Abstract
The development of DNA sequencing more than 30 years ago has profoundly impacted biological research. In the last couple of years, remarkable technological innovations have emerged that allow the direct and cost-effective sequencing of complex samples at unprecedented scale and speed. These next-generation technologies make it feasible to sequence not only static genomes, but also entire transcriptomes expressed under different conditions. These and other powerful applications of next-generation sequencing are rapidly revolutionizing the way genomic studies are carried out. Below, we provide a snapshot of these exciting new approaches to understanding the properties and functions of genomes. Given that sequencing-based assays may increasingly supersede microarray-based assays, we also compare and contrast data obtained from these distinct approaches.
Keywords: ChIP-Seq, high-throughput sequencing, massively parallel sequencing, microarray, RNA-Seq, transcriptome, yeast
Abbreviations: ChIP, chromatin immunoprecipitation; ChIP-on-chip, ChIP using microarrays; NRSF, neuron-restrictive silencer factor; STAT1, signal transducer and activator of transcription 1
Sequencing as never before
Just when the era of sequencing seemed to have passed its peak, technological breakthroughs are launching a new dawn with huge potential and broad applications that are already transforming biological research. Two pioneering papers reporting new sequencing developments in 2005 have provided the first glimpse of things to come [1,2]. The sequencing revolution is currently driven by three commercially available platforms: 454 (Roche), Genome Analyzer (Illumina/Solexa) and ABI-SOLiD (Applied Biosystems) [3,4]. The development of additional platforms is well under way. These new technologies are based on different principles than the classical Sanger-based method [5], and they are collectively referred to as either ‘next-generation’ sequencing, ‘high-throughput’ (or even ‘ultrahigh-throughput’) sequencing, ‘ultra-deep’ sequencing or ‘massively parallel’ sequencing. (Makes you wonder what terms they will come up with once even more powerful technologies become available…) These novel technologies apply distinct principles, resulting in differences in sequence read lengths and numbers, which may provide distinct advantages and disadvantages for different applications. All technologies have in common, however, that they generate sequences on an unprecedented scale, without the requirement for DNA cloning, and at a fraction of the costs required for traditional sequencing. These features are the basis for the current revolution and provide the inspiration to apply sequencing approaches to biological questions that would not have been economically or logistically practical before. Next-generation sequencing should also democratize science in the sense that ambitious sequencing-based projects can now be tackled by individual laboratories or institutes, whereas before such projects would only have been possible in genome centres.
From genomes to function
Early studies applied next-generation sequencing to sampling microbial diversities in a deep mine and in oceans [6–8], launching the field of ‘meta-genomics’ where entire biological communities are sequenced, en masse, to survey the variety of all organisms living together in particular ecosystems [9–13]. Other interesting applications are in the field of ancient DNA research, where next-generation sequencing has been successfully applied to analyse genomes of woolly mammoths [14] and Neanderthals [15,16]. Naturally, next-generation sequencing is also used to decode modern genomes, from bacteria [17,18] and viral isolates [19,20] to James Watson [21]. The latter example illustrates that the power of next-generation sequencing is increasingly exploited to re-sequence strains and individuals for which reference genome sequences are available to sample genomic diversity. Such studies have identified mutations in bacterial strains [22,23], polymorphisms in worm [24], structural variation in the human genome [25] and specific alleles involved in cancer [26].
In addition to established analyses of genome sequences, next-generation sequencing is triggering new assays and applications that should greatly advance our understanding of genome function (Figure 1) [27]. The principle behind these alternative applications, which have been termed ‘sequence census’ methods, is simple: complex DNA or RNA samples are directly sequenced to determine their content. With reference genomes available, short sequence reads are sufficient to map their locations (except for repeated regions), and once mapped, millions of sequence hits are simply counted to determine their genomic distribution (Figure 2). This concept is based on previous approaches such as serial analysis of gene expression [28] and massively parallel signature sequencing [29]. Next-generation sequencing, however, delivers much more information at affordable costs, and it is easy to implement for a wider range of applications. Below, we will survey initial studies that analyse genome function exploiting sequence census methods, which will increasingly supersede microarray-based approaches (Figure 1).
Figure 1. Sequence census technologies have added a new dimension to the analysis of gene expression regulation, which has been dominated by hybridization-based methods.
