38.Gu Y, Wu SW, Hancock W, Karger B, Meyer J, Hanlon D, Linder J, Burg L. ASMS. Seattle, WA: 2006. Proteomic Analysis of High-Grade Dysplastic Cells Obtained From ThinPrep Cervical Slides Using Laser Capture Microdissection and Mass Spectrometry. [DOI] [PubMed] [Google Scholar]