Abstract
Attachment of proteins to the 3′ end of DNA increases stability of the DNA in serum and retards clearance of DNA by major organs, thereby enhancing in vivo half-life and therapeutic potential of DNA. Unfortunately, the length of DNA molecules that can be produced with 3 ′ modifications by solid-phase synthesis for protein attachment is limited to 45–60 nucleotides due to uncertainties about sequence fidelity for longer oligonucleotides. Here we describe selective covalent coupling of proteins or other molecules to the 3′-adenine overhang of unlabeled and fluorophore-labeled double-stranded polymerase chain reaction products putatively at the N6 position of adenine using 2.5% glutaraldehyde at pH 6.0 and 4°C for at least 16 h. Gel mobility shift analyses and fluorescence analyses of the shifted bands supported conjugate formation between double-stranded polymerase chain reaction products and β2-microglobulin. In addition, blunt-ended DNA ladder fragments treated with glutaraldehyde at 4°C showed no evidence of DNA–DNA or DNA–protein conjugate formation. With the present cold glutaraldehyde technique, longer DNA–3′-protein conjugates might be easily mass-produced. The protein portion of a DNA–3′-protein conjugate could possess functionality as well, such as receptor binding for cell entry, cytotoxicity, or opsonization.
Keywords: 3 ′ terminus, adenine, aldehyde, aptamer, aryl amine, conjugate, EMSA, gel shift assay
There is broad interest in facile industrial-scale means for stable attachment of peptides and proteins to nucleic acids.1–5 There is specific interest in attachment of blocking molecules such as peptides or proteins to the 3′ end of DNA molecules, such as aptamers and antisense oligonucleotides, because blockage of the 3′ end, even with small molecules such as biotin, has been shown to significantly decrease serum exonuclease activity.6–11 In addition, blockage of the 3 ′ end with larger molecules, such as proteins or polyethylene glycol,8 has been shown to retard clearance of such conjugates by the kidneys and other major organs in vivo.9
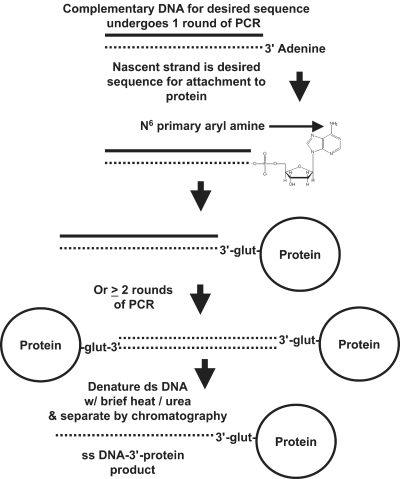
Taq DNA polymerase is known to add an adenine to the 3′ end of its nascent strands during polymerase chain reaction (PCR).12 Similarly, Deep Vent (exo-) polymerase adds a 3′-adenine overhang to about 30% of its PCR products (New England BioLabs, Ipswich, MA). This unpaired overhanging 3′-adenine has been cleverly used for facile incorporation of double-stranded PCR products into cloning vectors (e.g., TOPO TA cloning kits produced by Invitrogen, Carlsbad, CA). The unpaired and unprotected 3′-adenine is targeted as a site for coupling to proteins in the present work (Figure 1). While it may seem to be a relatively routine matter to synthesize a DNA oligonucleotide with a 3′-amino group attached to it for conjugation, one is generally limited to small oligonucleotides (< 60 bases), the reason being that the process is prone to errors and sequence “infidelity” as progressively longer oligonucleotides are synthesized (personal communications with DNA synthesis vendors). Therefore, 3′ modification of oligonucleotides is often not guaranteed by DNA synthesis vendors. While significant progress has been made in the area of complete solid-phase synthesis of oligonucleotide–3′-peptide conjugates,6,10,11 such methods are expensive on a large scale and are limited to peptide (not protein) synthesis. Most commercial peptide synthesis is limited to < 100 amino acids without ligation to form larger polypeptides and even if longer polypeptides are synthesized, correct conformational folding is an issue for many proteins or protein subunits along with correct quaternary structure of the interacting subunits. Hence, we sought a straightforward, general, and cost-effective means to conjugate peptides or proteins to the 3′ end of DNA which would not be limited by DNA or protein size and might someday permit whole genes or gene fragments to be PCR-amplified and coupled directly to proteins for serum nuclease protection and facilitated entry into target cells.3,6
FIGURE 1.
