Abstract
Background/objectives
The polymer conjugate enhanced enzyme immunoassay (IDEIA) and Cobas Amplicor polymerase chain reaction Chlamydia trachomatis (CT) (Amplicor PCR) are two commonly used assays for the diagnosis of CT infection. The performance of these assays was compared for the diagnosis of genital CT infection among 1000 consecutive patients attending a genitourinary medicine (GUM) clinic. Confirmation of positive results and the clinical significance of the absence of cryptic plasmid in chlamydia on the diagnosis of infection by Amplicor PCR were also investigated.
Methods
IDEIA, Amplicor PCR, and two nested in‐house PCR assays targeting cryptic plasmid and omp1 gene were performed on all samples. DNA from Amplicor PCR negative samples was pooled for in‐house PCR assays. Each pool contained DNA from seven Amplicor PCR negative samples.
Results
Sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and efficiency of IDEIA in the diagnosis of genital CT infection were 80%, 97%, 80%, 97%, and 95%, respectively. Sensitivity, specificity, PPV, NPV and efficiency of Amplicor PCR were 99%, 98%, 89%, 100%, and 98%, respectively. 16 (11%) of 144 Amplicor PCR positive results were identified as false positive by in‐house PCR assays. No isolate of plasmid free CT was detected among the study population.
Conclusions
IDEIA should not be used for the diagnosis of CT infection because of its poor sensitivity. Although the analytic specificity of Amplicor PCR was 98%, because of the adverse medical, social, and psychological impact of false positive results for patients, confirmation of Amplicor PCR positive results by a different assay with comparable sensitivity is essential. Amplification assays targeting cryptic plasmid are appropriate for the diagnosis of genital CT infections.
Keywords: Chlamydia trachomatis , polymerase chain reaction, IDEIA PCE , cryptic plasmid, omp1 gene
The organism Chlamydia trachomatis (CT) is the most common sexually transmitted bacterium in the United Kingdom.1 The enzyme immunoassay (EIA) is a commonly used front line assay for the diagnosis of CT infection. EIA reactive results are confirmed mainly by nucleic acid amplification tests (NAATs). Amplicor PCR (polymerase chain reaction) and the polymer conjugate enhanced enzyme immunoassay (IDEIA) are two commonly used assays for the diagnosis of genital CT infection. IDEIA has a distinct characteristic, dual amplification of signal, to increase the sensitivity of the assay. The increased sensitivity of IDEIA had been reported when compared to conventional EIA.2 The sensitivity of IDEIA in comparison with ligase chain reaction (LCx Chlamydia, Abbott) was reported to be 92%.3 Two small studies, performed mainly on female commercial sex workers reported comparable sensitivity for IDEIA and Amplicor PCR.4,5 A recent review in this journal suggested further critical evaluation of IDEIA for the diagnosis of genital CT infection.6 This is the first large study on the comparative performance of IDEIA and Amplicor PCR in the diagnosis of CT infection among patients attending a genitourinary medicine (GUM) clinic.
CT has 7–10 copies of cryptic plasmid that are highly conserved in sequence and size.7 Many in‐house and commercial assays including Amplicor PCR have chosen it as a target for amplification to enhance the sensitivity of detection. A number of studies have described clinical8,9,10,11 and laboratory12 isolates of CT that lack cryptic plasmid, suggesting that it is not essential for the growth of the organism. The impact of this phenomenon on the diagnosis of CT infection was investigated in this study. This issue of confirmatory testing of positive results, generated by Amplicor PCR, was also investigated.
Methods
Patients
A total of 1000 consecutive patients attending the GUM clinic, from June to November 2003, at Addenbrooke's Hospital, Cambridge participated in this study. The study population comprised 437 males, median age 26 years (range 15–77) and 563 females, median age 23 years (range 15–58).
