Abstract
Many of the protein–protein interactions that are essential for eukaryotic intracellular signal transduction are mediated by protein binding modules including SH2, SH3, and LIM domains. Nck is a SH3- and SH2-containing adaptor protein implicated in coordinating various signaling pathways, including those of growth factor receptors and cell adhesion receptors. We report here the identification, cloning, and characterization of a widely expressed, Nck-related adaptor protein termed Nck-2. Nck-2 comprises primarily three N-terminal SH3 domains and one C-terminal SH2 domain. We show that Nck-2 interacts with PINCH, a LIM-only protein implicated in integrin-linked kinase signaling. The PINCH-Nck-2 interaction is mediated by the fourth LIM domain of PINCH and the third SH3 domain of Nck-2. Furthermore, we show that Nck-2 is capable of recognizing several key components of growth factor receptor kinase-signaling pathways including EGF receptors, PDGF receptor-β, and IRS-1. The association of Nck-2 with EGF receptors was regulated by EGF stimulation and involved largely the SH2 domain of Nck-2, although the SH3 domains of Nck-2 also contributed to the complex formation. The association of Nck-2 with PDGF receptor-β was dependent on PDGF activation and was mediated solely by the SH2 domain of Nck-2. Additionally, we have detected a stable association between Nck-2 and IRS-1 that was mediated primarily via the second and third SH3 domain of Nck-2. Thus, Nck-2 associates with PINCH and components of different growth factor receptor-signaling pathways via distinct mechanisms. Finally, we provide evidence indicating that a fraction of the Nck-2 and/or Nck-1 proteins are associated with the cytoskeleton. These results identify a novel Nck-related SH2- and SH3-domain–containing protein and suggest that it may function as an adaptor protein connecting the growth factor receptor-signaling pathways with the integrin-signaling pathways.
INTRODUCTION
Protein–protein nteractions play central roles in signal transduction leading to cell proliferation, differentiation, survival, migration, and cytoskeleton organization. Many of the protein–protein interactions are mediated by adaptor proteins, noncatalytic proteins comprising multiple protein-binding modules such as Src homology (SH) and LIM domains. Nck is a SH2/SH3-containing protein (Lehmann et al., 1990) implicated in transducing signals from a variety of cell surface receptor kinases (Li et al., 1992; Meisenhelder and Hunter, 1992; Park and Rhee, 1992; Lee et al., 1993; Holland et al., 1997; Stein et al., 1998), integrin-associated focal adhesion kinase (Choudhury et al., 1996; Schlaepfer et al., 1997), and a number of other catalytic and noncatalytic proteins (see, for instance,
Lee et al., 1993; Chou and Hanafusa, 1995; Hu et al., 1995; Rivero-Lezcano et al., 1995; Birge et al., 1996; Bokoch et al., 1996; Galisteo et al., 1996; Kitamura et al., 1996; Quilliam et al., 1996; Lu et al., 1997; Lussier and Larose, 1997; Su et al., 1997; Anton et al., 1998). Nck is capable of physically associating with the signaling proteins through direct or indirect protein–protein interactions. For example, Nck binds directly to the ligand-stimulated PDGF receptor-β (Nishimura et al., 1993). On the other hand, Nck appears to associate with the activated EGF receptor indirectly via the GTPase-activating protein (GAP)–associated phosphotyrosine protein p62 (Tang et al., 1997). Studies using monoclonal anti-phospholipase C-γ1 antibodies have shown that Nck shares a common epitope with phospholipase C-γ1 (Park and Rhee, 1992; Meisenhelder and Hunter, 1992), and a mouse Nck cDNA has been isolated using an anti-phospholipase C-γ1 monoclonal antibody (Park, 1997). Nck is ubiquitously expressed in tissues (Li et al., 1992; Park and Rhee, 1992). A number of studies have shown that Nck plays important roles in control of fundamental cellular processes including cell proliferation, differentiation, migration, and cytoskeleton organization. Overexpression of Nck in mammalian cells induced anchorage-independent growth in culture and tumor formation in vivo, demonstrating that Nck is an oncoprotein (Chou et al., 1992; Li et al., 1992). Mutations in the Drosophila Nck homologue Dreadlocks (Dock), on the other hand, disrupted growth cone guidance and targeting in photoreceptor (Garrity et al., 1996). Moreover, Rao and Zipursky (1998) have demonstrated that different SH domains of Dock are utilized in different ways in different neurons. Finally, expression of mutated Nck in Xenopus laevis embryos respecified mesodermal cell fate in embryonic development (Tanaka et al., 1997). The molecular mechanisms by which Nck functions in signal transduction, however, are not completely understood and are likely complex, as each of the four Nck SH domains could potentially mediate one or more protein–protein interactions.
