Abstract
Major bottlenecks in systems biology studies arise from limitations of current sample preparation techniques. Multiple mutually exclusive sample preparation methods, which are often required to extract distinct classes of molecules from cells and tissues, are incompatible with studies of precious or very limited samples. Moreover, the strong detergents and chaotropic agents commonly required to solubilize sample constituents often interfere with subsequent separation and analysis. Here we describe a rapid, detergent-free sample preparation technique that allows efficient concurrent isolation and fractionation of protein, DNA, RNA, and lipids from biological samples, eliminating the need for multiple replicates. The method relies on a synergistic combination of physical disruption of the cellular material by hydrostatic pressure (pressure cycling technology) and novel extraction conditions to dissolve and partition distinct classes of molecules into separate fractions. We demonstrate parallel recovery of proteins, lipids, and intact DNA and RNA, from animal cells and tissues, for proteomic, lipidomic, and genomic analyses. The protein extracts require minimal cleanup and are compatible with 1D and 2D PAGE, liquid chromatography coupled with tandem mass spectrometry, and Western blotting. The lipid fractions have been profiled by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry without further processing. The isolated DNA and RNA were shown to be intact by agarose gel visualization, and the presence of intact mRNA was confirmed by real time reverse transcription polymerase chain reaction. Analysis and comparison of samples extracted using this method and a more traditional extraction technique revealed several protein species preferentially extracted by the new method.
Keywords: : Proteomics, genomics, metabolomics, lipidomics, sample preparation, pressure cycling technology (PCT), proteins, DNA, RNA, lipids, ProteoSolve-LRS
Systems biology studies are gaining momentum, driven by recent successes in genomics, transcription profiling, proteomics, and rapidly emerging metabolomic technologies, including shotgun lipidomics and profiling of small molecule metabolites. While powerful and sensitive methods are available for the analysis of nucleic acids, proteins, and small molecules, major bottlenecks arise from the limitations of current sample preparation techniques which frequently require the utilization of the entire sample for isolation of only one class of analytes. When limited quantity of sample is available and parallel sampling is not possible, attempts to correlate gene expression with protein data and metabolic status of the model typically fail.
Most previously reported methods of simultaneous isolation of DNA, RNA, and protein were originally developed for isolation of nucleic acid, and subsequently adapted to capture protein fractions. The most popular technique to date is the original phenol-chloroform extraction method published by Chomczynsky and Sacchi in 19871,2 and subsequently improved by several groups.3–7,8,9 The emerging field of lipidomics imposes new requirements on tissue sample preparation.10–13 For example, studies of lipid peroxidation require rapid and careful isolation of lipids to avoid artifacts caused by oxidation and breakdown of the sample components. Similar requirements have always existed for sample preparation steps preceding the analysis of redox-active small molecule messengers such as neurotransmitters and vitamins.14,15
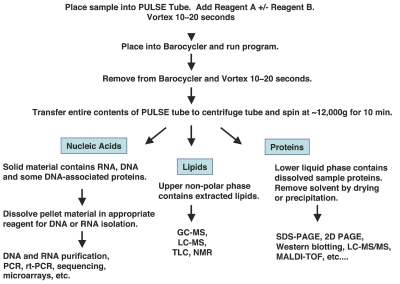
We have developed a detergent-free sample preparation technique that allows concurrent isolation and fractionation of proteins, DNA, RNA, and lipids from cells and tissues (Figure 1). This novel method takes advantage of a synergistic combination of cell disruption by alternating hydrostatic pressure (pressure cycling technology; PCT) and a carefully chosen reagent system that dissolves and partitions distinct classes of molecules into separate fractions.
Figure 1.
Pressure-mediated extraction with ProteoSolve-SB, workflow. Extraction of cells and tissues by alternating hydrostatic pressure in optimized organic solvent-based reagent systems (fluorinated alcohols and aliphatic hydrocarbons) leads to sample dissolution and fractionation into two liquid phases and an insoluble pellet. Three fractions contain lipids, proteins together with other polar molecules, and nucleic acids, respectively. Further purification may not be necessary for the downstream analysis of resulting fractions by popular analytical methods.
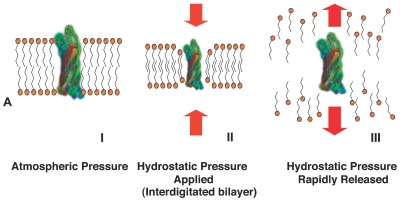
PCT destabilizes intermolecular interactions by rapidly and repeatedly raising and lowering pressure in the reaction vessel from ambient to high levels (up to 35,000 psi [240 MPa]).16,17 High hydrostatic pressure acts preferentially on the compressible constituents of the sample. Lipids, as the most compressible sample components, are affected most, and dissociate upon depressurization (Figure 2A). Thus, selective energy distribution results in destabilization of molecular interactions in the lipid bilayers and other cellular components, but not in the disruption of covalent bonds.
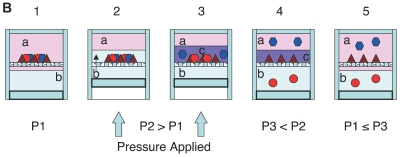
Figure 2.