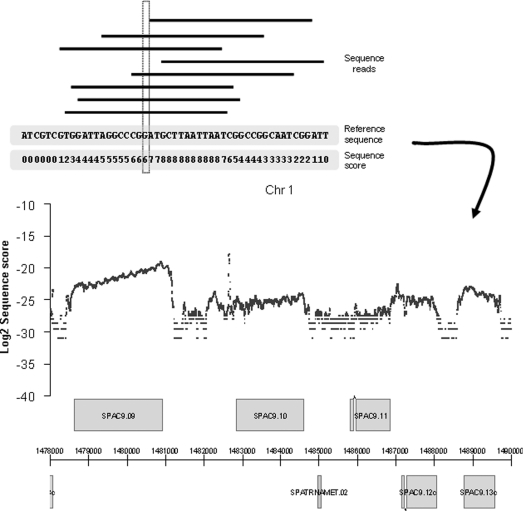
Figure 2. Sequence census data applied to cDNA allow genome-wide measurements of transcript levels (RNA-Seq).
The sequence score defines the number of times each base of the reference genome sequence is hit by a sequence read (top panel). Sequence scores (based on normalized read numbers) are then plotted along the genome (bottom panel). Based on data from our fission yeast transcriptome analysis [48].
Mapping of DNA-binding proteins and chromatin
ChIP-on-chip [ChIP (chromatin immunoprecipitation) using microarrays] [30,31] is a key approach to globally mapping the in vivo binding sites of various DNA-binding proteins across genomes. Instead of using the DNA that is precipitated with the protein of interest to interrogate microarrays, recent studies have directly sequenced this DNA to analyse the protein-binding sites at high resolution. This approach, termed ‘ChIP-Seq’, should produce a huge windfall, in particular for studies in multicellular eukaryotes where whole genome coverage has generally required the use of several arrays.
Initial studies looking at the binding sites of human NRSF (neuron-restrictive silencer factor) and STAT1 (signal transducer and activator of transcription 1) [32,33] indicate that the resolution of ChIP-Seq is far better than that of ChIP-on-chip. NRSF, a well-documented zinc-finger repressor that negatively regulates gene expression of neuronal genes in non-neuronal cells, has >80 previously validated targets, providing a well-defined test set to define false-positive and -negative rates [32]. The vast majority of previously known target sites have been confirmed among the ∼2000 targets identified through ChIP-Seq. Moreover, this analysis has exploited the deep sampling and high resolution of ChIP-Seq to identify a novel class of genomic NRSF-binding sites, suggesting the existence of different subclasses of genes regulated by the same factor [32]. Another ChIP-Seq study has mapped the binding sites of STAT1, a transcription factor that regulates genes involved in cell differentiation, survival and proliferation [33]. The dynamic behaviour of STAT1 is of interest as it usually localizes in the cytoplasm, but translocates to the nucleus on stimulation by an extracellular signal. As expected, the authors have observed a large increase in STAT1 binding sites after stimulation of cells with interferon-γ (from 11000 to 41000), and the results also agree well with previously published data.
Approaches to map the genomic protein-binding sites are not limited to transcription factors. One of the first papers demonstrating the utility of ChIP-Seq for whole-genome location analysis [34] mapped the genome-wide sites of 20 histone methylation marks, along with CTCF (CCCTC-binding factor), the histone variant H2A.Z and RNA polymerase II in human cells. The unprecedented detail of these data has led to several valuable conclusions about the association of specific sets of histone modifications with either active or repressed promoters. Such comprehensive and predictive patterns can be used not only to confirm annotated promoters but also to identify new ones [34]. Another comprehensive survey of two types of histone modifications has been reported for pluripotent and lineage-committed mouse cells, revealing how these modifications change during development [35].
In another adaptation of array-based methods, next-generation sequencing has also been applied to map regions with few or no chromatin proteins, namely DNase-hypersensitive sites in human cell lines to identify locations with regulatory elements [36]. Using this approach, ∼95000 DNase-hypersensitive sites have been uncovered, a surprising majority (∼80%) of which are not associated with promoter regions. This finding strongly suggests that the genome is replete with regions of open DNA, many of which may have unrecognized roles in genome function [36].