PINCH (Rearden, 1994) is a widely expressed and evolutionarily conserved protein comprising primarily five (the most among all LIM-containing proteins) LIM domains, which are cysteine-rich consensus sequences implicated in mediating protein–protein interactions (Schmeichel and Beckerle, 1994; Dawid et al., 1995; Gill, 1995). A mutation in the Caenorhabditis elegans PINCH gene homologue unc-97 causes locomotory defects resulting in an uncoordinated movement phenotype, indicating that the PINCH homologue is functionally important for muscle attachment assembly and touch neuron functions in C. elegans (Hobert, personal communication). At the molecular level, PINCH interacts with integrin-linked kinase (ILK) (Tu, Li, Goicoechea, and Wu, unpublished data), an ankyrin repeat-containing serine/threonine protein kinase that has been implicated in integrin (Hannigan et al., 1996), growth factor (Delcommenne et al., 1998), and Wnt (Novak et al., 1998) signaling pathways. ILK regulates integrin-mediated cell adhesion (Hannigan et al., 1996), the activation of the LEF-1/β-catenin–signaling pathway (Novak et al., 1998), and E-cadherin expression and pericellular fibronectin matrix assembly (Wu et al., 1998). Moreover, overexpression of ILK in epithelial cells promoted cell cycle progression in an anchorage-independent manner (Radeva et al., 1997), resulting in anchorage-independent cell growth in culture and tumor formation in vivo (Wu et al., 1998). Recent in vivo studies have shown that the expression of ILK could be regulated by erbB-2, a member of the erbB growth factor receptor tyrosine kinase family (Xie et al., 1998).
ILK comprises three structurally and functionally distinctive domains (Hannigan et al., 1996; Delcommenne et al., 1998). The C-terminal domain is highly homologous to the catalytic domains of a large number of protein kinases and is responsible for the kinase activity. In addition, it includes a binding site for the integrin β1 cytoplasmic domain (Hannigan et al., 1996). The kinase activity of ILK activity is regulated by cell–extracellular matrix interaction (Hannigan et al., 1996) as well as by insulin (Delcommenne et al., 1998). N-terminal to the kinase domain is a pleckstrin homology-like domain that likely binds phosphatidylinositol-(3,4,5) triphosphate and participates in the regulation of the kinase activity (Delcommenne et al., 1998). The ILK N-terminal-most domain comprises primarily four ankyrin repeats, which define a structure mediating the interaction with PINCH. We recently found that only one (LIM1) of five LIM domains of PINCH is required for mediating the PINCH–ILK interaction (Tu, Li, Goicoechea, and Wu, unpublished data), leaving the other LIM domains free to interact with other proteins. To understand the ILK/PINCH-signaling pathway at the molecular level, we have begun to identify and characterize additional PINCH interactive proteins. We report here the identification, cloning, and characterization of a novel PINCH interactive, SH2- and SH3-domain–containing protein that is structurally related to Nck (termed as Nck-2, and the human Nck that was initially cloned by Lehmann et al. [Lehmann et al., 1990] will be referred as Nck-1 in this paper for clarity). In addition to interacting with PINCH, we show that Nck-2 is capable of recognizing several key components of growth factor receptor kinase-signaling pathways including EGF receptors, PDGF receptor-β, and IRS-1. These results suggest that Nck-2 likely functions as an adaptor protein physically connecting the growth factor receptor-signaling pathways with the ILK- and integrin-signaling pathways.
MATERIALS AND METHODS
Cells, Antibodies, cDNAs, and Other Reagents
Human 293 embryonal kidney cells were from American Type Culture Collection (Manassas, VA). Human A431 epidermoid carcinoma cells and mouse NIH 3T3 cells were kindly provided by Drs. Jeffrey E. Kudlow, Stuart J. Frank, and Louise T. Chow (University of Alabama at Birmingham). Anti-IRS-1 (C-20), anti-PDGF receptor-β (P-20), anti-EGF receptor (1005), and anti-phosphotyrosine (PY20) antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal anti-HA antibody was from Zymed Laboratories (South San Francisco, CA). Full-length human Nck-1 cDNA was generated from a human lung cDNA library by PCR using primers corresponding to Nck-1 sequences (5′-GTCGAATTCATGGCAGAAGAAGTGGTGGTGG-3′ and 5′-GTGCTCGAGTCATGATAAATGCTTGACAAGA-3′) and the sequence of the product was verified by DNA sequencing. A cDNA encoding SAP97 (Muller et al., 1995) was a gift from Dr. Craig C. Garner (University of Alabama at Birmingham). EGF was purchased from Calbiochem-Novabiochem (La Jolla, CA). PDGF and insulin were from Life Technologies (Gaithersburg, MD).
Yeast Two-Hybrid Assays
A cDNA fragment encoding the human PINCH LIM4 domain (amino acid residues 192–249) was amplified by PCR and inserted into the EcoRI/XhoI site in the pLexA vector (CLONTECH, Palo Alto, CA). The sequence of the bait construct (pLexA/LIM4) was verified by DNA sequencing and introduced into EGY48[P8OP-lacZ] yeast cells by transformation. The transformants were used to screen a human lung MATCHMAKER LexA cDNA library (>5.7 × 106 independent clones, CLONTECH). Briefly, the EGY48[p8op-lacZ; pLexA/LIM4] cells transformed by the library plasmids were selected by plating on the SD/-His/-Ura/-Trp medium (CLONTECH). The expression of proteins encoded by the pB42AD/library vectors was induced by growing the cells in the presence of galactose (SD/Gal/Raf/-His/-Ura/-Trp medium, CLONTECH). Twenty four positive clones, as indicated by activation of both reporter genes (LEU2 and lacZ), were identified. The plasmids were isolated from the positive yeast clones and used to transform E. coli KC8 cells. The KC8 cells containing the pB42AD vectors were selected by growing in medium lacking tryptophan. The pB42AD plasmids were isolated from the KC8 cells and the sequences of the inserts were determined by DNA sequencing.