A: Representation of PCT-mediated extraction: Rapidly cycling hydrostatic pressure leads to destabilization of molecular interactions. I: At equilibrium transmembrane proteins reside in the lipid bilayer. II: When high hydrostatic pressure is applied, the membrane lipids compress forming an interdigitated state. III: upon rapid depressurization molecular interactions are destabilized and sample components are solubilized. B: PCT-assisted liquid-liquid extraction. 1: Solvents a and b are placed in a PuLSE tube with sample. At atmospheric pressure (P1) solvents a and b are immiscible. 2: Hydrostatic pressure is applied and causes compression of the liquid and the solid sample and mixing of components. 3: High hydrostatic pressure (P2) alters the mutual solubility of the solvents leading to the formation of a temporary solvent c with properties of solvents a and b, and to the dissolution of the sample. 4: As the mixture depressurizes (P3) it expands, resulting in reseparation of solvents a and b and the partitioning of the sample components between the two solvents according to their solubility at atmospheric pressure. 5: The system returns to its original equilibrium at pressure P1. The extracted sample components are now fractionated into different phases.
PCT-assisted liquid-liquid extraction (patent pending) uses high hydrostatic pressure to alter solvation energy and solubility of various compounds. Several liquids, which are poorly miscible at atmospheric pressure, interact under high pressure in such a way that the phase boundary presents less of a barrier for partitioning of molecules between solvent phases. As a result, partitioning occurs in the entire volume of the vessel, rather than just at the interface (Figure 2B).
Previously we reported the performance of the PCT-driven detergent-free protein isolation system,18,19 which laid the foundation for the development of the Proteo-Solve-LRS reagent kit, currently available from Pressure Biosciences. Proteins and lipids are solubilized under pressure and are maintained in solution by amphipathic organic solvents, such as fluorinated alcohols20,21 (e.g., hexafluoroisopropanol, a key ingredient of reagent A in the ProteoSolve-SB kit). Nucleic acids do not remain in solution after depressurization. Combined treatment by hydrostatic pressure and amphipathic solvent systems results in rapid cell disruption, dissolution of lipids, and dissolution and denaturation of proteins.
In the current study we report the expansion of the ProteoSolve-LRS strategy, and the development of the new ProteoSolve-SB kit for rapid simultaneous isolation of DNA, RNA, proteins, and lipids from biological samples. Gel electrophoresis and real-time reverse transcription polymerase chain reaction (RT-PCR) confirm that high recoveries of intact genomic DNA and RNA are obtained using this novel technique. Additionally, high yields of proteins and lipids are obtained from the same sample for proteomic and lipidomic analyses. Due to the unique conditions that favor the extraction of more lipophilic proteins, several protein species uniquely extracted by the new method have been identified by in-gel tryptic digestion and liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). The lipids extracted by this new method can be subjected to direct analysis using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) without additional cleanup and separation steps such as chromatography or enzymatic digestion. Further development of this analytical strategy will enable rapid profiling of the lipid complement of samples and potential structure elucidation by tandem mass spectrometry.
MATERIALS AND METHODS
ProteoSolve-LRS and ProteoSolve-SB kits were from Pressure BioSciences (South Easton, MA); Trizol was from Invitrogen (Carlsbad, CA); precast Reliant Gel System RNA and DNA mini-gels were from Lonza (Rock-land, ME); SDS-PAGE Criterion 4–12% and 8–16% gradient gels were from Bio-Rad (Hercules, CA); AllPrep kit, RNeasy Mini kit, and DNeasy Blood and Tissue kit were from Qiagen (Valencia, CA); and PARIS kit was from Ambion/Applied Biosystems (Austin, TX). Chemicals were purchased from Sigma (St Louis, MO) or VWR International (West Chester, PA).
PC12 rat pheochromocytoma and human fibroblast (HF) cell cultures were kindly provided by Dr. Susan Doctrow (Proteome Systems, Woburn, MA), and mouse adipose tissue was kindly provided by Dr. Haiming Cao (Harvard School of Public Health, Boston, MA)
Cell and Tissue Disruption by Pressure Cycling
Twenty pressure cycles were applied to each sample using a Barocycler (model NEP3229 or NEP2320, Pressure BioSciences, South Easton MA.). Each pressure cycle consisted of 20 sec at high pressure (35,000 psi) followed by 20 sec at low (atmospheric) pressure.22
Protein, lipid, RNA, and DNA extraction used the modified ProteoSolve-SB kit protocol: Unhomogenized fragments of tissue were combined in a PULSE tube with 0.9–1.1 mL ProteoSolve-SB reagent A and brought to 1.4 mL using reagent B. For cell culture, 1–5 × 106 pelleted cells were suspended in 0.9–1.0 mL ProteoSolve-SB reagent A, transferred to PULSE tubes and brought up to 1.4 mL with reagent B. All samples were vortexed briefly before and after pressure cycling.
After pressure cycling, samples were transferred to centrifuge tubes and centrifuged for 15 min at 12,000 × g to promote phase separation. The lipid-containing upper phase was transferred to a clean tube for subsequent lipid analysis by MALDI-TOF. The protein-containing lower phase was transferred to a clean tube for protein analysis. For 1D SDS-PAGE, aliquots of protein lysate were dried by centrifugation under vacuum and dissolved in Laemmli sample buffer at 60–65°C to facilitate dissolution. For 2D PAGE, aliquots of protein lysate were dried by centrifugation under vacuum and dissolved in the isoelectric focusing (IEF buffer), containing 7 M urea, 2 M thiourea, and 4% CHAPS.
The pellet and any solid interface layer, containing the bulk of the sample’s DNA and RNA, were then processed for nucleic acid extraction. Nucleic acid recovery was measured with a Qubit Fluorometer (Invitrogen), using the Quant-iT RNA assay kit for quantification of RNA and the Quant-iT dsDNA BR assay kit for quantification of DNA.