Conceptual diagram illustrating glutaraldehyde linkage to the unpaired 3′ adenine overhang added by some polymerases during PCR. In theory, near neutral pH values, hydrogen bonds between complementary strands of the PCR product would remain intact and the product would remain double-stranded so that the only possible point of linkage would be the N6 primary aryl amine of the 3 ′ adenine. Covalent coupling to a protein on the 3′ end would provide protection from serum exonuclease and retard clearance by major organs to enhance pharmacokinetics. The double-stranded DNA might be converted to single-stranded DNA by brief heating and treatment with high concentrations of urea and other chaotropic agents to disrupt hydrogen bonds between the DNA strands followed by chromatographic purification to yield active aptamer- or antisense-3′-protein constructs.
Since the 3′-adenine of double-stranded PCR products is unpaired, we hypothesized that it might be more susceptible to attack from certain types of chemical coupling reactions than other nucleotides which are “protected” by hydrogen bonding between the strands at nearly neutral pH values. Many well-known linkage chemistries are readily available for attaching to primary amines. Unfortunately, the N6 of adenine is a primary aryl amine which is considered to be virtually unreactive with succinimides and other popular amine coupling reagents due to stabilizing resonance structures within the adenine ring. Yet several reports exist in the literature describing covalent attachment of aldehydes and other bifunctional linkers to the N6 amino group of adenine.5,13–18
The impact of a simple DNA–3′-protein conjugation method in the pharmaceutical industry is potentially significant, because such a method might enable inexpensive mass production of therapeutic DNA–3′-protein conjugates. The conjugates could be chimeras of active antisense oligonucleotides or aptamers and functional proteins. For example, if the protein or peptide is a ligand for a specific receptor, it could facilitate entry of DNA into cells,3,6 trigger cellular responses, opsonize encapsulated bacteria (e.g., via an Fc fragment of IgG or C3b component of complement on the 3′ end), or activate the complement system to kill bacteria, parasites, and cancer cells (e.g., fixation of a C1qrs complex on the 3′ end). Mass production of DNA–3′-antibody reagents for immuno-PCR constitutes another possible diagnostic application of this approach to DNA–3′-protein conjugate synthesis.
MATERIALS AND METHODS
Generation of Unlabeled and Fluorophore-Labeled PCR Products
All oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). For each conjugation experiment, 5–10 EasyStart Micro 100 premade PCR tubes (Molecular BioProducts, San Diego, CA) were used. The following components were added to each PCR tube: 2 μL of template DNA (a 72-base aptamer template designated “MPA1” for binding to methylphosphonic acid; MPA)19 at a stock concentration of 5.6 nmoles/mL, 5 μL of forward and reverse primer (5′-ATACGGGAGCCAACACCA-3′ and 5′-ATCCGTCACACCTGCTCT-3′, respectively) each at stock concentrations of 85 nmoles/mL, 1 μL of recombinant rTaq polymerase (TakaRa, Otsu, Shiga, Japan), 10 μL of 10X PCR buffer (TakaRa) and 27 μL of nuclease-free deionized water. PCR was conducted under the following conditions: 95°C for 5 min followed by 30 cycles of 95°C for 30 sec, 53°C for 30 sec, and 72°C for 30 sec, followed by a 7-min extension period at 72°C and storage at 4°C. In one experiment, the MPA aptamers were labeled with 4 μM AlexaFluor (AF) 546-14-dUTP (Invitrogen) by 30 cycles of PCR with 5 U of Deep Vent (exo-) polymerase (New England Bio Labs, Ipswich, MA) per PCR reaction.20
PCR Product–3′-Protein Coupling Method
Blunt-ended Amplisize (50–2000 bp) DNA ladder was purchased from Bio-Rad Laboratories (Hercules, CA). Glutaraldehyde (Grade I, 25% solution), human β2-microglobulin, and wide-range molecular weight markers for SDS-PAGE were purchased from Sigma-Aldrich (St. Louis, MO).