Sample collection and testing for Chlamydia trachomatis
This study was performed on routine diagnostic samples for genital CT infection. IDEIA chlamydia collection kit, S600730, was used for the collection of endocervical and urethral swabs from females and urethral swabs from males. Two swabs (endocervical and urethral) from every female were placed in a single tube and were treated as a single sample. A single urethral swab was analysed from each male patient. Both IDEIA and Amplicor PCR were performed on the same sample according to the manufacturers' protocols, DakoCytomation Ltd, Ely, UK and Roche Molecular Systems, Inc, CA, USA respectively. After completion of the diagnostic work, samples were anonymised, coded, and stored at −20°C for further quality assurance work. The Cambridge local research ethics committee approved this study.
DNA extraction
DNA for in‐house PCR was extracted from 200 μl of each sample using the MagNA Pure LC total nucleic acid isolation kit and MagNA Pure LC Robot according to the manufacturer's protocol (Roche Molecular Systems, Inc, CA, USA). DNA was concentrated during extraction and was eluted in 60 μl of elution buffer.
In‐house plasmid and MOMP nested PCR
Two in‐house nested PCR assays targeting cryptic plasmid (plasmid PCR) and omp1 gene (MOMP PCR) were performed on all samples. In all, 122 pools from 854 of 856 Amplicor PCR negative samples were prepared. The volume of a pool was 35 μl containing 5 μl of DNA from each of seven Amplicor PCR negative samples. The two remaining Amplicor PCR negative samples were processed separately. Individual in‐house plasmid and MOMP PCR assays were performed on all samples in a pool which gave a positive result and 144 Amplicor PCR positive samples.
Nested PCR
Two rounds of PCR were performed on each sample or pool of DNA in 50 μl volume. A volume of 5 μl of DNA from a single sample or 35 μl of DNA from each pool was used in the first round PCR. The reactions in the second round contained 2 μl of amplicons from the first round PCR. The amplification reactions also contained 200 μM of each of dNTP, 25 pmol of each primer, 4 mM MgCl2, 5 μl of 10× PCR buffer and 1.5 units of Taq DNA polymerase recombinant (Invitrogen life technologies, Paisley, UK). DNA from CT LGV‐II, strain 434 (ATCC VR‐902B) was used as a positive control and nuclease free water was used as a negative control for amplification. Six molecules of 100 bases long synthetic DNA and 2.5 pmol of forward and reverse primers to amplify this DNA were also added in the reaction to identify inhibition of amplification. Tenfold serial dilutions of CT positive control DNA were used to investigate the sensitivity of MOMP and plasmid PCR assays. Samples and controls were denatured at 95°C for 3 minutes, followed by 30 cycles of amplification in the first round and 25 cycles in the second round in a PTC‐200 Peltier Thermal Cycler (Genetic Research Instrumentation Ltd, Essex, UK). Each cycle consisted of denaturation at 95°C for 30 seconds, annealing of primers at 50°C for 1 minute, and extension at 72°C for 1 minute followed by final extension at 72°C for 7 minutes. The sequence of primers and internal control are shown in table 1. The size of amplicons was ascertained in reference to PCR Markers (Promega, Madison, USA) by gel electrophoresis using 3% agarose (NuSieve 3:1 Agarose, Bio Whittaker Molecular Applications, Rockland, ME, USA).
Table 1 Primers for plasmid and MOMP PCR.
| Sequence | Position* | Amplicon size (bps) | |
|---|---|---|---|
| First round MOMP PCR | |||
| Outer forward primer | 5′‐TTGTTTTCGACCGTGTTTTG‐3′ | 203 | |
| Outer reverse primer | 5′‐AGCRTATTGGAAAGAAGCBCCTAA‐3′ | 657 | 455 |
| Second round MOMP PCR | |||
| Inner forward primer | 5′‐AAACWGATGTGAATAAAGARTT‐3′ | 223 | |
| Inner reverse primer | 5′‐TCCCASARAGCTGCDCGAGC‐3′ | 617 | 395 |
| First round plasmid PCR | |||
| Outer forward primer | 5′‐TTGGCYGCTAGAAAAGGCGATT‐3′ | 7153 | |
| Outer reverse primer | 5′‐TCCGGAACAYATGATGCGAAGT‐3′ | 7365 | 212 |
| Second round plasmid PCR | |||
| Inner forward primer | 5′‐AACCAAGGTCGATGTGATAG‐3′ | 7179 | |
| Inner reverse primer | 5′‐TCAGATAATTGGCGATTCTT‐3′ | 7328 | 150 |
| DNA sequence of internal control | |||
| 5′‐GTGCTCACACCAGTTGCCGCGGAAAGTATGTGGAATGTTAACACACCCACACCACACCCACACACGTGTTGGATCAATTTCGAGATGCGAGCTGCCAAGC‐3′ | |||
| Forward primer for internal control | 5′‐GTGCTCACACCAGTTGCCGC‐3′ | ||
| Reverse primer for internal control | 5′‐GCTTGGCAGCTCGCATCTCG‐3′ | ||
Criteria for a true positive result
A positive or equivocal result by IDEIA was considered as a true positive if it was confirmed by two of the three PCR assays. A positive result by Amplicor PCR, plasmid PCR, or MOMP PCR was considered as a true positive if it was confirmed by two of the three PCR assays.