In addition to library screening, we performed yeast two-hybrid binding assays to determine the interactions between specific domains of Nck-2, PINCH, and other proteins. Yeast cells were cotransformed with purified pB42AD and pLexA expression vectors encoding various Nck-2, PINCH, and Nck-1 sequences or other control proteins as specified in each experiment. The transformants were selected as described above and plated on leucine-deficient selection medium containing 80 μg/ml X-gal (SD/Gal/Raf/-His/-Ura/-Trp/-Leu/X-Gal medium, CLONTECH). The growth of blue colonies in the leucine-deficient medium indicates a positive interaction. Additionally, the β-galactosidase activities of a number of yeast transformants were quantified using o-nitrophenyl β-d-galactopyranoside as a substrate in a liquid culture assay (Yeast Protocols Handbook, CLONTECH), and the results were consistent with the blue colony growth assay (our unpublished results).
5′-RACE Reactions and Generation of Full-length Nck-2 cDNA
The 5′-cDNA fragment of Nck-2 was obtained by 5′-RACE PCR using Marathon-Ready cDNA from human fetal lungs (22–23 wk, CLONTECH). The full-length Nck-2 cDNA was generated by end-to-end PCR using 5′-CGGAATTCAAGCTTATGACAGAAGTTATTGTGATAGCC3-′ and 5′-GACGTACTCGAGTCACTGCAGGGCCCTGACGAGGTAGA-3′ as primers.
DNA Sequencing
Sequences of DNA fragments were determined manually using a Sequenase Version 2.0 kit (United States Biochemicals, Cleveland, OH) and a Thermo Sequenase kit (CLONTECH).
Northern Blot
A 32P-labeled human Nck-2 cDNA probe was prepared by labeling the full-length human Nck-2 cDNA using a random-primed DNA-labeling kit (Boehringer Mannheim, Indianapolis, IN). A blot containing equal amounts of polyA+ RNA (2 μg/lane) from human heart, brain, spleen, lung, liver, skeletal muscle, kidney, and testis (CLONTECH) was hybridized with the 32P-labeled Nck-2 cDNA probe following the manufacturer’s protocol. The mRNA bands hybridized with the radioactively labeled Nck-2 cDNA probe were visualized by autoradiography.
Nck-2 and PINCH Mutations
DNA fragments encoding Nck-2 and PINCH deletion mutants were generated by PCR, and the amino acid residues encoded were specified in each experiment. 5′-EcoRI and 3′-XhoI restriction sites were incorporated into the amplified products via PCR primers to facilitate the insertion of the Nck-2 and PINCH DNA fragments into the pB42AD and pLexA expression vectors. A QuickChange site-directed mutagenesis system (Stratagene, La Jolla, CA) was used to change the conserved W (amino acid residue 234) to K in the third SH3 domain of Nck-2. Correct reading frame and sequences of all the constructs were verified by DNA sequencing.
Expression of Recombinant GST- and His-tagged Fusion Proteins
To generate GST-Nck-2 and GST-Nck-1 fusion proteins, human Nck-2 and Nck-1 cDNA sequences (as specified in each experiment) were amplified by PCR and inserted into the EcoRI/XhoI site of a pGEX-5x-1 vector (Pharmacia, Piscataway, NJ). The recombinant vectors were then used to transform E. coli cells (M20). The expression of the GST-Nck-2 and GST-Nck-1 fusion proteins were induced with isopropyl β-d-thiogalactopyranoside, and the proteins were purified with glutathione-Sepharose 4B beads. To produce His-tagged PINCH proteins, human PINCH cDNA sequences (as specified in each experiment) were amplified by PCR and inserted into the NdeI/BamHI site of a pET-15b vector (Novagen, Madison, WI). The recombinant vectors were then used to transform E. coli BL21(DE3) cells, and the recombinant proteins were purified with His-Bind Resin (Novagen) following the manufacturer’s protocol.
Coprecipitation Assays Using Mammalian Proteins
Human 293 embryonal kidney cells were cultured in Eagle’s MEM supplemented with 10% FBS. Human A431 epidermoid carcinoma cells and NIH 3T3 cells were grown in DMEM supplemented with 10% FBS. For stimulation with EGF, PDGF, or insulin, cells were seeded in 100-mm cell culture plates and grown until approximately 70–80% confluent. The cells were then serum starved overnight, followed by stimulation with EGF, PDGF, or insulin as specified in each experiment. Cells were washed once with cold PBS and lysed with the lysis buffer (0.5% Nonidet P-40 in 10 mM Tris-HCl buffer, pH 7.1, containing 50 mM NaCl, 30 mM sodium pyrophosphate, 50 mM sodium fluoride, 200 μm sodium orthovanadate, 1 mg/ml BSA, 0.2 mM 4-(2-aminoethyl)benzenesulfonylfluoride, HCl, 10 μg/ml aprotinin, 1 μg/ml pepstatin A, and 5 μg/ml leupeptin). The lysates were clarified by centrifugation at 10,000 × g for 15 min and preincubated with glutathione-Sepharose 4B beads (Pharmacia) at 4°C for 0.5 h. The beads were removed by centrifugation at 3,000 × g for 5 min, and the clarified cell lysates were incubated with equal amounts (as specified in each experiment) of GST-fusion proteins containing the full-length or various domains of Nck-2, Nck-1 or other proteins, or GST alone as a negative control, for 30 min at 4°C. At the end of the incubation, the solutions were mixed with glutathione-Sepharose 4B beads, incubated for 1 h or longer, and the GST fusion proteins were then precipitated with glutathione-Sepharose 4B beads by centrifugation. The precipitates were washed five times with washing buffer (20 mM Tris-HCl buffer, pH 7.6, containing 300 mM NaCl, 2 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS). After washing, IRS-1, PDGF receptor-β, and EGF receptors that were associated with the full length or specific SH domains of Nck-2 or Nck-1 were detected by immunoblotting with specific antibodies as indicated in each experiment, a horseradish peroxidase-conjugated anti-rabbit IgG antibody (27 ng/ml), and the SuperSignal chemiluminescent substrate (Pierce Chemical, Rockford, IL).