The MPA aptamer19 PCR product was diluted to 4 μg/mL in 0.1 M phosphate buffered saline (PBS) which had been adjusted to pH 6.0 with acetic acid to slightly “acidify” or protonate the N6 of 3′ terminal adenines in the PCR product. One hundred microliters of the double-stranded MPA aptamer PCR product, 150 μL of β2-microglobulin, and 20 μL of blunt-ended Amplisize DNA ladder were added in various combinations to a series of microfuge tubes. Half of these tubes received 25 μL of cold 25% glutaraldehyde and all tubes were equalized to a final volume of 250 μL in 0.1 M PBS at pH 6.0. Tubes were refrigerated at 4°C for a minimum of 16 h.
Gel-Shift Analyses
Up to 20 μL of conjugates or other samples were added to an equal volume of Novex 2X SDS-PAGE loading buffer (Invitrogen) and loaded into 4–20% gradient SDS-polyacrylamide Precise mini-gels (Pierce Chemical, Rockford, IL) and run in cold BupH Tris-HEPES-SDS running buffer at 100–200 V. Samples were routinely loaded alongside 5–10 μL of 20 bp DNA ladder (Bio-Rad Laboratories) and SigmaMarker SDS-PAGE wide-range protein molecular weight standards (6.5–205 kD, No. 4038, Sigma-Aldrich). Lanes containing only the unlabeled or AF 546-dUTP-labeled aptamer PCR products and the protein by themselves were also run as controls to determine the migration patterns of each. Following electrophoresis, if gels contained AF 546-labeled DNA, they were rinsed briefly in deionized water and then photographed with Polaroid type 667 film on a UV transilluminator to determine the positions of the AF 546-labeled PCR products. All gels were sequentially stained to determine the locations of DNA and protein bands in the same gels21 by first staining with 4 μL of 1% ethidium bromide in 100 mL of Tris-Borate-EDTA (TBE) buffer for 10 min on an orbital shaker and then washing in 100 mL of deionized water for 10 min and photographing on a UV transilluminator. Next, the same gels were stained in 20–30 mL of Simply-Blue (Coomassie blue G-250, Invitrogen) stain for 1 h followed by washing in 100 mL of deionized water for at least 1 h and photography on a white light box. Finally, some of the previously ethidium bromide–stained and Coomassie blue-stained gels were fixed in an aqueous solution of 40% ethanol and 10% acetic acid at room temperature overnight and stained with an Invitrogen SilverQuest silver stain kit according to manufacturer’s instructions and again photographed on a white light box.
Fluorescence Measurements
Control bands and bands which appeared shifted to a higher molecular weight were excised from some of the polyacrylamide gels to determine if they contained AF 546 dye which would be indicative of DNA in the band and conjugation to β2-microglobulin, since AF 546-dUTP does not contain any primary amines or other functional groups known to be reactive with glutaraldehyde (Figure 3C). Excised test and control gel slices were eluted in 2 mL of PBS (0.1 M PBS, pH 7.2) overnight at 4°C and their fluorescence emission spectra were acquired by excitation at 555 nm with 5-nm slits and a 900-V photomultiplier setting on a Cary Eclipse spectrofluorometer (Varian, Palo Alto, CA).
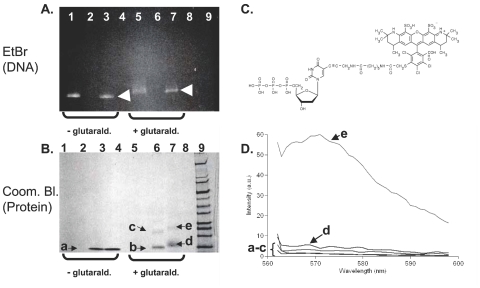
FIGURE 3.
A: Unstained 4–20% SDS-HEPES polyacrylamide gel showing various combinations of the methylphosphonic acid aptamer (amplified by Deep vent (exo-) polymerase to optimally incorporate AF 546-dUTP) and β2-microglobulin components alone and together in the presence and absence of 2.5% glutaraldehyde for 16 h at 4°C. Shifted bands are indicated by arrow heads. B: Same gel stained with Coomassie blue. Contents of each lane: lane 1, AF 546-labeled MPA aptamer; lane 2, β2-microglobulin; lane 3, AF-546-aptamer plus β2-microglobulin; lane 4, vacant; lanes 5–7, same contents and order as lanes 1–3 but with 2.5% glutaraldehyde at pH 7.2 added for 16 h; lane 8, vacant; lane 9, wide-range molecular weight markers (Sigma-Aldrich). C: Structure of AF-546-dUTP which contains no primary amines, but does contain a carboxylic acid (arrow) which may aid in protonating the N6 of adenine for enhanced reactivity. D: Spectrofluorometric assessment of gel slices excised from the gels shown in A and B. Spectra “a–e” correspond to the bands shown in B. Excitation was at 555 nm (peak absorbance of AF 546-dUTP).