Deep sampling of transcriptomes
Next-generation sequencing is also changing the ways in which gene expression is studied, which is likely to have much future impact. Complex RNA mixtures can be analysed using sequence census methods, an approach termed ‘RNA-Seq’. Initial applications include the accelerated discovery of small RNAs [37–40]. Other studies have used early RNA-Seq approaches to quantify expression levels using a modified paired-end ditagging method [41], to detect rare cardiac mRNAs in mouse by ‘polony multiplex analysis of gene expression’ [42,43], or to directly sequence cDNAs of human tumour and fly cells [44–46]. Another RNA-Seq study has moved beyond simply describing the expression levels of transcripts towards assigning functions to the observed expression differences [47]. Together, such studies pave the way for complete transcriptome coverage, providing the ultimate resolution to analyse the levels as well as the structures of both processed and unprocessed transcripts under different conditions.
We have recently applied RNA-Seq, complemented with high-resolution tiling arrays, to obtain a detailed picture of the fission yeast transcriptome, independently of available gene annotations, at the best possible resolution [48]. The transcriptome has been interrogated under multiple conditions, including rapid proliferation, meiotic differentiation and environmental stress, as well as in splicing and exosome mutants, to analyse the dynamic adaptation of the transcriptional landscape as a function of environmental, developmental and genetic factors. These results provide rich, condition-specific information on widespread transcription, on novel, mostly non-coding transcripts, as well as on untranslated regions and gene structures, thus improving the existing genome annotation. Perhaps most interestingly, sequence reads spanning exon–exon or exon–intron junctions have given a unique and direct insight into a surprising variability in splicing efficiency across introns, genes and conditions. This analysis has revealed that splicing efficiency is largely co-ordinated with transcript levels, and hundreds of introns show regulated splicing during cellular proliferation or differentiation. These results suggest a global co-ordination between splicing efficiency and transcription, which may help to optimize and streamline gene expression programmes. As elaborated in the next section, the combined RNA-Seq and array data have also allowed comparing and contrasting of the relative performance and properties of sequencing- and hybridization-based approaches.
Next-generation sequencing compared with microarrays
Currently, global analysis of gene expression relies largely on hybridization-based platforms such as microarrays, which are routinely used for determining relative expression levels or changes in gene expression between different biological conditions. Unlike hybridization data, which consist of continuous signals, sequence census data are made of absolute numbers of reads (Figure 2). The countable, almost digital, nature of these results makes them highly suitable for the analysis of gene expression levels. Applying sequence census approaches to cDNAs (RNA-Seq), we can therefore estimate the relative abundance of given transcripts by counting the number of times they are hit by sequence reads. Recent studies have shown that, indeed, scores based on the number of sequence reads hitting a transcript, or on the average number of hits per base and per transcript, provide accurate measurements of relative RNA levels [45,46,48]. We have shown that sequencing-based estimates of transcript abundance are in good agreement with estimates acquired with microarrays, provided that sequencing depth is sufficient [48]. In addition, unlike microarray data, which are affected by the dynamic range of the scanner, sequence data have a linear dynamic range only limited by the sequencing depth. This aspect is attractive, because the dynamic range of different RNAs in a cell is almost certainly larger than the range provided by microarray scanners. In addition, and unlike hybridization-based techniques, sequencing-based approaches produce little or no noise, allowing detection of even very minimally expressed transcripts [48]. In the short term, however, the costs and amount of data produced make it unlikely that sequence census approaches will completely replace microarrays as the routine tool for expression profiling.
The structure of eukaryotic transcriptomes has received considerable attention with the availability of high-density tiling arrays [49]. These arrays consist of probes tiled evenly across the genome allowing characterization of transcript structure without prior knowledge of genome annotation. Sequence census methods are likely to provide an exciting new twist in the structural analysis of transcriptomes. Unlike tiling arrays whose resolution is limited by the number of probes on the platform, sequencing provides, by default, the best possible resolution. This feature may prove particularly powerful for dense genomes with small gene features or for large genomes that would otherwise require a substantial number of tiling arrays to provide adequate resolution. In addition, sequencing of the fission yeast transcriptome has proved sensitive enough to detect widespread transcription in >90% of the genome, including traces of RNAs that are not robustly transcribed or rapidly degraded [48], such that very rare isoforms expressed at levels below the detection threshold of tiling arrays can be identified. However, the sequencing costs incurred to reach a sufficiently deep coverage of large genomes remain currently an issue. Fortunately, the costs are likely to decrease further, allowing a wider and more extensive use of these technologies.
