Abstract
Linear amplification is a method of synthesizing single-stranded DNA from either a single-stranded DNA or one strand of a double-stranded DNA. In this protocol, molecules of a single primer DNA are extended by multiple rounds of DNA synthesis at high temperature using thermostable DNA polymerases. Although linear amplification generates the intended full-length single-stranded product, it is more efficient over single-stranded templates than double-stranded templates. We analyzed linear amplification over single- or double-stranded mouse H-ras DNA (exon 1–2 region). The single-stranded H-ras template yielded only the intended product. However, when the double-stranded template was used, additional artifact products were observed. Increasing the concentration of the double-stranded template produced relatively higher amounts of these artifact products. One of the artifact DNA bands could be mapped and analyzed by sequencing. It contained three template-switching products. These DNAs were formed by incomplete DNA strand extension over the template strand, followed by switching to the complementary strand at a specific Ade nucleotide within a putative hairpin sequence, from which DNA synthesis continued over the complementary strand.
Keywords: linear amplification, template-switching, artifact, hairpin structure
In linear amplification, single-stranded DNA is synthesized by repeatedly extending molecules of a single primer over single-stranded or double-stranded DNA templates. Linear amplification is used for cycle sequencing1 and mapping DNA sequences at which carcinogens form adducts with various degrees of specificity.2,3
The experimental conditions of linear amplification are similar to the polymerase chain reaction, except that a single primer is used. Despite using a high temperature of DNA synthesis, linear amplification works better with single-stranded templates than with double-stranded templates, which can present difficulties in optimizing linear amplification for some DNA sequences. One of the difficulties of linear amplification over double-stranded DNA is premature termination of DNA synthesis.3,4 It was found that premature chain termination is partly due to incorporation of wrong bases and strand extension problems at DNA secondary structures.5 Premature chain termination may be minimized by conducting the DNA synthesis with a nick-translating DNA polymerase supplemented with a small amount of a proofreading DNA polymerase.3,6
Linear amplification of some double-stranded DNA sequences can form artifact products that are not removed by the above combination of DNA polymerases. We have studied the mouse H-ras exon 1–2 region DNA to describe such artifacts, and report that some of them are produced by template-switching during primer extension.
MATERIALS AND METHODS
Preparation of H-ras Templates
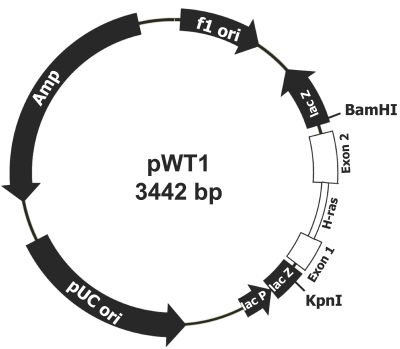
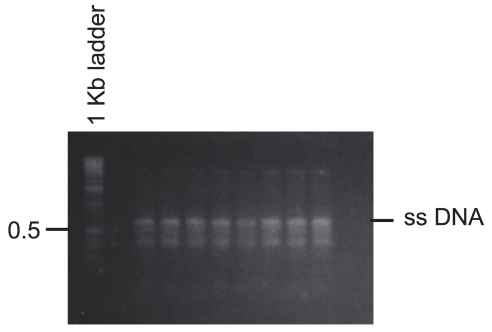
Double-stranded H-ras template DNA (548 bp) was prepared by polymerase chain reaction (PCR) amplification of a BamHI-digested plasmid (pWT1) containing the mouse H-ras exon 1–2 region (Figure 1) with primers MRF (5′-CCG CTG TAG AAG CTA TGA CA) and MRR (5′-TGC CAG GGC TCA CGG GCT AG) using previously described experimental conditions.5 The single-stranded H-ras template DNA (Figure 2) was prepared by linear amplification of the BamHI-digested pWT1 with MRF. Both double-stranded and single-stranded H-ras DNAs were separated by 1% low-melting-point agarose gel and extracted with GELase (Epicentre Biotechnologies, Madison, WI).
FIGURE 1.
Structure of pWT1.
FIGURE 2.
Synthesis of single-stranded (ss) H-ras DNA by linear amplification of BamHI-digested pWT1. The indicated DNA band indicated was extracted for use as single-strand H-ras template.