For protein re-extraction from the lipid phase, 350 mg of bovine adipose tissue was processed by PCT in 1.05 mL reagent A without reagent B. After extraction and centrifugation, the solvent phase was removed, the lipid phase was transferred to a clean test tube, and the pellet and interface were pooled. The pellet/interface fraction and the lipid fraction were then separately re-extracted with fresh reagent A and centrifuged for 10 min at 12,000 × g. The solvent was removed by evaporation under vacuum, and the resulting samples were dissolved in Laemmli sample buffer and subjected to SDS-PAGE.
Protein Electrophoresis, Image Analysis, and In-Gel Digestion
SDS PAGE was performed on 4–12% polyacrylamide gradient gels. For 2D-PAGE separation, the simultaneous reduction and alkylation by tributylphosphine/ acrylamide23 was employed. Immobilized pH gradient strips pH 3–10 were hydrated with samples for 6 h, followed by IEF for 100,000 volt-h at 10,000 V. All precast electrophoresis supplies and Criterion vertical gel electrophoresis system were from Bio-Rad Laboratories, while the IsoelectrIQ2 integrated IEF instrument was from Proteome Systems (Woburn, MA). Gels were stained with colloidal CBB or SYPRO Ruby,24 scanned, and analyzed with PDQuest software to determine statistically significant differentially extracted proteins. Selected gel spots were excised and processed using a conventional in-gel digestion protocol.25 Sequencing grade modified porcine trypsin (Promega, Madison, WI) was used for digestion.
Protein Identification by Nano-LC Coupled to MS/MS
Protein digests (5–10 μL) were separated using a C18 solid phase extraction trapping column (300 μm i.d. × 5 mm; Dionex, Sunnyvale, CA) and a 100-μm i.d. × 12 cm nano-LC reversed-phase self-packed fused silica column (PicoFrit, pulled tip of 8 μm i.d.; New Objective, Woburn MA); stationary phase: Magic C18AQ, 3 μm, 100 Å (Michrom Bioresources, Auburn, CA) using a linear gradient of acetonitrile in 0.1% formic acid. The eluate was introduced into either an LTQ Orbitrap or LCQ Deca XP Plus mass spectrometer (Thermo Fisher Scientific, San Jose, CA) by nanoelectrospray. Data analysis was conducted on the Sorcerer (Sage-N Research, San Jose, CA) search engine using the SEQUEST-Sorcerer algorithm. The search was performed against a concatenated “forward” and “reverse” FASTA database. Identification results were filtered and validated using Protein Prophet and Peptide Prophet platforms. The balance between the reliability and sensitivity of protein identification data was set by adjusting the estimated false positive identification rate to ≤ 1%.
Lipidomics Analysis
Phospholipid and triglyceride profiling of lipid phase by MALDI-TOF were performed as described previously,26 with the following modifications: aliquots of lipid phase fraction (0.5 μL) were spotted directly onto a 2 μL droplet of 0.5 M 2,5-dihydroxy-benzoic acid (DHB) matrix solution in 50% acetonitrile/water immediately after droplet deposition onto the MALDI target. This method of sample application prevented spreading of the droplet across multiple spot locations of the MALDI target due to very low viscosity and surface tension of the lipid solution in organic solvent. Additionally, application of an amphipathic solvent onto the crystallizing matrix mixture resulted in formation of relatively uniform matrix/sample spots. Data were collected in positive ionization mode on an ABI 4700 Proteomics Analyzer (Applied Biosystems, Foster City, CA).
Distribution of Proteins and Nucleic Acids After PCT-Mediated Extraction
For RNA and protein distribution analysis ~4 × 107 PC12 cells were washed once with PBS and suspended in 0.9 mL reagent A. The suspension was transferred to a PULSE tube and 0.5 mL of reagent B was added. Pressure cycling was performed as described above; subsequently, the entire sample was split evenly into two tubes for protein and RNA replicate samples and centrifuged for 15 min at ~12,000 × g to separate phases. Following centrifugation, the nonpolar top phase layers were removed. The interface layers were transferred to clean tubes, centrifuged briefly, and any carryover of solvent was aspirated off. The solvent phases were transferred to clean tubes and dried in a SpeedVac to remove solvent. The pellets were centrifuged briefly to facilitate aspiration of residual solvent.
For RNA isolation, 1 mL Trizol reagent was added to each fraction and the standard Trizol protocol for extraction of RNA from cells was followed.
For protein visualization by SDS-PAGE, the pellet, interface, and dried solvent fractions were dissolved in 1 mL Laemmli sample buffer with 50 mM DTT. For DNA distribution analysis, 200 mg of frozen mouse liver was extracted as described above with 1.0 mL reagent A and 250 μL Reagent B. The samples were split into equal replicates and phases were separated as described above. DNA was extracted from the pellet, interface, and dried solvent fraction of one replicate using the Qiagen DNeasy Blood and Tissue kit according to manufacturer’s instructions for extraction of DNA from cells.
Comparison of DNA Recovery Using the DNeasy Kit With or Without Proteosolve-SB
Equal aliquots of cultured mammalian cells were processed by pressure cycling using the ProteoSolve-SB kit for extraction of proteins and DNA. Protein extract was dried, dissolved in IEF buffer and subjected to 2D PAGE.
Frozen mouse liver (23 mg per sample) was processed as above. After centrifugation, the protein phase was dried and subjected to SDS-PAGE. DNA was extracted from the solid phase using the DNeasy kit. Control tissue was digested with Proteinase K prior to DNA isolation with the DNeasy kit according to manufacturer’s instructions.