Sequence census technologies have been developed to analyse double-stranded DNA. When it comes to the analysis of transcriptomes, converting RNA into double-stranded cDNA is required, which may, in many protocols, result in the loss of strand-specific information. Given the large extent of overlapping and antisense transcription reported even in simple eukaryotes, this is clearly an issue. Moreover, classical protocols for cDNA synthesis do not produce samples allowing unambiguous detection of overlapping transcription. Therefore, in addition to classical cDNA analysis, techniques allowing the specific analysis of the 5′- and 3′-ends of transcripts combined with sequencing census approaches should prove useful for unambiguously determining the extent and structure of different transcripts.
Finally, a unique feature of sequence census technologies is their ability to identify, without prior knowledge, transcripts made of sequences that are not adjacent in the genome but that are connected when they are expressed. For instance, spliced transcripts can be uniquely detected through the presence of sequence reads spanning exon–exon junctions. Such positive evidence for splicing is not available from tiling array data, and although spliced transcripts could be probed with specially designed arrays, it would require a priori knowledge of the splice sites. For these reasons, sequence census technologies will provide powerful and versatile tools to study post-transcriptional processing of genetic sequences.
Concluding remarks
Next-generation sequencing technologies have had an enormous impact on research within a short time frame, and this impact appears certain to increase further, as many institutions are now acquiring these prevailing new sequencing platforms. Beyond conventional sampling of genome content, wide-ranging applications are rapidly evolving for next-generation sequencing. Sequence census methods such as ChIP-Seq and RNA-Seq are becoming powerful and quantitative approaches to analyse the structures and functions of both genomes and transcriptomes at maximal resolution. At this time, the huge amount of data generated by next-generation sequencing creates an informatics challenge. The establishment of routine data analysis methods, together with future decreases in sequencing costs and increases in the numbers and lengths of sequence reads, will help to unleash the full potential of next-generation sequencing.
Note added in proof (received 4 August 2008)
Since submission of this paper, six additional papers have been published reporting various applications of RNA-Seq [50–55].
Acknowledgments
We thank Josette-Renée Landry, Vera Pancaldi and Falk Schubert for comments on this paper. S.M. was supported by a Fellowship for Advanced Researchers from the Swiss National Science Foundation, and B.T.W. by Sanger postdoctoral and Canadian NSERC (Natural Sciences and Engineering Research Council) fellowships. Work in our laboratory is funded by Cancer Research UK grant number C9546/A6517.
References
- 1.Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.-J., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shendure J., Porreca G.J., Reppas N.B., Lin X., McCutcheon J.P., Rosenbaum A.M., Wang M.D., Zhang K., Mitra R.D., Church G.M. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 3.Rusk N., Kiermer V. Primer: sequencing: the next generation. Nat. Methods. 2008;5:15. doi: 10.1038/nmeth1155. [DOI] [PubMed] [Google Scholar]
- 4.Schuster S.C. Next-generation sequencing transforms today's biology. Nat. Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- 5.Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angly F.E., Felts B., Breitbart M., Salamon P., Edwards R.A., Carlson C., Chan A.M., Haynes M., Kelley S., Liu H., et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards R.A., Rodriguez-Brito B., Wegley L., Haynes M., Breitbart M., Peterson D.M., Saar M.O., Alexander S., Alexander E.C., Jr, Rohwer F. Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics. 2006;7:57. doi: 10.1186/1471-2164-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sogin M.L., Morrison H.G., Huber J.A., Mark Welch D., Huse S.M., Neal P.R., Arrieta J.M., Herndl G.J. Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox-Foster D.L., Conlan S., Holmes E.C., Palacios G., Evans J.D., Moran N.A., Quan P.L., Briese T., Hornig M., Geiser D.M., et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318:283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 10.Huber J.A., Mark Welch D.B., Morrison H.G., Huse S.M., Neal P.R., Butterfield D.A., Sogin M.L. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- 11.Tringe S.G., Rubin E.M. Metagenomics: DNA sequencing of environmental samples. Nat. Rev. Genet. 2005;6:805–814. doi: 10.1038/nrg1709. [DOI] [PubMed] [Google Scholar]