Results
IDEIA and Amplicor PCR
According to the manufacturer's criteria, 113 samples were positive, 15 samples were equivocal, and 872 were negative for chlamydia antigen by IDEIA; 144 samples were positive and 856 samples were negative for cryptic plasmid DNA by Amplicor PCR. Internal control was detected in all samples identified as negative by Amplicor PCR. All equivocal IDEIA results were grouped with IDEIA positive results to investigate the performance of this assay. The results are shown in table 2.
Table 2 Results of IDEIA, Amplicor PCR, and in‐house PCR assays.
| Results | Assays | |||
|---|---|---|---|---|
| IDEIA | Amplicor PCR | Plasmid PCR | MOMP PCR | |
| Positive | 128* | 144 | 140 | 117 |
| Negative | 872 | 856 | 860 | 883 |
| Total | 1000 | 1000 | 1000 | 1000 |
*15 of 128 results were equivocal.
In‐house nested plasmid and MOMP PCR
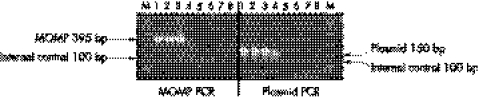
MOMP PCR was able to detect up to a single genome copy of CT in the amplification reaction. The sensitivity of plasmid PCR was 10‐fold higher than MOMP PCR (fig 1); 128 (89%) and 116 (81%) of 144 Amplicor PCR positive samples produced positive results with plasmid PCR and MOMP PCR, respectively. All 116 samples, which were positive by MOMP PCR, were also positive by plasmid PCR. One and seven of 122 pools from Amplicor PCR negative samples generated a signal with MOMP PCR and plasmid PCR respectively. Both MOMP and plasmid PCR detected one Amplicor PCR negative sample as positive. Plasmid PCR detected 13 Amplicor PCR and MOMP PCR negative samples as positive. The internal control was amplified in all pools. The results are shown in table 2.
Figure 1 Sensitivity of in‐house nested MOMP and plasmid PCR assays. M, PCR markers. Lanes 1–8, serial 10‐fold dilutions of C trachomatis DNA. Arrows indicate the size of amplicons.
According to the criteria described in the methods section, the results of IDEIA, Amplicor PCR, plasmid, and MOMP PCR are summarised in tables 3 and 4.
Table 3 Interpretation of results according to the criteria.
| Results | Assays | |||
|---|---|---|---|---|
| IDEIA | Amplicor PCR | Plasmid PCR | MOMP PCR | |
| True positive | 103 | 128 | 129 | 117 |
| True negative | 846 | 855 | 860 | 871 |
| False positive | 25 | 16 | 11 | 0 |
| False negative | 26 | 1 | 0 | 12 |
| Total | 1000 | 1000 | 1000 | 1000 |
Table 4 Performance of IDEIA, Amplicor PCR, and in‐house plasmid and MOMP PCR.