Comparison of Four Methods for Simultaneous Extraction of RNA and Protein
Equal aliquots of 106 PC12 cells were processed for RNA and protein extraction using four commercially available kits; the ProteoSolve-SB kit from Pressure BioSciences, Trizol, the AllPrep RNA/Protein kit, and the PARIS kit. After PCT extraction with ProteoSolve-SB, the protein-containing solvent phase was dried, dissolved in sample buffer, and submitted to SDS-PAGE analysis. The pellet and solid interface layers were pooled and processed for RNA extraction by adding 0.5 mL of Trizol and extracting by the standard Trizol protocol. The other three samples were processed according to the manufacturer’s instructions. Equivalent aliquots of protein extract from each sample were separated by SDS-PAGE. Final volume of each RNA sample was 100 μL. Equal aliquots of each RNA sample were run on a gel to confirm that RNA was not degraded. Total RNA recovery was measured by Qubit assay, and real time RT-PCR with rat β-actin primers was performed for mRNA quantification.
Protein, DNA, and RNA Extraction From Rat Tissue
Flash frozen rat tissues (264 mg kidney, 330 mg abdominal fat pad, 310 mg liver, 264 mg brain, 200 mg cardiac muscle) were processed with the ProteoSolve-SB kit using 1.0 mL reagent A and 150–200 μL of reagent B per sample. After PCT and centrifugation, 10% aliquots of each protein fraction were either evaporated or precipitated using reagent C, and reconstituted in Laemmli sample buffer with 50 mM DTT for SDS-PAGE. The pellet and interface fractions from each sample were pooled, dissolved in 0.5 mL Trizol, vortexed thoroughly and processed according to the standard Trizol protocol for extraction of RNA and DNA from cells.27
RESULTS
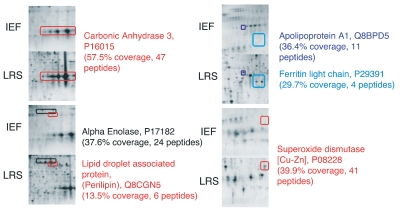
In order to characterize the ability of the PCT-mediated liquid-liquid extraction method to delipidate and solubilize some hydrophobic proteins which are typically under-represented in standard proteomic samples, we compared extraction of murine white adipose tissue using two different extraction buffers followed by 2D-PAGE separation, in-gel digestion, and analysis by LC-MS/MS. Replicate samples of murine abdominal fat were processed under similar conditions by pressure cycling using either the ProteoSolve-SB kit or a popular detergent-based extraction buffer (7 M urea, 2 M thiourea, 4% CHAPS). Several proteins were identified on 2D gels of mouse adipose tissue extracted with ProteoSolve-SB, which were absent or less abundant in samples extracted with detergent (Figure 3).
Figure 3.
Paired images of 2D gels showing pronounced differences in the protein spots detected in samples extracted either in ProteoSolve-SB or in IEF buffer. Differentially extracted proteins were identified by nano-LC coupled with electrospray ionization tandem mass spectrometry. Peptide coverage is shown for each protein extracted by the PCT-mediated ProteoSolve-SB method.
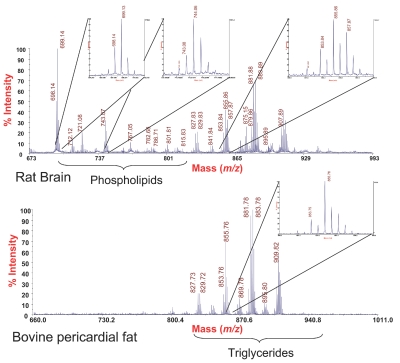
We demonstrate that direct application of the lipid fractions derived from the pressure cycling-mediated ProteoSolve-SB extraction are compatible with MALDI-TOF analysis using DHB matrix in positive ionization mode. These results indicate that lipids extracted by the new PCT-based method represent a broad range of lipid types and are sufficiently pure to be analyzed directly without additional purification steps. Tissue-specific lipid spectra are shown in Figure 4. Phospholipids characteristic of brain samples are apparent in the rat brain extract but absent from the lipid composition of the bovine adipose tissue, which is consistent with previously published materials.26
Figure 4.
Examples of phospholipids, di- and triacylglyceride profiles obtained by direct analysis of lipid phase fractions by MALDI-ToF mass spectrometry. Beef fat samples were processed in pure reagent A, while rat brain phospholipids were extracted by addition of reagent B, which does not exhibit any significant impact on ionization efficiency and signal intensity.
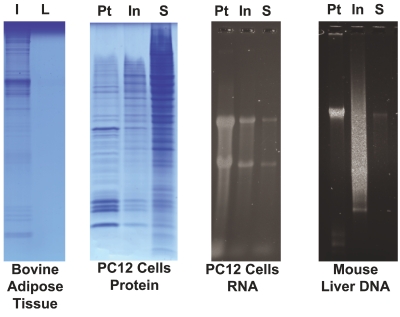
Examination of individual fractions after pressure-cycling-mediated extraction with the ProteoSolve-SB kit (Figure 5) has demonstrated the following: (1) there is no detectable extractable protein in the lipid layer; (2) the polar solvent phase contains the bulk of the sample proteins, and only traces of RNA and DNA; and (3) the pellet and the interface both contain RNA and DNA with the pellet containing ~70% of recovered nucleic acids and the interface containing ~20% (the remaining ~10% can be recovered from the solvent phase after evaporation).
Figure 5.