RESULTS AND DISCUSSION
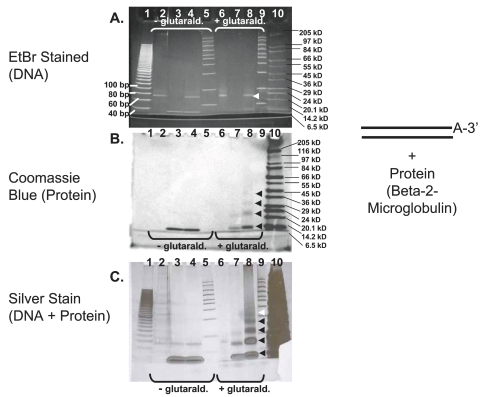
Figure 2 illustrates gel-shift analysis of the DNA aptamer glutaraldehyde β2-microglobulin system. The ethidium bromide-stained gel in Figure 2A suggests a very slight upward shift in the position of the DNA aptamers in lanes 6 and 8 (glutaraldehyde-treated) versus lanes 2 and 4 (untreated). The slight gel shift and mild higher molecular weight smearing in lanes 6 and 8 versus lanes 2 and 4 might be expected since one of the byproducts should be end-to-end linked double-stranded aptamer–3′-glutaraldehyde–3′-aptamer “concatamer” conjugates that shift molecular weight upward. The same gel is shown in Figure 2B after Coomassie blue staining and is more revealing, because there is clear evidence of band shifts for the β2-microglobulin in lanes 7 and 8 (glutaraldehyde-treated) versus lanes 3 and 4 (untreated) and the bands in lane 8 (aptamer plus protein) are shifted upward relative to the bands in lane 7 (β2-microglobulin protein only). The upward shift of bands in lane 8 relative to lane 7 which still stain with Coomassie blue is very suggestive of DNA–protein conjugate formation. These observations are further corroborated by the appearance of lanes 7 and 8 following a silver stain of the same gel in Figure 2C.
FIGURE 2.
A: Ethidium bromide–stained 4–20% SDS-HEPES polyacrylamide gel showing various combinations of the methylphosphonic acid aptamer (amplified by Taq polymerase) and β2-microglobulin components alone and together in the presence and absence of 2.5% glutaraldehyde for 16 h at 4°C. B: Same gel stained with Coomassie blue. C: Same gel following a silver stain procedure. Contents of each lane: lane 1, 20 bp DNA ladder (Bio-Rad); lane 2, methylphosphonic acid aptamer; lane 3, β2-microglobulin; lane 4, methylphosphonic acid aptamer plus β2-microglobulin; lane 5, Amplisize 50–2,000 bp DNA ladder (Bio-Rad); lanes 6–9, same contents and order as lanes 2–5 but with 2.5% glutaraldehyde added at pH 6.0; lane 10, wide-range molecular weight markers (Sigma-Aldrich). Shifted bands are indicated by arrow heads.
Unfortunately, the molecular weights of the double-stranded DNA aptamer (anticipated to be 2 × 22.5 kD or 45 kD) and the β2-microglobulin (11.6 kD) to yield a single conjugate weighing 56.6 kD do not seem to be additive or supported by visible banding in the stained gels except possibly in the silver-stained gel shown in Figure 2C (minor band in lane 8 around the 55 kD marker indicated by the white arrowhead in Figure 2C). This lack of clear verification by addition of band molecular weights may be due to false migration of certain bands. For example, the ethidium bromide-stained lanes containing DNA aptamers (lanes 2, 4, 6, and 8 in Figure 2A) appear to lie between 60 and 80 bp as expected for the 72-base MPA aptamer and stain with ethidium bromide as expected for double-stranded DNA, but they also seem to lie between 20.1 kD and 24 kD as would be expected for the single-stranded DNA aptamer reported by the supplier to weigh 22.5 kD, not 45 kD as expected for the double-stranded PCR product employed. Hence, we are placing less credence in addition of observed molecular weights and more credence is relative shifts in mobility such as those seen between bands in lanes 7 and 8 of Figures 2B and 2C or lanes 6 and 7 of Figure 3B which can only be accounted for by the coupling of a fraction of the DNA to β2-microglobulin.