Linear Amplification and Sequencing
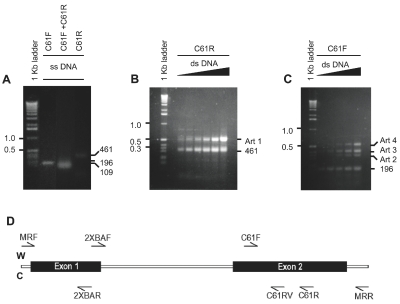
Linear amplification reactions were carried out in 20-mL reactions with a 10:1 mixture of Tth and Vent DNA polymerases at 80°C, as previously described.3 Additional primer sequences were 2XBAF (5′-GAT CCC TCT AGA GAG GTG GACT), 2XBAR (5′-ATA CTC GTC CAC AAA GTG GTT C), C61F (5′-GTG GTC ATT GAT GGG GAG ACA), C61RV (5′-ATA CTC TTC TTG ACC TGC TGT), and C61R (5′-AGA GGA AGC CCT CCC CTG TGC). The locations of the primers are indicated in Figure 3D. The linear amplification product Art 1 (2.5–10 ng/μL) was directly sequenced with the primer C61R at the Genomics Core Research Facility, University of Nebraska, Lincoln, NE. Nucleotide numbers of the H-ras exon 1–2 region correspond to GenBank Accession No. U89950.
FIGURE 3.
Formation of artifact DNA products by linear amplification. Single-stranded H-ras template yielded the desired products with primers C61F or C61R (a), whereas double-stranded H-ras template yielded the desired products as well as artifact DNAs (Art 1–4) (B, c). D : The sites of annealing of the primers used in this study. The top strand was designated as the Watson strand (W) and the bottom strand was designated the Crick strand (C). An alternative definition also exists in which the antisense or the transcription template strand is designated as the Watson strand and the sense strand is designated as the Crick strand. Art, artifact.
RESULTS
Linear Amplification of Double-Stranded and Single-Stranded H-ras DNA
Linear amplification reactions were conducted either with single-stranded (Figure 3A) or double-stranded DNAs (Figure 3B, C) containing the H-ras exon 1–2 region. When using the single-stranded DNA as template, linear amplification with primer C61F or C61R (196 and 461 nt) or PCR reaction with C61F and C61R (109 bp) produced the intended products. On the other hand, when double-stranded DNA was used as template, artifact (Art) products were observed. Furthermore, the relative amounts of the artifact products (Art 1–4 in Figure 3B, C) appeared to increase with the amount of the double-stranded template.
Primer-Walking and Sequence Analysis of Artifact 1
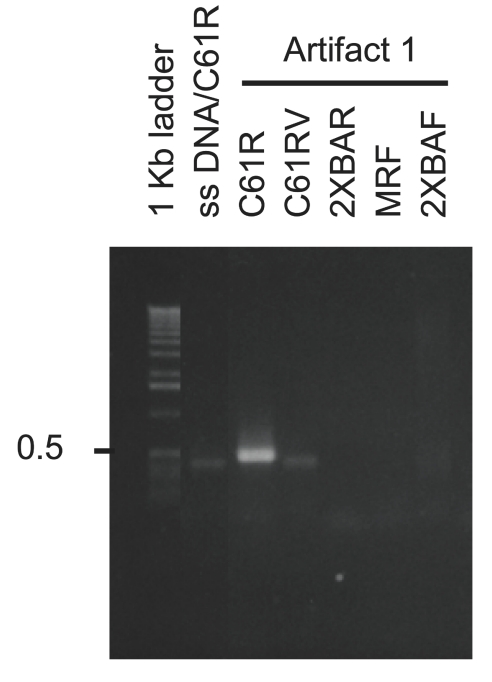
A primer-walking experiment (Figure 4) of the single-stranded artifact product obtained from linear amplification of double-stranded DNA with C61R (Art 1 in Figure 3B) was conducted with primers shown in Figure 3D. For size comparison, the linear amplification product of single-stranded H-ras gene with C61R (461 nt) was included. MRF and 2XBAF did not produce a linear amplification product from Art 1, indicating that Art 1 did not extend to exon 1 (Figure 4). However, C61R and C61RV were successful in linear amplifying Art 1. This suggested that during the formation of Art 1, DNA synthesis stopped copying the intended Watson (top) strand towards exon 1, and switched to copying the Crick (bottom) strand in the exon 2 direction. In this way, Art 1 could contain annealing sites for C61RV and C61R. On the other hand, 2XBAR did not linear amplify Art 1, which meant that Art 1 did not contain the sequences for annealing 2XBAR. These results suggest that while forming Art 1, DNA synthesis switched templates somewhere between C61RV and 2XBAF. Linear amplification of Arts 2–4 with these primers did not generate readily identifiable and reproducible bands (not shown). It was therefore concluded that Arts 2–4 were more complex in structure and not examined further.