Distribution of proteins, DNA, and RNA after extraction by PCT with ProteoSolveLRS. Proteins were re-extracted from the insoluble fraction (I) and the lipid fraction (L) of bovine adipose tissue to determine whether any additional proteins could be recovered from the lipid phase after PCT. The re-extracted samples were visualized by SDS-PAGE and confirmed that while a small amount of additional protein can be recovered from the solid residue, no detectable protein could be recovered from the lipid phase. RNA and protein were extracted from PC12 cells, and DNA was extracted from mouse liver. RNA and DNA recovery from the three fractions was compared by agarose gel electrophoresis. Proteins from all three fractions were dissolved in an equal volume of Laemmli buffer and separated by SDS-PAGE. The bulk of the protein was recovered from the soluble phase (S). Nucleic acid recovery quantified by qubit assay confirmed that the pellet (Pt) and interface (In) together accounted for ~90% of both RNA and DNA.
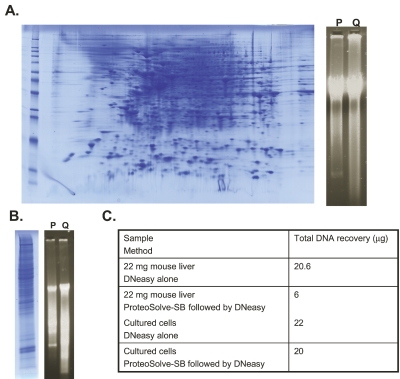
Most of the simple and efficient methods for extraction of genomic DNA from tissues rely upon extensive enzymatic digestion of proteins to release intact DNA. We demonstrate that after extraction of proteins from cells or tissue by PCT with ProteoSolve-SB, the nucleic acid fraction can be processed for DNA isolation and that high yields of intact genomic DNA can be recovered. After protein extraction from a sample of cultured mammalian cells with ProteoSolve-SB, the solid fraction was processed for DNA isolation using the DNeasy kit (Qiagen) and recovery was compared with a control aliquot of cells extracted directly with the DNeasy kit. This method normally is not compatible with recovery of intact tissue proteins since the protocol calls for extensive proteinase K digestion to obtain maximal DNA yield. We found that the combination of the PCT/ProteoSolve-SB method for cell disruption and protein extraction, together with the DNeasy kit for DNA purification, resulted in very good simultaneous recovery of DNA and protein from cells (Figure 6A). In addition, the same sequential protocol was successfully applied to protein and DNA extraction from liver tissue (Figure 6B). The protein extracts were analyzed by 1D or 2D PAGE and confirmed that a broad range of proteins are recovered from the ProteoSolve-SB extract. DNA recovery from cell culture using the combined method was comparable to DNeasy control with proteinase K digestion, while DNA recovery from the tissue sample was about 30% of that obtained from the control (Figure 6C).
Figure 6.
A: DNA and protein recovery from the cell culture. one aliquot of cells was processed with PCT and ProteoSolve-SB (P). DNA was extracted from the solid phase using the DNeasy kit. The control aliquot of cells was processed directly with the qiagen DNeasy kit according to manufacturer’s instructions (q). No intact protein was recovered from the control sample due to extensive proteinase K digestion of the proteins. DNA quantification by qubit assay indicates that the yield of DNA from the new method is comparable to the yield of DNA after extensive proteinase K digestion. B: DNA and protein recovery from mouse liver with PCT and Proteosolve-SB (P). DNA was extracted from the solid phase using the DNeasy kit. Control tissue was processed directly with the DNeasy kit according to manufacturer’s instructions (q). No protein was recovered from the control sample due to extensive proteinase K digestion of the tissue. DNA quantification by qubit assay indicates that the new method yields about 30% of DNA that can be recovered after extensive tissue digestion. c: Table of DNA recovery from liver tissue and cell culture.
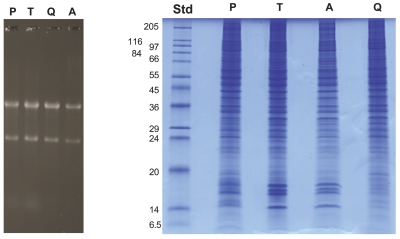
In order to compare the new, expanded PCT-based method with other commercially available kits for simultaneous extraction of RNA and protein, we processed PC12 cells (106 cells per sample) using the ProteoSolve-SB kit, Trizol reagent, the Qiagen AllPrep RNA/Protein kit, and the PARIS kit from Ambion (Figure 7 and Table 1). Trizol, which is an optimized reagent designed primarily for RNA extraction, gave excellent RNA recovery, but the multistep protocol for protein extraction and cleanup from the organic phase was slow, requiring two precipitation steps, three 30-min washes in 300 mM guanidine HCl in 95% ethanol, and a final 20-min wash in ethanol in order to remove the phenol and dye from the Trizol reagent. This extensive cleanup may result in substoichio-metric in vitro protein modifications (N-termini, Lys), which could potentially interfere with downstream quantitative analysis of protein posttranslational modifications. While the other three kits were easy to use, RNA recovery from the Ambion PARIS kit was consistently lower than that of the other reagents (Table 1). This was likely due to the fact that after sample lysis, only half the lysate is used for RNA extraction, while the other half is reserved for protein analysis. In addition, since at the end of the procedure the protein extract is still in the cell disruption buffer, which contains salts and detergent, additional protein cleanup steps, such as dialysis or filtration, may be required. RNA recovery using the Qiagen kit was similar to that obtained with Trizol and ProteoSolve-SB, but due to the low binding capacity of the columns, the sample (106 PC12 cells) had to be split onto two columns, increasing the amount of work and the cost of each sample. Also, due to the nature of the AllPrep spin column used to separate the protein fraction from the RNA, many proteins can remain with the bound RNA fraction and may be absent from the recovered protein fraction (see bottom of protein gel, Figure 7A right panel). In addition, since the protein fraction collected by the AllPrep protocol contains RNA stabilizing buffer, which is not compatible with SDS-PAGE, the proteins must be acetone-precipitated prior to SDS-PAGE analysis, which can lead to potential losses.