| Assay | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Efficiency (%) |
|---|---|---|---|---|---|
| IDEIA | 80 | 97 | 80 | 97 | 95 |
| Amplicor PCR | 99 | 98 | 89 | 100 | 98 |
| Plasmid PCR | 100 | 99 | 91 | 100 | 99 |
| MOMP PCR | 91 | 100 | 100 | 99 | 99 |
Discussion
The only two previously published reports to have compared the performance of IDEIA and Amplicor PCR for the diagnosis of genital CT infection reported comparable sensitivity.4,5 However, this study demonstrated that IDEIA is 20% less sensitive than Amplicor PCR. The study population in the previous two studies mainly comprised high risk commercial female sex workers and separate samples were collected in two different transport media for IDEIA and Amplicor PCR. This study was performed on 1000 consecutive patients attending a GUM clinic, on a single sample from each patient in only one type of transport medium for both assays, hence removing a number of confounding factors that may have affected the previous studies.
A number of studies have reported reproducibility problems with Amplicor PCR for the diagnosis of CT infection.13,14,15 Sixteen (11%) of 144 Amplicor PCR positive results were identified as false positive in this study. This finding is consistent with a previous study, which was performed on 733 endocervical swabs, reporting 13 (15%) of 87 Amplicor PCR positive results as false.16 Because of the significant number of false positive results by Amplicor PCR, the need for confirmatory testing on Amplicor PCR positive samples is evident. Data presented in this study suggest that the combination of Amplicor PCR on all samples and plasmid PCR on Amplicor PCR reactive samples is adequate to identify all false positive results without affecting the sensitivity of diagnosis. MOMP PCR did not have a significant impact on either sensitivity or specificity.
A number of studies have used chromosome based PCR assays to confirm the reactive results generated by Amplicor PCR, which is essentially a plasmid based PCR. These studies have reported discrepant results between these two types of PCR assays in the range of 6%–12%.17,18,19 MOMP PCR used in this study was 8% less sensitive than Amplicor PCR. With multiple copies of cryptic plasmid in CT, plasmid based PCR assays have shown up to 1000‐fold higher sensitivity in comparison with chromosome based PCR assays.20,21 Hence, chromosome based PCR assays should not be used to confirm the reactive results of plasmid based PCR assays. MOMP PCR, because of its nested format, detected up to a single genome copy/reaction, while sensitivity of plasmid PCR was 10‐fold higher than MOMP PCR. Twelve samples did not appear to have even a single copy of CT genome copy/reaction but were positive with both plasmid PCR and Amplicor PCR. These were regarded as true positives. The clinical significance of positive results generated by plasmid based PCR from samples, which do not have any chromosomal DNA of CT, needs further investigation.
IDEIA is not an appropriate test for the diagnosis of genital CT infection, because of its poor sensitivity in comparison to Amplicor PCR. Reactive results generated by Amplicor PCR should be confirmed by a different amplification assay. The sensitivity of the confirmatory assay should be comparable with the front line assay. Chromosomal based confirmatory assays for plasmid based PCR may lead to false negative confirmation. The absence of cryptic plasmid in CT was not identified in the population studied. The in‐house assays reported in this study are nested PCR with gel electrophoresis. The assays in this format are not suitable for large scale routine diagnostic use owing to risk of contamination and their laborious nature. However, in‐house real time PCR assays using closed systems will be useful to confirm positive results generated by commercial NAATs.
Acknowledgements
We thank DakoCytomation Ltd and Roche Molecular Systems, Inc for providing support for this study. The sponsors of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of this paper.
Contributors
HJ, CS, and CC participated in study planning and design; HJ, HS, and AA performed practical work and data analysis; HJ was the lead writer; and all authors provided revision and commented on the paper.