The definitive method to determine if DNA–protein conjugation has occurred involves prior radiolabeling of DNA with 32P followed by conjugation, electrophoresis and autoradiography to track radiolabeled DNA in conjugates as Chu et al.5 did to prove that they had developed a single-stranded DNA aptamer–recombinant gelonin protein conjugate by an electrophoretic mobility shift assay (EMSA). Unfortunately, working with radioisotopes was not possible for the experiments presented herein, but fluorescence-based electrophoretic mobility shift assays have been used as acceptable substitutes for radioisotopic methods.22,23 Therefore, a fluorescence method was used in the present work to demonstrate that higher molecular weight shifted bands from our gels contained AF 546 dye suggesting that DNA–3′-protein conjugates were formed. In most unstained gels we were able to visualize fluorescence in some of the AF 546-labeled and shifted bands when placed on a UV transilluminator (Figure 3A). To further prove that the fluorescence was emanating from AF 546, we cut and eluted the bands and demonstrated that there was fluorescence which peaked around 570–575 nm (characteristic of AF 546 emission) associated with these shifted bands relative to lower molecular weight bands as in spectrum “e” in Figure 3D from band “e” in Figure 3B. Not all shifted bands exhibited intense fluorescence. For example, the band labeled “d” corresponding to spectrum “d” in Figures 3B and 3D did not fluoresce visibly (Figure 3A, lane 7) nor did it fluoresce when eluted (Figure 3D). We hypothesize that this lack of fluorescence may be due either to a lack of DNA in the “d” band or to quenching of AF 546 by an abundance of β2-microglobulin in band “d.”
There is only one source of AF 546 dye in our experiments, namely the AF 546-dUTP-PCR-labeled DNA aptamers. AF 546-dUTP cannot directly couple to the β2-microglobulin because it does not contain a free primary amine or other known reactive group for coupling to glutaraldehyde as illustrated in Figure 3C. Therefore, detection of fluorescence at >570 nm in a shifted gel band in our system must indicate the presence of AF 546-labeled DNA (spectrum “e” in Figure 3D) and DNA–3′-β2-microglobulin conjugate formation. Gel band “e” in the Coomassie blue-stained gel in Figure 3B contains both higher molecular weight DNA (see Figure 3A, lane 7) and protein (Figure 3B, lane 7), yet it fluoresces when excited at 555 nm relative to any of the excised and eluted bands labeled “a–d” in Figures 3B and 3D. It is of interest to note that this band shift occurred after using Deep Vent (exo-) polymerase which, according to the manufacturer, is thought to produce 3′ adenine overhangs in only 30% of its PCR products. Even with this reduced abundance of 3′-adenine overhangs (versus nearly 100% 3′-adenine addition with Taq),12 we were still able to detect a gel shift indicative of a DNA–protein conjugate in lane 7 of Figures 3A and 3B.
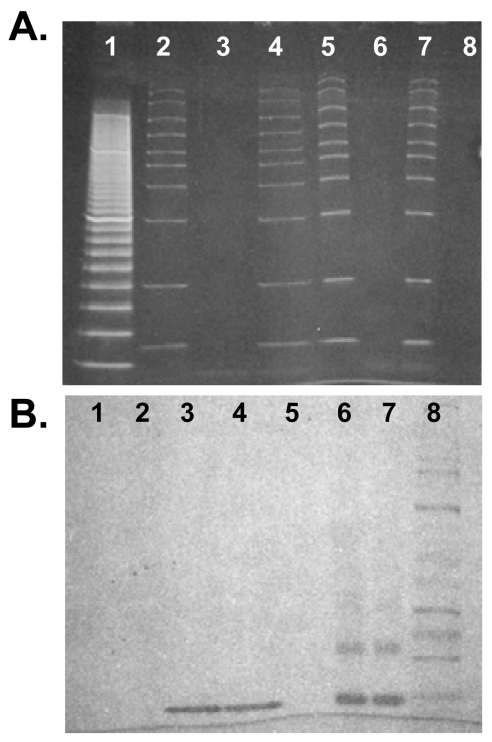
Besides the band mobility retardation and fluorescence evidence, a strong line of evidence for selective conjugation to the 3 ′ end exists in the negative control experiments. We repeatedly saw no evidence of conjugation among the glutaraldehyde-treated Amplisize DNA ladder bands at pH 6.0, because according to their producer (Bio-Rad) these bands are blunt-ended (e.g., compare lanes 5 and 9 of Figs. 2A and C which show no difference in banding pattern). Likewise, at pH 6.0 we were not able to induce blunt-ended Amplisize DNA ladder fragments to couple with β2-microglobulin via glutaraldehyde for lack of a primary amine on the 3′ end (Figure 4B, lanes 6 and 7 are identical). While clear protein–protein dimers and trimers of β2-microglobulin can be seen in lanes 6 and 7 or Figure 4B, these protein–protein conjugates are not further shifted upward by the presence of blunt-ended DNA in the sample run in lane 7. This experiment indicates that a 3′-adenine is necessary for coupling of double-stranded DNA to the protein.