Figure 7.
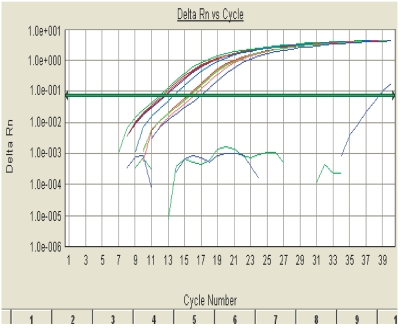
Isolation of RNA and protein from a single sample—comparison of four methods. a: Equal aliquots of 106 PC12 cells were processed for RNA and protein extraction using ProteoSolve-SB kit (P) from Pressure BioSciences, Trizol (T) from Invitrogen, AllPrep RNA/Protein kit from qiagen (q), and PARIS kit from Ambion (A). Molecular weight standards (Std) are provided as a reference. Samples were processed according to manufacturer’s instructions. B: Real-time RT-PCR analysis of PC12 RNA. RNA recovery from 106 PC12 cells, quantification by real-time RT-PCR using primers for rat β-actin. RNA recovered after PCT extraction in ProteoSolve-SB is intact and contains the expected concentration of mRNA.
TABLE 1.
Comparison of Four Reagents for Simultaneous RNA and Protein Recovery
| Method | RNA Recovery From 106 PC12 Cells by qubit Assay of Total RNA | RNA Recovery From 106 PC12 Cells by Real Time RT-PCR |
|---|---|---|
| ProteoSolve-LRS | 11.9 μg | 16.4 μg |
| Ambion PARIS kit | 5.3 μg | 9.0 μg |
| qiagen AllPrep kit | 11.4 μg | 14.0 μg |
| Trizol | 16.2 μg | 14.0 μg |
To confirm that the RNA fraction recovered using the new PCT-based method was not only intact, as indicated by the presence of 28S and 18S RNA on gels (Figures 5 and 7A), but also contained mRNA and was compatible with RT-PCR, the four RNA samples described above were subjected to real time-RT-PCR amplification using β-actin primers. As shown in Figure 7B and Table 1, the RNA extracted from PC12 cells using the new method compared very favorably with the three standard methods tested, indicating that the sample contained amplifiable mRNA at the expected concentration.
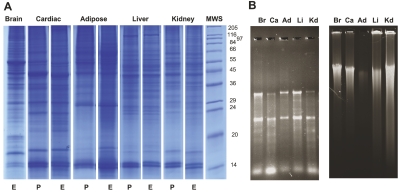
To demonstrate the utility of the new method, sequential extraction of proteins, RNA, and DNA from five flash-frozen rat tissues was performed (Figure 8). The protein-containing fractions separated by SDS-PAGE revealed tissue-specific protein patterns in each of the samples. Using the Trizol protocol for isolation of RNA and DNA from the solid phase, we confirmed that both genomic DNA and RNA can be isolated intact after protein extraction (Figure 8 and Table 2).
Figure 8.
Protein, RNA, and DNA recovery from rat tissues. Samples of flash-frozen tissue were processed using 1.0 mL reagent A and 150–200 μL of reagent B. A: Solvent from the protein-containing fraction was either evaporated (E) or removed by precipitating the protein (P). B: Trizol was used to sequentially purify RNA and DNA from the solid phase. The results confirm that PCT-mediated tissue disruption and extraction with Proteo-SolveLRS allows for the efficient recovery of intact RNA and genomic DNA from a variety of tissues. Br, brain; Ca, cardiac; Ad, adipose; Li, liver; Kd, kidney; MWS, Molecular Weight Standard.
TABLE 2.
RNA and DNA Recovery From Cells and Tissues
| Tissue | RNA Recovery per mg Tissue or 106 Cells | DNA Recovery per mg Tissue or 106 Cells |
|---|---|---|
| Rat liver | 2.40 μg | 34.5 ng (140 nga) |
| Rat kidney | 0.70 μg | 35.50 ng |
| Rat adipose | 0.02 μg | 0.67 ng |
| Rat brain | 0.55 μg | 37.08 ng |
| Rat cardiac muscle | 0.34 μg | 12.80 ng |
| PC12 cells | 11.90 μg | 10.70 μga |
Pressure cycling technology/ProteoSolve-SB protein extraction was usually followed by RNA and DNA isolation using Trizol reagent (or the DNeasy kit where indicated by).
DISCUSSION
Pressure-cycling-mediated extraction in combination with the unique chemistry of the ProteoSolve-SB kit has been demonstrated to be an efficient method to extract proteins from lipid-rich samples such as adipose and brain tissue.18 Here we expand the utility of this PCT-based sample preparation system, by demonstrating that four major components of biological samples, proteins, lipids, RNA and DNA, can all be easily and efficiently isolated from a single sample of cells or tissue using the new ProteoSolve-SB kit. In addition, we confirm that protein and RNA recovery is comparable to, or better than, that achieved using currently available kits and reagents, such as Trizol, the AllPrep kit, and the PARIS kit (Figure 7).
The new expanded application of the pressure-cycling-enhanced ProteoSolve-SB kit can provide efficient simultaneous extraction of proteins, lipids, and nucleic acids. This is especially important when working with samples that are precious or unique, such as human or wild animal biopsy tissue or samples that are difficult to duplicate, such as small cell populations, like early stem cell cultures. Another advantage of ProteoSolve-SB, is in more accurate analysis of nonhomogenous samples. Since splitting samples for separate protein, lipid, and nucleic acid analyses is not necessary, artifacts due to uneven distribution of components in the sample are avoided.