- 12.Warnecke F., Luginbühl P., Ivanova N., Ghassemian M., Richardson T.H., Stege J.T., Cayouette M., McHardy A.C., Djordjevic G., Aboushadi N., et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 13.Woyke T., Teeling H., Ivanova N.N., Huntemann M., Richter M., Gloeckner F.O., Boffelli D., Anderson I.J., Barry K.W., Shapiro H.J., et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature. 2006;443:950–955. doi: 10.1038/nature05192. [DOI] [PubMed] [Google Scholar]
- 14.Poinar H.N., Schwarz C., Qi J., Shapiro B., Macphee R.D., Buigues B., Tikhonov A., Huson D.H., Tomsho L.P., Auch A., et al. Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- 15.Green R.E., Krause J., Ptak S.E., Briggs A.W., Ronan M.T., Simons J.F., Du L., Egholm M., Rothberg J.M., Paunovic M., Paabo S. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- 16.Noonan J.P., Coop G., Kudaravalli S., Smith D., Krause J., Alessi J., Chen F., Platt D., Paabo S., Pritchard J.K., Rubin E.M. Sequencing and analysis of Neanderthal genomic DNA. Science. 2006;314:1113–1118. doi: 10.1126/science.1131412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofreuter D., Tsai J., Watson R.O., Novik V., Altman B., Benitez M., Clark C., Perbost C., Jarvie T., Du L., Galan J.E. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 2006;74:4694–4707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh J.D., Kling-Backhed H., Giannakis M., Xu J., Fulton R.S., Fulton L.A., Cordum H.S., Wang C., Elliott G., Edwards J., et al. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc. Natl. Acad. Sci. U.S.A. 2006;103:9999–10004. doi: 10.1073/pnas.0603784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann C., Minkah N., Leipzig J., Wang G., Arens M.Q., Tebas P., Bushman F.D. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 2007;35:e91. doi: 10.1093/nar/gkm435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C., Mitsuya Y., Gharizadeh B., Ronaghi M., Shafer R.W. Characterization of mutation spectra with ultra-deep pyrosequencing: application to HIV-1 drug resistance. Genome Res. 2007;17:1195–1201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wheeler D.A., Srinivasan M., Egholm M., Shen Y., Chen L., McGuire A., He W., Chen Y.-J., Makhijani V., Roth G.T., et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 22.Andries K., Verhasselt P., Guillemont J., Gohlmann H.W., Neefs J.M., Winkler H., Van Gestel J., Timmerman P., Zhu M., Lee E., et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 23.Velicer G.J., Raddatz G., Keller H., Deiss S., Lanz C., Dinkelacker I., Schuster S.C. Comprehensive mutation identification in an evolved bacterial cooperator and its cheating ancestor. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8107–8112. doi: 10.1073/pnas.0510740103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hillier L.W., Marth G.T., Quinlan A.R., Dooling D., Fewell G., Barnett D., Fox P., Glasscock J.I., Hickenbotham M., Huang W., et al. Whole-genome sequencing and variant discovery in C. elegans. Nat. Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 25.Korbel J.O. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas R.K., Baker A.C., Debiasi R.M., Winckler W., Laframboise T., Lin W.M., Wang M., Feng W., Zander T., Macconnaill L.E., et al. High-throughput oncogene mutation profiling in human cancer. Nat. Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 27.Wold B., Myers R.M. Sequence census methods for functional genomics. Nat. Methods. 2008;5:19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- 28.Velculescu V.E., Zhang L., Vogelstein B., Kinzler K.W. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 29.Brenner S., Johnson M., Bridgham J., Golda G., Lloyd D.H., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 30.Iyer V.R., Horak C.E., Scafe C.S., Botstein D., Snyder M., Brown P.O. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 31.Ren B., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 32.Johnson D.S., Mortazavi A., Myers R.M., Wold B. Genome-wide mapping of in vivo protein–DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 33.Robertson G., Hirst M., Bainbridge M., Bilenky M., Zhao Y., Zeng T., Euskirchen G., Bernier B., Varhol R., Delaney A., et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 34.Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyle A.P., Davis S., Shulha H.P., Meltzer P., Margulies E.H., Weng Z., Furey T.S., Crawford G.E. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu C., Kulkarni K., Souret F.F., MuthuValliappan R., Tej S.S., Poethig R.S., Henderson I.R., Jacobsen S.E., Wang W., Green P.J., Meyers B.C. MicroRNAs and other small RNAs enriched in the Arabidopsis RNA-dependent RNA polymerase-2 mutant. Genome Res. 2006;16:1276–1288. doi: 10.1101/gr.5530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruby J.G., Jan C., Player C., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 39.Stark A., Bushati N., Jan C.H., Kheradpour P., Hodges E., Brennecke J., Bartel D.P., Cohen S.M., Kellis M. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. 2008;22:8–13. doi: 10.1101/gad.1613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyler D.M., Okamura K., Chung W.J., Hagen J.W., Berezikov E., Hannon G.J., Lai E.C. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 2008;22:26–36. doi: 10.1101/gad.1615208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng P., Tan J.J., Ooi H.S., Lee Y.L., Chiu K.P., Fullwood M.J., Srinivasan K.G., Perbost C., Du L., Sung W.K., et al. Multiplex sequencing of paired-end ditags (MS-PET): a strategy for the ultra-high-throughput analysis of transcriptomes and genomes. Nucleic Acids Res. 2006;34:e84. doi: 10.1093/nar/gkl444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J.B., Porreca G.J., Song L., Greenway S.C., Gorham J.M., Church G.M., Seidman C.E., Seidman J.G. Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science. 2007;316:1481–1484. doi: 10.1126/science.1137325. [DOI] [PubMed] [Google Scholar]
- 43.Velculescu V.E., Kinzler K.W. Gene expression analysis goes digital. Nat. Biotechnol. 2007;25:878–880. doi: 10.1038/nbt0807-878. [DOI] [PubMed] [Google Scholar]
- 44.Bainbridge M.N., Warren R.L., Hirst M., Romanuik T., Zeng T., Go A., Delaney A., Griffith M., Hickenbotham M., Magrini V., et al. Analysis of the prostate cancer cell line LNCaP transcriptome using a sequencing-by-synthesis approach. BMC Genomics. 2006;7:246. doi: 10.1186/1471-2164-7-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugarbaker D.J., Richards W.G., Gordon G.J., Dong L., De Rienzo A., Maulik G., Glickman J.N., Chirieac L.R., Hartman M.L., Taillon B.E., et al. Transcriptome sequencing of malignant pleural mesothelioma tumors. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3521–3526. doi: 10.1073/pnas.0712399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torres T.T., Metta M., Ottenwalder B., Schlotterer C. Gene expression profiling by massively parallel sequencing. Genome Res. 2008;18:172–177. doi: 10.1101/gr.6984908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toth A.L., Varala K., Newman T.C., Miguez F.E., Hutchison S.K., Willoughby D.A., Simons J.F., Egholm M., Hunt J.H., Hudson M.E., Robinson G.E. Wasp gene expression supports an evolutionary link between maternal behavior and eusociality. Science. 2007;318:441–444. doi: 10.1126/science.1146647. [DOI] [PubMed] [Google Scholar]
- 48.Wilhelm B.T., Marguerat S., Watt S., Schubert F., Wood V., Goodhead I., Penkett C.J., Rogers J., Bähler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 49.Kapranov P., Willingham A.T., Gingeras T.R. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 50.Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lister R., O'Malley R.C., Tonti-Filippini J., Gregory B.D., Berry C.C., Millar A.H., Ecker J.R. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 53.Cloonan N., Forrest A.R., Kolle G., Gardiner B.B., Faulkner G.J., Brown M.K., Taylor D.F., Steptoe A.L., Wani S., Bethel G., et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 54.Morin R., Bainbridge M., Fejes A., Hirst M., Krzywinski M., Pugh T., McDonald H., Varhol R., Jones S., Marra M. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Bio Techniques. 2008;45:81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- 55.Sultan M., Schulz M.H., Richard H., Magen A., Klingenholff A., Scherf M., Seifert M., Borodina T., Soldatov A., Parkhomchuk D., et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]