In control experiments, IRS-1, PDGF receptor-β, and EGF receptors were precipitated with antibodies recognizing IRS-1, PDGF receptor-β, and EGF receptor, respectively. The tyrosine phosphorylation of IRS-1, PDGF receptor-β, or EGF receptors upon stimulation with insulin, PDGF, or EGF was determined by immunoblotting of the precipitates with a mouse monoclonal anti-phosphotyrosine antibody (PY20, 0.2 μg/ml), a horseradish peroxidase-conjugated anti-mouse IgG antibody (40 ng/ml), and the SuperSignal chemiluminescent substrate (Pierce Chemical).
Coprecipitation Assays Using Purified GST- and His-tagged Fusion Proteins
Affinity-purified GST-Nck-2 (2.5 μg), or GST (2.5 μg) as a negative control, was mixed with 2.5 μg of His-tagged PINCH LIM1–4 (residues 1–249) or His-tagged PINCH LIM1 (residues 1–70) in binding buffer (0.2% Triton X-100, 10 mM Tris, and 100 mM NaH2PO4, pH 8.0; total mixture volume = 0.6 ml). The mixtures were incubated with 50 μl His-Bind resin for 3 h on a rocker at 4°C. At the end of incubation, the resin was washed six times with the binding buffer, and the bound proteins were eluted with 40 μl of the binding buffer supplemented with 50 mM EDTA. The proteins eluted from the resin were analyzed by immunoblotting (each lane contained 10 μl of the eluted proteins mixed with 5 μl of SDS-PAGE sample buffer) with a polyclonal rabbit anti-GST-Nck-2 antibody (see below). In separate control experiments, we have determined that the polyclonal rabbit anti-GST-Nck-2 antibody recognized both GST and the GST-Nck-2 fusion protein in immunoblotting assays (our unpublished results).
Rabbit Polyclonal and Mouse Monoclonal anti-Nck-2 Antibodies
Rabbit polyclonal anti-Nck-2 antibodies were produced by immunizing New Zealand white rabbits with a GST fusion protein containing the C-terminal three SH domains of Nck-2 (residues 115–380) using a standard protocol. To generate monoclonal antibodies recognizing Nck-2, BALB/C mice were immunized with 100 μg of the GST fusion protein containing the C-terminal region of Nck-2 (residues 115–380) in complete Freund’s adjuvant administered subcutaneously. Subsequent boosts, given at 3-wk intervals, consisted of the same amount of antigen in incomplete Freund’s adjuvant. To facilitate antibody screening, we produced a MBP (maltose binding protein) fusion protein containing the full-length Nck-2 by inserting the full-length human Nck-2 cDNA into the EcoRI/SalI site of the pMAL-C2 vector (New England BioLabs, Beverly, MA). The MBP-Nck-2 fusion protein was expressed in E. coli (DH5α) and purified by affinity chromatography using amylose-agarose beads (New England BioLabs). Mouse sera were collected and tested by ELISA and immunoblot using the affinity-purified MBP-Nck-2 fusion proteins. Three days after the final boost, lymph node and spleen cells were collected and fused with the P3X63-Ag8.653 myeloma line using standard procedures. Fourteen days after fusion, supernatants were screened for antibody activity with MBP-Nck-2 by ELISA. Confirmation of specificity was obtained by immunoblot analysis using affinity-purified MBP-Nck-2 fusion proteins. The epitopes recognized by the monoclonal antibodies were mapped by immunoblotting using recombinant proteins containing individual SH domains of Nck-2.
RESULTS
Identification, Cloning, and Primary Structure of a Novel PINCH Interactive Protein Nck-2
We used a yeast two-hybrid system to identify proteins that interact with the LIM domains of PINCH. A bait construct (pLexA/LIM4) encoding the fourth LIM domain of PINCH (amino acid residues 192–249) was used to screen a human lung LexA cDNA library (>5.7 × 106 independent clones). Twenty four positive clones were obtained. DNA sequencing showed that plasmids from 16 of the 24 positive clones contained an open reading frame encoding an identical protein sequence (266 residues). Clone 4 from this positive group was used for further analyses. Transformation of yeast cells with purified pB42AD-Clone4 and the pLexA constructs encoding PINCH and the LIM4 domain confirmed that it specifically interacted with PINCH as well as the PINCH LIM4 domain (Table 1). In control experiments, elimination of either the PINCH LIM4 sequence or the Clone4 coding sequence resulted in inactivation of both reporter genes, indicating that neither protein can activate the reporter genes in the absence of the other binding partner. In addition, replacement of the PINCH sequence with those of irrelevant proteins (e.g., lamin C) abolished the interaction (Table 1), further confirming the specificity of the interaction.