The new PCT-based method is advantageous not only for small and precious samples, but also for larger samples where a single convenient method for purification of multiple components is desired. Since the Proteo-Solve-SB protocol is easily scalable for large samples, it has many advantages over other currently available methods. Popular and easy column-based methods are limited by the binding capacity of the columns, often making it necessary to divide samples over several columns, resulting in more work and less efficient recovery. Other reagents, such as Trizol, only work well when the sample to be extracted comprises 10% or less of the total reaction volume (requiring 1 mL reagent for every 100 mg of tissue). ProteoSolve-SB can be used with sample-to-solvent ratios as high as 250–300 mg/mL, and possibly higher in some cases. Higher sample-to-solvent ratios are important when subsequent purification steps involve precipitation reactions, since these are always less efficient in more dilute solutions. After ProteoSolve-SB extraction of the bulk of the sample’s proteins and lipids, the nucleic acid-enriched fraction can be brought up in a relatively small volume of reagent for subsequent RNA or DNA purification. This would allow for much more efficient extraction from large samples that contain little RNA and DNA (e.g., soil, yogurt, skin).
The expanded ProteoSolve-SB method provides excellent recovery of proteins, high yields of intact DNA and RNA, and simplified recovery of lipids. In addition, the method is easy and rapid, requiring fewer steps and less hands-on time than other currently available techniques. Since simultaneous sample homogenization and extraction is performed in the Barocycler instrument, labor-intensive and inconsistent tissue disruption steps like sonication and grinding in liquid nitrogen are avoided. For most types of samples, pieces of tissue are placed directly into a PULSE Tube, reagents A and B are added, and the sample is inserted into the Barocycler and subjected to pressure cycling for 10–15 min. Subsequently, the homogenate is transferred to a centrifuge tube and centrifuged for 10–15 min. Once the three phases are separated, the protein fraction can be dried, eliminating the need for protein precipitation, washing, desalting, ultra-filtration, and other time-consuming cleanup steps. More importantly, there is no need to use aggressive chemistry, which allows avoidance of unwanted in vitro modifications and subproducts. The lipid fraction does not require further cleanup methods such as chromatography, additional extraction steps, or enzymatic digestion. We have shown that lipid fraction samples can be used directly for analysis by MALDI-TOF MS. DNA and RNA can be easily extracted from the solid phase by a number of available reagents, again without the need for labor-intensive sample homogenization. In addition, since the bulk of the proteins have already been extracted from the nucleic acid fraction and enzyme activity in the reagent A is minimal or absent, the likelihood of RNA and/or DNA degradation during extraction is very low.
ProteoSolve-SB is compatible with most common downstream applications. Following solvent removal by evaporation or precipitation, protein pellets can be dissolved directly in an appropriate buffer such as SDS/ Laemmli or a urea/thiourea/CHAPS IEF buffer23 and subjected to further analyses. The lipid fractions are available for direct profiling by MS or for separation and enzymatic digestion for structural analysis. The DNA/RNA fraction is compatible with many commonly used reagents and kits for isolation of DNA and/or RNA, such as Trizol and the Qiagen DNeasy and RNeasy kits.
The combination of sample disruption by PCT and extraction in ProteoSolve-SB relies on nonenzymatic and detergent-free dissolution and partitioning of sample components to efficiently and easily extract lipids, proteins, RNA, and DNA from many types of samples without the need for multiple replicates, inconvenient and time-consuming tissue homogenization methods, or extensive post-extraction cleanup. Thus, this novel PCT-based method may help enable unique systems biology studies, where correlation of transcription profiles with protein expression, analyses of posttranslational protein modifications, and tissue lipid composition were previously considered impractical due to the limited quantities of available material, or high variability between individual sample replicates.
Future efforts to scale down this method further will lead to opportunities for systems biology experiments on very small samples such as needle biopsies, as well as primary cell cultures derived from individual stem cells.