FIGURE 4.
Negative control gels using blunt-ended Amplisize DNA ladder fragments with or without β2-microglobulin in the presence and absence of 2.5% glutaraldehyde at 4°C for 16 h. A: ethidium bromide-stained 4–20% SDS-HEPES polacrylamide gel. B: Coomassie blue stain of same gel. Lane 1, 20 bp DNA ladder beginning at 40 bp since the ladder was defective and started at 40 bp instead of 20 bp as it should have; lane 2, Amplisize DNA; lane 3, β2-microglobulin; lane 4, Amplisize DNA plus β2-microglobulin; lanes 5–7 are the same lanes 2–4 but with 2.5% glutaraldehyde; lane 8, wide-range molecular weight standards.
If the pH of the Amplisize standards is lowered to ≤ 5.0, significant smearing occurs in both the untreated and glutaraldehyde-treated Amplisize markers (data not shown), presumably due to disruption of hydrogen bonding between complementary strands of double-stranded DNA molecules. The double-stranded DNA must remain intact in order for our selective 3′-adenine protein conjugation to be valid, but Williams et al. have elegantly shown that pH 6.0 will not cause separation of the strands of double-stranded DNA.24 The AF 546-dUTP-labeled PCR products may also have been more reactive due to the carboxylic acid group in AF 546 (Figure 3C, arrow) acting locally to acidify the N6 of adenine, because the pH 6.0 PBS buffer was not necessary to achieve a gel shift from the AF 546-labeled DNA plus protein. AF 546-labeled aptamers reacted readily with β2-microglobulin at pH 7.2.
While DNA–DNA and protein–protein dimer, trimer and other byproducts surely form as well, the DNA–protein conjugates of interest clearly appear and may be separated by chromatography on a small scale or in a larger scale industrial purification process. Prior to that process, the double-stranded DNA–protein conjugates could be separated into single-stranded DNA–protein conjugates by rapid heating and high concentrations of chaotropic denaturing agents (e.g., urea, formamide, guanidine salts).
The major differences between the present conjugation method and that of Chu et al.5 are the use of glutaraldehyde and the fact that the protein is forced to locate at the 3 ′ end, because N 6 of the unpaired 3′-adenine is the only vulnerable primary amine available for nucleophilic attack by a linker in the predominately double-stranded and protected DNA. In the work of Chu et al.,5 the bifunctional linker SPDP (N-succinimidyl 3-(2-pyridyldithio)-propionate) appeared to attach to free primary arylamines on adenine, cytosine, or guanine at various locations in the single-stranded aptamer used in those studies. In the present work, all of the adenine, cytosine, and guanine primary amines are involved in hydrogen bonding to the complementary strand of the double-stranded PCR product at neutral pH, so that the only point of attachment for a protein must be the N6 primary amine of the 3′-adenine overhang. Glutaraldehyde is a bifunctional aldehyde linker known to spontaneously attack the N6 of adenine,13,15,18 but others have shown that metal-based catalysts such as Pt(II) complexes can “acidify” the N6 of adenine, thereby making it more reactive.25 We considered platinum-containing compounds, but compounds such as cis-platinum cancer drugs are known to cross-link DNA internally at random and we wished to selectively couple to the 3′ end. Still, the concept of protonating the N6 of adenine to increase its reactivity was intriguing because our coupling system did not work well at neutral pH and low pH (≤5.0) tended to breakdown the double-stranded structure of the Amplisize and aptamer DNA so that smears resulted in the gels (data not shown). Fortunately, pH 6.0 appears to be ideal for mild coupling to the 3 ′ end, because pH 6.0 does not disrupt the hydrogen bonds between the strands of double-stranded DNA.24 Another possible mild, yet effective strategy for DNA–3′-adenine-protein coupling that we envision is the use of natural transferase enzymes to attach ligands with a linker functionality to the N6 of terminal 3′-adenines.26 Nature is replete with methyltransferases for methylation of the N6 of adenine and other transferases that might efficiently place lengthier ligands and linkers onto the N6 atom of 3′-adenines.26
While single-stranded DNA–aptamer-3′-peptide or antisense-3′-peptide conjugates can be made in one elegant solid-phase process, or 3′-functional groups can be attached to single-stranded DNA during solid-phase synthesis for later attachment to functional proteins and other macromolecules, such processes are relatively expensive on a large-scale basis and require a high level of quality control.27 The facile method of direct PCR amplicon-3′-adenine attachment described herein may provide a cost-effective alternative for mass-producing therapeutic DNA–3′-protein conjugates with enhanced pharmacokinetic properties.