Table 1.
Identification of a PINCH interactive protein by yeast two-hybrid binding assays
| pB42AD construct | pLexA construct | Reporter gene
|
|
|---|---|---|---|
| LEU2 | lacZ | ||
| pB42AD-Clone4 | pLexA-PINCH | + | + |
| pB42AD-Clone4 | pLexA-LIM4 | + | + |
| pB42AD-Clone4 | pLexA | − | − |
| pB42AD-Clone4 | pLexA-Lamin C | − | − |
| pB42AD | pLexA-LIM4 | − | − |
The pB42AD-Clone4 plasmid encodes a protein fragment containing two SH3 domains and one SH2 domain (residues 115–380, Figure 1A). The yeast two-hybrid binding assays using purified DNA constructs were performed as described in MATERIALS AND METHODS.
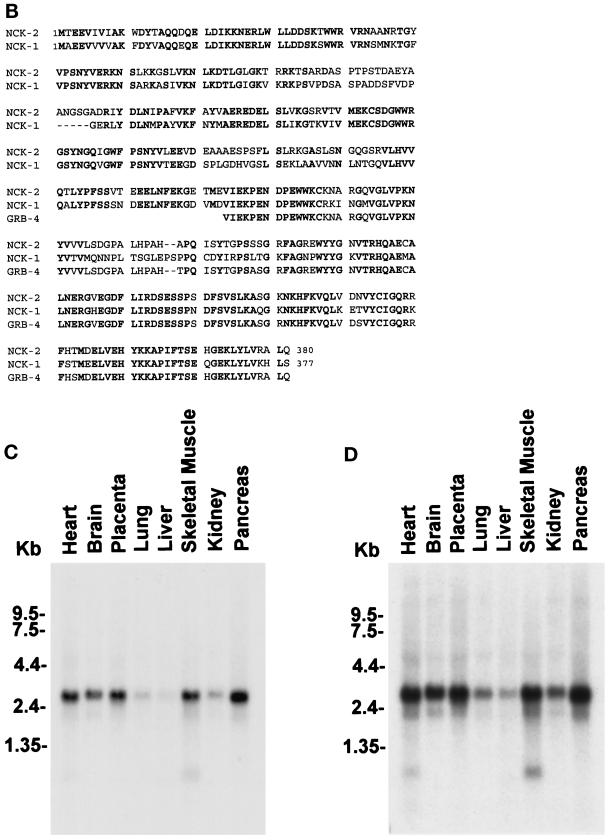
The full-length cDNA encoding the PINCH interactive protein was obtained by 5′-RACE and end-to-end PCR using Marathon-Ready cDNA from human fetal lungs (CLONTECH). The full-length protein comprises primarily three N-terminal SH3 domains and one C-terminal SH2 domain (Figure 1A). Sequence analysis indicated that it was structurally related to human Nck-1 (68% identical at the protein level) (Figure 1B) and thus was designated as Nck-2. In addition, the C-terminal sequence (residues 224–380) of Nck-2 is highly homologous (96% identical) to the C-terminal sequence of GRB-4 (Margolis et al., 1992)(Figure 1B).
Figure 1.
Primary structure and tissue distribution of human Nck-2. (A) Nucleotide and deduced amino acid sequences of human Nck-2. The Nck-2 amino acid sequence is shown below the nucleotide sequence, and the stop codon is indicated by a star. The sequence data reported here have been deposited in the GenBank (accession number AF047487). (B) Human Nck-2 protein sequence is aligned with protein sequences of human Nck-1 and mouse GRB-4, and the identical residues are in bold. (C and D) Northern blot analysis of Nck-2 mRNA in human tissues. Two micrograms of polyA+ RNA from human tissues as indicated in the figure were hybridized with a 32P-labeled Nck-2 cDNA probe, and the hybridized mRNA bands were visualized by autoradiography after exposure for 3 (C) or 13 (D) d.
Nck-2 Is Widely Expressed in Human Tissues
Previous studies have shown that Nck-1 is ubiquitously expressed (Li et al., 1992; Park and Rhee, 1992). To determine tissue expression of Nck-2, we analyzed the distributions of Nck-2 mRNA in human tissues by Northern blot. A predominant Nck-2 transcript (3 kilobase [kb]) was detected in all the human tissues that were analyzed, with stronger signals in the heart, brain, placenta, skeletal muscle, and pancreas (Figure 1C). Two additional, smaller transcripts, which were more apparent upon longer exposure (Figure 1D), were also observed. One of the minor transcripts has an apparent size of 2.2 kb. The 2.2 transcript was widely expressed in the tissues and most likely represented the human Nck-1 mRNA. The other transcript was relatively small (<1.35 kb) and was exclusively expressed in the skeletal muscle and the heart among the tissues analyzed. The nature of this small transcript is not known.