References
- 1.Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat Protoc. 2006;1(2):581–585. doi: 10.1038/nprot.2006.83. [DOI] [PubMed] [Google Scholar]
- 2.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 3.Gauthier ER, Madison SD, Michel RN. Rapid RNA isolation without the use of commercial kits: Application to small tissue samples. Pflugers Arch. 1997;433(5):664–668. doi: 10.1007/s004240050328. [DOI] [PubMed] [Google Scholar]
- 4.Gilchrist M, MacDonald AJ, Neverova I, Ritchie B, Befus AD. Optimization of the isolation and effective use of mRNA from rat mast cells. J Immunol Methods. 1997;201(2):207–214. doi: 10.1016/s0022-1759(96)00230-x. [DOI] [PubMed] [Google Scholar]
- 5.Gill SS, Aubin RA, Bura CA, Curran IH, Matula TI. Ensuring recovery of intact RNA from rat pancreas. Mol Biotechnol. 1996;6(3):359–362. doi: 10.1007/BF02761714. [DOI] [PubMed] [Google Scholar]
- 6.Verhofstede C, Fransen K, Marissens D, et al. Isolation of HIV-1 RNA from plasma: Evaluation of eight different extraction methods. J Virol Methods. 1996;60(2):155–159. doi: 10.1016/0166-0934(96)02062-9. [DOI] [PubMed] [Google Scholar]
- 7.Monstein HJ, Nylander AG, Chen D. RNA extraction from gastrointestinal tract and pancreas by a modified Chomczynski and Sacchi method. Biotechniques. 1995;19(3):340–344. [PubMed] [Google Scholar]
- 8.Louveau I, Chaudhuri S, Etherton TD. An improved method for isolating RNA from porcine adipose tissue. Anal Biochem. 1991;196(2):308–310. doi: 10.1016/0003-2697(91)90471-5. [DOI] [PubMed] [Google Scholar]
- 9.Riol H, Jeune B, Moskovic A, Bathum L, Wang E. Optimized lymphocyte protein extraction performed simultaneously with DNA and RNA isolation: Application to the study of factors affecting DNA, RNA, and protein recovery from lymphocytes of the oldest individuals. Anal Biochem. 1999;275(2):192–201. doi: 10.1006/abio.1999.4328. [DOI] [PubMed] [Google Scholar]
- 10.Yang K, Zhao Z, Gross RW, Han X. Shotgun lipidomics identifies a paired rule for the presence of isomeric ether phospholipid molecular species. PLoS ONE. 2007;2(12):e1368. doi: 10.1371/journal.pone.0001368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrdwell WC. Dual parallel liquid chromatography with dual mass spectrometry (LC2/MS2) for a total lipid analysis. Front Biosci. 2008;13:100–120. doi: 10.2741/2663. [DOI] [PubMed] [Google Scholar]
- 12.Han X. Neurolipidomics: Challenges and developments. Front Biosci. 2007;12:2601–2615. doi: 10.2741/2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adibhatla RM, Hatcher JF, Dempsey RJ. Lipids and lipidomics in brain injury and diseases. A APS J. 2006;8(2):E314–321. doi: 10.1007/BF02854902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang G, Terry AV, Jr, Bartlett MG. Sensitive liquid chromatography/tandem mass spectrometry method for the simultaneous determination of olanzapine, risperidone, 9-hydroxyrisperidone, clozapine, haloperidol and ziprasidone in rat brain tissue. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;858(1–2):276–281. doi: 10.1016/j.jchromb.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang G, Terry AV, Jr, Bartlett MG. Sensitive liquid chromatography/tandem mass spectrometry method for the determination of the lipophilic antipsychotic drug chlorpromazine in rat plasma and brain tissue. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;854(1–2):68–76. doi: 10.1016/j.jchromb.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 16.Tao F, Li C, Smejkal G, Lazarev A, Lawrence N, Schumacher RT. Pressure cycling technology (PCT) applications in extraction of biomolecules from challenging biological samples. The 4th International Conference on High Pressure Bioscience and Biotechnology; 2007; Kyoto, Japan: Wiley International; 2007. pp. 166–173. [Google Scholar]
- 17.Tao F, Behnke J, Li C, Saravis C, Schumacher RT, Lawrence NP. Applications of pressure cycling technology (PCT) in proteomics. In: Smejkal G, Lazarev A, editors. Separation Methods in Proteomics. Boca Raton, FL: Taylor and Francis; 2006. pp. 3–18. [Google Scholar]
- 18.Lazarev AV, Smejkal G, Romanovsky I, Cao H, Hotamisligil GS, Ivanov AR. Proteomic analysis of adipose tissue using detergent-free protein extraction by pressure cycling and high resolution tandem mass spectrometry. J Am Soc Mass Spectrom. 2007 May;:93S. [Google Scholar]
- 19.Romanovsky I, Smejkal G, Lazarev A, Lawrence N, Schumacher RT. Pressure cycling technology (PCT): A novel sample preparation approach to biomarker discovery and drug development in lipid-rich samples. Next Generation Pharmaceutical. 2007:270872. [Google Scholar]
- 20.Redeby T, Emmer A. Membrane protein and peptide sample handling for MS analysis using a structured MALDI target. Anal Bioanal Chem. 2005;381(1):225–232. doi: 10.1007/s00216-004-2854-0. [DOI] [PubMed] [Google Scholar]
- 21.Redeby T, Roeraade J, Emmer A. Simple fabrication of a structured matrix-assisted laser desorption/ionization target coating for increased sensitivity in mass spectrometric analysis of membrane proteins. Rapid Commun Mass Spectrom. 2004;18(10):1161–1166. doi: 10.1002/rcm.1466. [DOI] [PubMed] [Google Scholar]
- 22.Smejkal GB, Witzmann FA, Ringham H, et al. Sample preparation for two-dimensional gel electrophoresis using pressure cycling technology. Anal Biochem. 2007;363(2):309–311. doi: 10.1016/j.ab.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smejkal GB, Li C, Robinson MH, Lazarev AV, Lawrence NP, Chernokalskaya E. Simultaneous reduction and alkylation of protein disulfides in a centrifugal ultrafiltration device prior to two-dimensional gel electrophoresis. J Proteome Res. 2006;5(4):983–7. doi: 10.1021/pr050439w. [DOI] [PubMed] [Google Scholar]
- 24.Smejkal GB, Robinson MH, Lazarev A. Comparison of fluorescent stains: relative photostability and differential staining of proteins in two-dimensional gels. Electrophoresis. 2004;25(15):2511–2519. doi: 10.1002/elps.200406005. [DOI] [PubMed] [Google Scholar]
- 25.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1(6):2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 26.Woods AS, Jackson SN. Brain tissue lipidomics: direct probing using matrix-assisted laser desorption/ionization mass spectrometry. AAPS J. 2006;8(2):E391–E395. doi: 10.1007/BF02854910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Invitrogen. Trizol Technical Note. On-line technical literature. 2005 http://tools.invitrogen.com/content/sfs/productnotes/F_071215_Trizol%20and%20Trizol%20LS.pdf.