Abbreviations
CT - Chlamydia trachomatis
EIA - enzyme immunoassay
GUM - genitourinary medicine
IDEIA - polymer conjugate enhanced enzyme immunoassay
NAATs - nucleic acid amplification tests
NPV - negative predictive value
PCR - polymerase chain reaction
PPV - positive predictive value
References
- 1.Chief Medical Officer's Expert Advisory Group Main report of the CMO's Expert Advisory Group on Chlamydia trachomatis. London: Department of Health, 1998
- 2.Hirose T, Iwasawa A, Satoh T.et al Clinical study of the effectiveness of a dual amplified immunoassay (IDEIA PCE Chlamydia) for the diagnosis of male urethritis. Int J STD AIDS 19989414–417. [DOI] [PubMed] [Google Scholar]
- 3.Chernesky M, Jang D, Copes D.et al Comparison of a polymer conjugate‐enhanced enzyme immunoassay to ligase chain reaction for diagnosis of Chlamydia trachomatis in endocervical swabs. J Clin Microbiol 2002392306–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka M, Nakayama H, Yoshida H.et al Detection of Chlamydia trachomatis in vaginal specimens from female commercial sex workers using a new improved enzyme immunoassay. Sex Transm Dis 199874435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka M, Nakayama H, Sagiyama K.et al Evaluation of a new amplified enzyme immunoassay (EIA) for the detection of Chlamydia trachomatis in male urine, female endocervical swab, and patient obtained vaginal swab specimens. J Clin Pathol 200053350–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chernesky M A. Chlamydia trachomatis diagnostics. Sex Transm Infect 200278232–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas N S, Lusher M, Storey C C.et al Plasmid diversity in chlamydia. Microbiol 19971431847–1854. [DOI] [PubMed] [Google Scholar]
- 8.Peterson E M, Markoff B A, Schachter J.et al The 7.5‐Kb plasmid present in Chlamydia trachomatis is not essential for the growth of this microorganism. Plasmid 199023144–148. [DOI] [PubMed] [Google Scholar]
- 9.Qi A N, Radcliffe G, Vassallo R.et al Infection with a plasmid –free variant chlamydia related to Chlamydia trachomatis identified by using multiple assays for nucleic acid detection. J Clin Microbiol 1990302814–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stothard D A, Williams J A, Van Der Pol B.et al Identification of a Chlamydia trachomatis serovar E urogenital isolate which lacks the cryptic plasmid. Infect Immun 1998666010–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farencena A, Comanducci M, Donati M.et al Characterization of a new isolate of Chlamydia trachomatis which lacks the common plasmid and has properties of biovar trachoma. Infect Immun 1997652965–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumoto A, Izutsu H, Miyashita N.et al Plaque formation by and plaque cloning of Chlamydia trachomatis biovar trachoma. J Clin Microbiol 1998363013–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bauwens J E, Clark A M, Stamm E. Diagnosis of Chlamydia trachomatis endocervical infections by a commercial polymerase chain reaction. J Clin Microbiol 1993313023–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Barbeyrac B, Rodriguez P, Dutilh B.et al Detection of Chlamydia trachomatis by ligase chain reaction compared with polymerase chain reaction and cell culture in urogenital specimens. Genitourin Med 199571382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peterson E M, Darrow V, Blanding J.et al Reproducibility problems with the Amplicor PCR Chlamydia trachomatis test. J Clin Microbiol 199735957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wylie J L, Moses S, Babcock R.et al Comparative evaluation of Chlamydiazme, PACE 2, and AMP‐CT assays for detection of Chlamydia trachomatis in endocervical specimens. J Clin Microbiol 1998363488–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lister N A, Tabrizi S N, Fairley C K.et al Validation of Roche cobas Amplicor assay for detection of Chlamydia trachomatis in rectal and pharyngeal specimens by an omp1 PCR assay. J Clin Microbiol 200442239–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Der Pol B, Quinn T C, Gaydos C A.et al Multicenter evaluation of the Amplicor and automated Cobas Amplicor CT/NG tests for detection of Chlamydia trachomatis. J Clin Microbiol 2000381105–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vincelette J, Schirm J, Bogard M.et al Multicenter evaluation of the fully automated cobas Amplicor PCR test for detection of Chlamydia trachomatis in urogenital specimens. J Clin Microbiol 19993774–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ossewaarde J M, Rieffe M, Rozenberg‐Arska M.et al Development and clinical evaluation of a polymerase chain reaction test for detection of Chlamydia trachomatis. J Clin Microbiol 1992302122–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahony J B, Luinstra K E, Sellors J W.et al Comparison of plasmid‐ and chromosome‐based polymerase chain reaction assays for detecting Chlamydia trachomatis nucleic acids. J Clin Microbiol 1993311753–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]