Nck-2 Interacts with PINCH
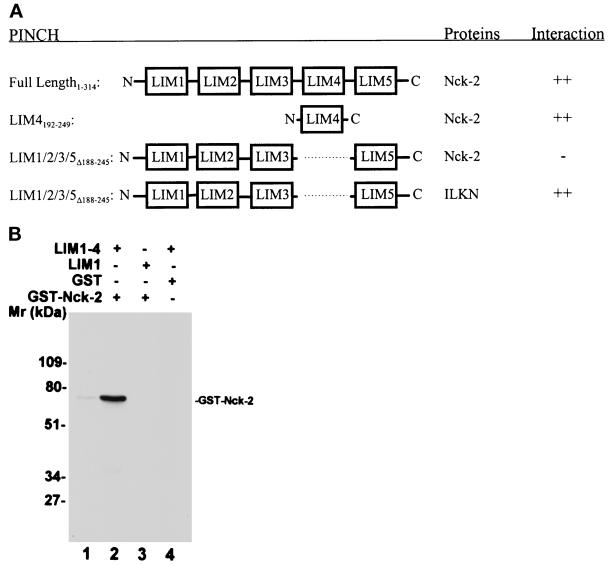
We next tested the ability of the full-length Nck-2 to interact with PINCH. The full-length Nck-2, like its C-terminal fragment containing two SH3 domains and one SH2 domain (Table 1, Clone4), readily interacted with PINCH in yeast two-hybrid binding assays (Figure 2A). To test whether Nck-2 could directly interact with PINCH in vitro, we generated a GST fusion protein containing the full-length Nck-2 and His-tagged fusion proteins containing either the LIM domains 1–4 or the LIM1 domain of PINCH and analyzed their ability to interact with each other in solution. The GST-Nck-2 fusion protein bound to the His-tagged fusion protein containing the LIM1–4 domains of PINCH (Figure 2B, lane 2). In control experiments, the GST-Nck-2 fusion protein failed to bind to the His-tagged fusion protein containing the LIM1 domain of PINCH (Figure 2B, lane 3). Additionally, no interaction was detected between GST and the His-tagged fusion protein containing the LIM1–4 domains of PINCH (Figure 2B, lane 4). Thus, the Nck-2 fusion protein is capable of interacting with specific LIM domains of PINCH in solution.
Figure 2.
Analyses of the PINCH-Nck-2 interaction. (A) Yeast two-hybrid binding assays. The cDNAs encoding PINCH sequences were inserted into the pB42AD vector. The cDNAs encoding Nck-2 and ILK sequences were inserted into the pLexA vector. The protein–protein interactions were analyzed by yeast two-hybrid assays as described in MATERIALS AND METHODS. ++, Growth of blue colonies on the leucine-deficient selection medium containing 80 μg/ml X-gal was detected within 1 d; −, no blue colony was detected after 5 d. LIM1/2/3/5▵188–245, PINCH mutant in which the LIM4 domain (residues 188–245) was deleted; LIM4192–249, PINCH mutant containing the LIM4 domain (residues 192–249); ILKN, the N-terminal domain of ILK (residues 1–163). (B) Coprecipitation assays. Affinity-purified GST-Nck-2 (lanes 2 and 3), or GST as a control (lane 4), was mixed with His-tagged PINCH LIM1–4 (residues 1–249)(lanes 2 and 4) or His-tagged PINCH LIM1 (residues 1–70)(lane 3). The GST-Nck-2 was coprecipitated with the His-tagged LIM1–4 and detected by immunoblotting with a polyclonal rabbit anti-GST-Nck-2 antibody as described in MATERIALS AND METHODS. Lane 1 was loaded with 10 ng of affinity- purified GST-Nck-2 fusion protein.
The LIM4 Domain of PINCH Mediates the Interaction with Nck-2
The results described in Table 1 indicate that the PINCH LIM4 domain is sufficient for interacting with the C-terminal fragment of Nck-2 (Clone4). We next determined whether the PINCH LIM4 domain is also sufficient for interacting with the full-length Nck-2 by yeast two-hybrid binding assays. The results showed that the LIM4 domain of PINCH readily interacted with the full-length Nck-2 (Figure 2A). Thus, the LIM4 domain of PINCH is sufficient for mediating the interaction with Nck-2. To determine whether the LIM4 domain is also required for the PINCH–Nck-2 interaction, we generated a PINCH mutant in which the LIM4 domain is deleted. Unlike the full-length PINCH or the LIM4 domain, the PINCH mutant lacking the LIM4 domain was unable to interact with Nck-2 (Figure 2A). In control experiments, the PINCH mutant lacking the LIM4 domain interacted with the N-terminal domain of ILK (Figure 2A), indicating that the PINCH mutant was expressed and functional. We conclude from these experiments that the LIM4 domain of PINCH is both sufficient and necessary for interacting with Nck-2.