FIGURE 4.
Linear amplification primer walking analysis of artifact 1. The artifact product generated by linear amplification of double-stranded H-ras DNA with the C61R primer was subjected to linear amplification with a number of primers that anneal to the H-ras sequence. Only C61R and C61Rv yielded linear amplification products with artifact 1.
Sequence Analysis of Artifact 1
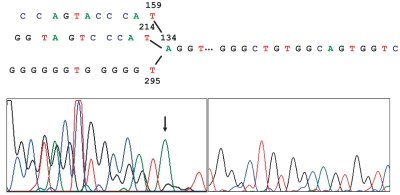
Direct sequence analysis of Art 1 with C61R revealed that it contained a mixture of template-switching products (Figure 5). Within the overlapping peaks of the electropherogram, it was possible to read 10 or more contiguous bases from three areas of the H-ras insert. A 10-nt-long unique sequence is thought to occur once every 524,800 bases,7 which is much longer than the H-ras insert in pWT1. Therefore, Art 1 contained at least three different template-switching products (Figures 5 and 6). It appeared that one of these products was generated as strand extension switched from the Watson strand at nt 159, another at nt 214, and the third at nt 295. However, all of these syntheses switched to the same base (nt 134) in the Crick strand. Art 1, despite containing various products, produced a single DNA band (Figure 3B). Therefore, it is likely that not all products in Art 1 extended all the way to the end of exon 2. On the other hand, Art 1 could be amplified by C61R; therefore, at least one of these template-switching products extended at least up to the C61R-annealing site. If all three products in Art 1 were to contain the C61R annealing site, the 159–134 switch product would be 593 nt, the 214–134 product would be 538 nt, and the 295–134 product would be 457 nt long. Direct sequencing of Arts 3 and 4 with C61F produced complex electropherograms with overlapping peaks, which did not permit easy analysis.
FIGURE 5.
Sequence analysis of artifact 1. Results of sequence analysis from two experiments are joined together. The electropherogram showed overlapping peaks in sequences previous to nt 134, but none at the downstream sequences. The indicated sequences were readable within the overlapping peaks. The nucleotide 133 should have been a g, which is not observed in the electropherogram.
FIGURE 6.
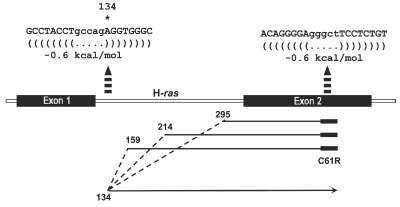
Illustration of the template-switched products of artifact 1. At 80ºC, the H-ras gene segment may contain two hairpin-type secondary structures (the locations shown with segmented columns, and the sequences indicated on top).
To explore whether the template-switching in Art 1 could be associated with DNA secondary structures, the H-ras DNA sequence was analyzed by the online mfold software (http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/dna-form1.cgi), using default parameters, except the temperature was set to 80°C, i.e., the temperature of DNA synthesis in these experiments. This analysis revealed that at 80°C, the H-ras segment would contain two hairpin structures (nt 120–141 and nt 423–443), both with a ΔG value of 0.6 kcal/mol (Figure 6). The first hairpin spanned the nt 134, which is the nucleotide at which the three template-switch products in Art 1 restarted strand extension after changing from the Watson template. The C61R primer anneals within the second hairpin structure.
DISCUSSION
Template-switching has been previously reported in PCR.8,9 Therefore, it can also be expected to occur during linear amplification. Template-switching is a well-known phenomenon for synthesizing the ends of retroviral genome by reverse transcriptases.10,11 A previous study concluded that template-switching for PCR polymerases may have a different mechanism than that for the reverse transcriptases,9 and it would frequently occur at inverted repeats.
In the present study, we noted that template-switch sequences where DNA synthesis left the Watson strand were T nucleotides preceded by a run of Gs or Cs. For two of the three template-switching products (159–134 and 214–134), there was an A residue between the T and the run of Cs.
Premature termination of DNA synthesis is a possible mechanism for the incomplete chain extension that induced these template-switching artifacts. The DNA polymerase combination used in our study minimizes premature termination, but does not completely eliminate their formation. If template-directed premature chain termination is a mechanism, it would be reasonable to speculate that the sequences at which the syntheses had switched would be hotspots of premature chain termination. Such ideas could not be expanded any further, because there were no identifiable nontemplate bases in the switch junctions, which argues against misincorporation as the reason for premature chain termination, and under our reaction conditions, these sequences did not appear to be prone to form secondary structures, which argues against the idea that the premature chain terminations occurred at structure-related pause sites.