ACKNOWLEDGMENTS
The authors gratefully acknowledge support of this project from U.S. Army Contract No. W911SR-06-C-0059, National Institutes of Health Grant No. 1R43AI058423-01, and National Science Foundation Grant No. DMI-0319766. The authors are also grateful for laboratory assistance from Lisa Walker.
References
- 1.Soukchareun S, Haralambidis J, Tregear G. Use of N-alpha-Fmoccysteine (S-thiobutyl) derivatized oligodeoxynucleotides for the preparation of oligodeoxynucleotide-peptide hybrid molecules. Bioconjug Chem. 1998;9:466–475. doi: 10.1021/bc980004h. [DOI] [PubMed] [Google Scholar]
- 2.Soukchareun S, Tregear GW, Haralambidis J. Preparation and characterization of antisense oligonucleotide-peptide hybrids containing viral fusion peptides. Bioconjug Chem. 1995;6:43–53. doi: 10.1021/bc00031a004. [DOI] [PubMed] [Google Scholar]
- 3.Chen CP, Li XX, Zhang LR, et al. Synthesis of antisense oligonucleotide-peptide conjugate targeting to GLUT-1 in HepG-2 and MCF-7 cells. Bioconjug Chem. 2002;13:525–529. doi: 10.1021/bc015540f. [DOI] [PubMed] [Google Scholar]
- 4.Gait MJ. Peptide-mediated cellular delivery of antisense oligonucleotides and their analogues. Cell Mol Life Sci. 2003;60:844–853. doi: 10.1007/s00018-003-3044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu TC, Marks JW, Lavery LA, et al. Aptamer:toxin conjugates that specifically target prostate tumor cells. Cancer Res. 2006;66:5989–5992. doi: 10.1158/0008-5472.CAN-05-4583. [DOI] [PubMed] [Google Scholar]
- 6.Juby CD, Richardson CD, Brousseau R. Facile preparation of 3′ oligonucleotide-peptide conjugates. Tetrahed Lett. 1991;32:879–882. [Google Scholar]
- 7.Gamper HB, Reed MW, Cox T. Facile preparation of nuclease resistant 3 ′ modified oligodeoxynucleotides. Nucl Acids Res. 1993;21:145–150. doi: 10.1093/nar/21.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaschke A, Furste JP, Nordhoff E, et al. Synthesis and properties of oligodeoxyribonucleotide-polyethylene glycol conjugates. Nucl Acids Res. 1994;22:4810–4817. doi: 10.1093/nar/22.22.4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dougan H, Lyster DM, Vo CV, et al. Extending the lifetime of anticoagulant oligodeoxynucleotide aptamers in blood. Nucl Med Biol. 2000;27:289–297. doi: 10.1016/s0969-8051(99)00103-1. [DOI] [PubMed] [Google Scholar]
- 10.Stetsenko DA, Gaitt MJ. A convenient solid-phase method for synthesis of 3′-conjugates of oligonucleotides. Bioconjug Chem. 2001;12:576–586. doi: 10.1021/bc000157g. [DOI] [PubMed] [Google Scholar]
- 11.Stetsenko DA, Malakhov AD, Gait MJ. Total stepwise solid-phase synthesis of oligonucleotide-(3′-N)-peptide conjugates. Org Lett. 2002;4:3259–3262. doi: 10.1021/ol026502u. [DOI] [PubMed] [Google Scholar]
- 12.Zhou MY, Gomez-Sanchez CE. Universal TA cloning. Curr Issues Mol Biol. 2000;2:1–7. [PubMed] [Google Scholar]
- 13.Henderson A, Bleasdale C, Clegg W, Golding BT. 2,6-Diarylaminotetrahydropyrans from reactions of glutaraldehyde with anilines: models of biomolecule cross-linking. Chem Res Toxicol. 2004;17:378–382. doi: 10.1021/tx034177t. [DOI] [PubMed] [Google Scholar]