The Third SH3 Domain of Nck-2 Mediates the Interaction with PINCH
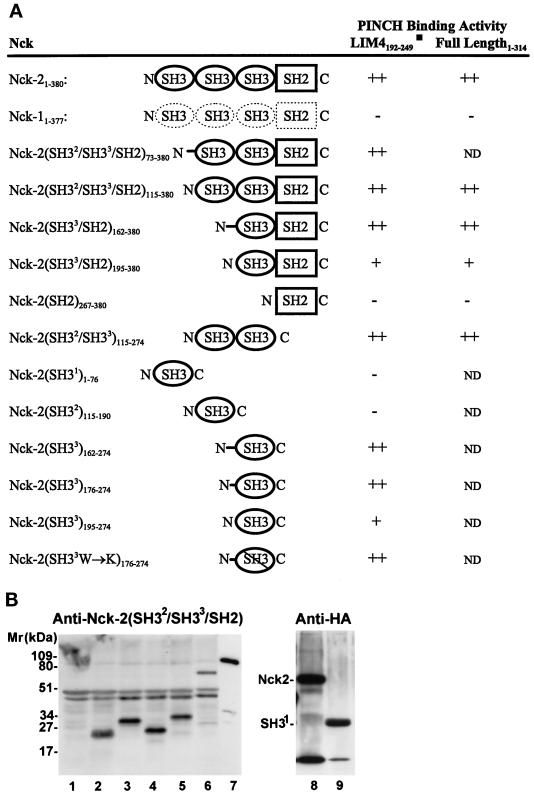
We next sought to identify the domain of Nck-2 that mediates the interaction with PINCH. To do this, we generated a series of Nck-2 mutants and determined their PINCH binding activities (Figure 3). As expected, the full-length Nck-2 readily interacted with PINCH and the LIM4 domain (Figure 3A). In addition, the Nck-2 mutants in which the first SH3 domain, the first and the second N-terminal SH3 domains, or the SH2 domain was (were) deleted retained PINCH binding activity (Figure 3A). By contrast, N-terminal deletion that removed all three Nck-2 SH3 domains completely abolished the PINCH binding activity (Figure 3A). In control experiments, we analyzed the yeast cells harboring the pB42AD expression vector that contains the PINCH binding-defective mutant with an antibody raised against a GST-fusion protein containing the C-terminal three SH domains of Nck-2 (anti-Nck-2(SH32/SH33/SH2)). The results showed that the PINCH binding-defective Nck-2 mutant was expressed in the yeast cells (Figure 3B, lane 5), suggesting that the lack of binding was not caused by a defect in protein expression. We conclude from these experiments that 1) the SH2 domain is neither required nor sufficient for the interaction with PINCH, and 2) the third SH3 domain, but not the first or the second SH3 domain, is required for the interaction with PINCH.
Figure 3.
Mapping of the PINCH binding site on Nck-2. The cDNA sequences encoding the full-length or various domains of Nck-2 and Nck-1 were inserted into the pB42AD vector. The cDNAs encoding the full-length PINCH or its LIM4 domain were inserted into the pLexA vector. (A) PINCH binding activity. The numbers in subscript indicate amino acid residues of Nck-2, Nck-1, and PINCH encoded by each construct. W234→K, the conserved W (amino acid residue 234) in the third SH3 domain of Nck-2 was changed to K. The PINCH binding activity was determined based on the interaction of each of the Nck proteins with the LIM4 domain and/or the full length PINCH in yeast two-hybrid assays as described in MATERIALS AND METHODS. ++, Growth of blue colonies on the leucine-deficient selection medium containing 80 μg/ml X-gal was detected within 1 d; +, growth of blue colonies on the leucine-deficient selection medium containing 80 μg/ml X-gal was detected within 2 or 3 d; −, no blue colony was detected after 5 d. ND, not determined. (B) Expression of B42AD fusion proteins containing various Nck-2 and Nck-1 sequences. The expression of the Nck fusion proteins in yeast cells harboring the pLexA-LIM4 and the pB42AD that contains the first SH3 domain of Nck-2 (residues 1–76)(lanes 1 and 9), the second SH3 domain of Nck-2 (residues 115–190)(lane 2), the third SH3 domain of Nck-2 (long form, residues 176–274)(lane 3), the third SH3 domain of Nck-2 (short form, residues 195–274)(lane 4), the SH2 domain of Nck-2 (residues 267–380)(lane 5), the full- length Nck-1 (lane 6), or the full-length Nck-2 (lane 8) were determined by immunoblotting. Yeast cells from 3-ml cultures were extracted with 200 μl of urea/SDS protein extraction buffer (40 mM Tris-HCl, pH 6.8, containing 8 M urea, 5% (wt/vol) SDS, 0.1 mM EDTA, and 0.4 mg/ml) and lanes 1–6, 8, and 9 were loaded with 20 μl of the yeast extracts per lane. Lane 7 was loaded with 1 ng MBP fusion protein containing the full-length Nck-2. Lanes 1–7 were probed with a rabbit anti-Nck antibody raised against a GST fusion protein containing the two C-terminal SH3 domains and the SH2 domain of Nck-2 (residues 115–380), and lanes 8 and 9 were probed with a rabbit anti-HA antibody (Zymed Laboratories; 1 μg/ml).