Low DNA polymerase processivity could also be a mechanism of the incomplete chain extension. We have used a mixture of Tth and Vent DNA polymerases (at 10:1 ratio) for these experiments. Tth DNA polymerase has nick-translating activity (5′-3′-exonuclease) but is deficient in proofreading (3′-5′-exonuclease); whereas Vent DNA polymerase has the opposite properties.3 In this cocktail, since Tth DNA polymerase is present at greater amounts, it is thought to be chiefly responsible for DNA synthesis, while Vent DNA polymerase provides the proofreading. The processivity of Tth (30–40 nt) is lower than AmpliTaq (50–60 nt), but higher than Pfu (15–20 nt), Vent (7–10 nt), and the Stoffel fragment (5–10 nt).3 Parallel experiments with a panel of DNA polymerases may help in determining whether processivity is a factor for the incomplete chain extension associated with template-switching. We did not observe artifacts by linear amplification of the single-stranded template, but in that case, incompletely extended products may only anneal to the same template and extend it to the end.
On the other hand, the observation that at increased double-strand template concentrations, linear amplification favored the formation of template-switching artifacts over the intended products (Figure 3B, C) can shed light on the mechanism of template-switching. It suggests that double-strand template concentration is an extremely important determinant for template-switching. On the other hand, incomplete DNA synthesis, whether due to premature chain termination at hotspot sequences and/or DNA polymerase processivity, was clearly involved. Even though we do not understand the exact mechanisms, these results suggest that when incompletely extended DNA molecules can find sufficient numbers of complementary strands to anneal with, they can function as primers for DNA synthesis in the opposite direction, and generate the template-switching artifacts. The reason why these linear amplification artifacts switched to the same Ade residue inside a putative hairpin structure is also not obvious, but this switch to sequence preference suggests that switching only required a minimal level of complementary homology between the incompletely extended (primer) strand, and would be favored at stable structural regions in the complementary (template) strand.
ACKNOWLEDGMENT
We acknowledge the support provided by Drs. Ercole L. Cavalieri and Eleanor G. Rogan, and help with drawing the figures by Mohamed F. Ali.
REFERENCES
- 1.Krishnan BR, Blakesley RW, Berg DE. Linear amplification DNA sequencing directly from single phage plaques and bacterial colonies. Nucleic Acids Res. 1991;19:1153. doi: 10.1093/nar/19.5.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ponti M, Forrow SM, Souhami RL, D’Incalci M, Hartley JA. Measurement of the sequence specificity of covalent DNA modification by antineoplastic agents using Taq DNA polymerase. Nucleic Acids Res. 1991;19:2929–2933. doi: 10.1093/nar/19.11.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakravarti D, Cavalieri EL, Rogan EG. Linear amplification mapping of polycyclic aromatic hydrocarbon-reactive sequences in H-ras gene. DNA Cell Biol. 1998;17:529–539. doi: 10.1089/dna.1998.17.529. [DOI] [PubMed] [Google Scholar]
- 4.Olsen DB, Eckstein F. Incomplete primer extension during in vitro DNA amplification catalyzed by Taq polymerase; exploitation for DNA sequencing. Nucleic Acids Res. 1989;17:9613–9620. doi: 10.1093/nar/17.23.9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakravarti D, Mailander P, Franzen J, Higginbotham S, Cavalieri EL, Rogan EG. Detection of dibenzo[a,l]pyrene-induced H-ras codon 61 mutant genes in preneoplastic SENCAR mouse skin using a new PCR-RFLP method. Oncogene. 1998;16:3203–3210. doi: 10.1038/sj.onc.1201853. [DOI] [PubMed] [Google Scholar]
- 6.Barnes WM. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dervan PB. Molecular recognition of DNA by small molecules. Bioorg Med Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 8.Odelberg SJ, Weiss RB, Hata A, White R. Template-switching during DNA synthesis by Thermus aquaticus DNA polymerase I. Nucleic Acids Res. 1995;23:2049–2057. doi: 10.1093/nar/23.11.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel R, Lin M, Laney M, Kurn N, Rose S, Ullman EF. Formation of chimeric DNA primer extension products by template switching onto an annealed downstream oligonucleotide. Proc Natl Acad Sci USA. 1996;93:2969–2974. doi: 10.1073/pnas.93.7.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilboa E, Mitra SW, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 11.Temin HM. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci U S A. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]