- 14.Hopwood D. The reactions of glutaraldehyde with nucleic acids. Histochem J. 1975;7:267–276. doi: 10.1007/BF01003595. [DOI] [PubMed] [Google Scholar]
- 15.Gacesa P, Whish WJD. The immobilization of adenine nucleotides on polysaccharides by using glutaraldehyde coupling and borohydride reduction. Biochem J. 1978;175:349–352. doi: 10.1042/bj1750349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rall TW, Lehne RA. Evidence for cross-linking of cyclic AMP constituents of brain tissue by aldehyde fixatives: potential utility in histochemical procedures. J Cyclic Nucleotide Res. 1982;8:243–265. [PubMed] [Google Scholar]
- 17.Alderson T. Formaldehyde-induced mutagenesis: a novel mechanism for its action. Mutat Res. 1985;154:101–110. doi: 10.1016/0165-1110(85)90022-3. [DOI] [PubMed] [Google Scholar]
- 18.Blackburn MG. Reactions with electrophiles. In: Blackburn MG, Gait MJ, editors. Nucleic Acids in Chemistry and Biology. 2. New York: Oxford University Press; 1996. p. 291. [Google Scholar]
- 19.Bruno JG, Carrillo MP, Phillips T, et al. J Fluoresc. In Press: 2008. Competitive FRET-aptamer-based detection of methylphosphonic acid: a common nerve agent metabolite. [DOI] [PubMed] [Google Scholar]
- 20.Bruno JG, Carrillo MP, Phillips T. Development of DNA aptamers to a foot-and-mouth disease peptide for competitive FRET-based detection. J Biomolec Techn. 2008;19:109–115. [PMC free article] [PubMed] [Google Scholar]
- 21.Bruno JG, Sincock SA, Stopa PJ. Highly selective acridine and ethidium staining of bacterial DNA and RNA. Biotechnic Histochem. 1996;71:130–136. doi: 10.3109/10520299609117149. [DOI] [PubMed] [Google Scholar]
- 22.Ruscher K, Reuter M, Kupper D, et al. A fluorescence based non-radioactive electrophoretic mobility shift assay. J Biotechnol. 2000;78:163–170. doi: 10.1016/s0168-1656(00)00207-8. [DOI] [PubMed] [Google Scholar]
- 23.Murphy K, Shimamura T, Bejeck BE. Use of fluorescently labeled DNA and a scanner for electrophoretic mobility shift assays. BioTechniques. 2001;30:504–508. doi: 10.2144/01303bm07. [DOI] [PubMed] [Google Scholar]
- 24.Williams MC, Wenner JR, Rouzina I, Bloomfield VA. Effect of pH on the overstretching transition of double-stranded DNA: evidence of force-induced DNA melting. Biophys J. 2001;80:874–881. doi: 10.1016/S0006-3495(01)76066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Añorbe MG, Lüth MS, Roitzsch M, et al. Perturbation of the NH2 pKa value of adenine in platinum(II) complexes: Distinct stereochemical internucleobase effects. Chem Eur J. 2004;10:1046–1057. doi: 10.1002/chem.200305509. [DOI] [PubMed] [Google Scholar]
- 26.Abe I, Tanaka H, Abe T, et al. Enzymatic formation of unnatural cytokinin analogs by adenylate isopentenyltransferase from mulberry. Biochem Biophys Res Commun. 2007;355:795–800. doi: 10.1016/j.bbrc.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 27.Cload ST, McCauley TG, Keefe AD, Healy JM, Wilson C. Properties of therapeutic aptamers. In: Klussmann S, editor. The Aptamer Handbook. Weinheim, Germany: Wiley-VCH Verlag GmbH; 2006. pp. 398–399. ch 17. [Google Scholar]