To determine whether the third SH3 domain of Nck-2 is sufficient for interacting with PINCH, we expressed each of the three Nck-2 SH3 domains individually in yeast cells (Figure 3B). Immunoblotting analyses with the anti-Nck-2(SH32/SH33/SH2) antibody showed that the second (Figure 3B, lane 2) and the third SH3 domain (Figure 3B, lanes 3 and 4), respectively, were expressed in the yeast cells. No specific band was recognized by the anti-Nck-2(SH32/SH33/SH2) antibody in yeast cells that were transformed with the pB42AD vector containing the first SH3 domain sequence (Figure 3B, lane 1), confirming the specificity of the antibody. Because the B42AD fusion proteins contain a hemagglutinin (HA) epitope tag, we probed the yeast cell extracts with an anti-HA tag antibody, and the results showed that the fusion protein containing the first SH3 domain was expressed in the yeast cells (Figure 3B, lane 9). The yeast cells were then used to analyze the PINCH binding activity of the SH3 domains (Figure 3A). The results showed that the fusion proteins containing the third Nck-2 SH3 domain bound to the LIM4 domain of PINCH (Figure 3A, Nck-2(SH33)162–274, Nck-2(SH33)176–274, and Nck-2(SH33)195–274), whereas those containing the first Nck-2 SH3 domain or the second SH3 domain did not (Figure 3A, Nck-2(SH31)1–76 and Nck-2(SH32)115–190). Thus, the third SH3 domain of Nck-2, but not the first or the second SH3 domain, is not only necessary but also sufficient for interacting with PINCH. Noticeably, the PINCH binding activity of the SH33 fusion protein lacking the N-terminal linker sequence (Nck-2(SH33)195–274) was significantly lower than those containing the complete (Nck-2(SH33)162–274) or partial (Nck-2(SH33)176–274) linker sequence.
Comparison of Nck-1 and Nck-2 sequences reveals that the two Nck proteins are structurally related (Figure 1B). To test the ability of Nck-1 to interact with PINCH, we expressed full-length Nck-1 in yeast cells. Consistent with the sequence homology with Nck-2, the Nck-1 fusion protein expressed by the yeast cells was recognized by the polyclonal anti-Nck-2 antibody (Figure 3B, lane 6) as well as the anti-HA antibody (our unpublished results). However, in contrast to Nck-2, Nck-1 failed to interact with either the full-length PINCH or the LIM4 domain of PINCH (Figure 3A). Thus, in spite of the significant structure similarity between Nck-1 and Nck-2 (Figure 1B), Nck-1 does not interact with PINCH, indicating that the PINCH binding is a specific property of Nck-2 protein.
To further analyze the structural requirement for the Nck-2-PINCH interaction, we generated a “traditional” SH3 mutant in which the first tryptophan (residues 234) of the highly conserved tryptophan doublet in the third SH3 domain of Nck-2 was changed to lysine. Similar mutations in other SH3 domains (e.g., the SH3 domains of Abl and Crk) eliminated the binding of the proline-rich sequences by the SH3 domains (Tanaka et al., 1995). Analysis of the Nck-2 SH33W→K mutant in the yeast two-hybrid binding assays showed that it was readily recognized by the PINCH LIM4 domain (Figure 3A). Thus, substitution of the conserved W with K in the third SH3 domain of Nck-2 does not disrupt the structure required for the recognition by the PINCH LIM4 domain.
Association of Nck-2 with EGF Receptor and Its Regulation by EGF Stimulation
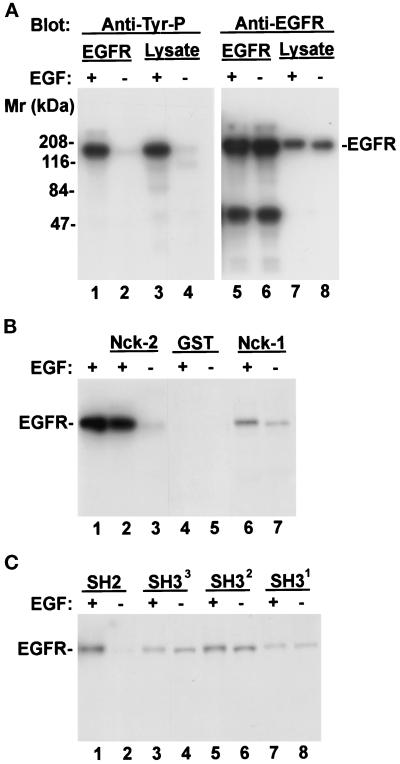
The primary structure of Nck-2 (Figure 1, A and B) suggests that Nck-2 is primarily involved in mediating protein-protein interactions. We have, therefore, sought to identify protein targets, in addition to PINCH, that are recognized by Nck-2. It has been well established that Nck-1 can associate with several components of receptor tyrosine kinase-signaling pathways including EGF receptor (Li et al., 1992; Park and Rhee, 1992), PDGF receptor (Li et al., 1992; Nishimura et al., 1993), and IRS-1 (Lee et al., 1993) upon ligand activation. To test whether Nck-2 could recognize EGF receptors, we analyzed the ability of GST-Nck-2 (Figure 4, lane 1) to associate with EGF receptors from human A431 cells that were either serum starved or stimulated with EGF (Figure 5). Stimulation of human A431 cells with EGF dramatically induced tyrosine phosphorylation of EGF receptor (Figure 5A, lanes 1–4), whereas the overall protein level of EGF receptor was not altered (Figure 5A, lanes 5–8). Coprecipitation experiments showed that only a very small amount of EGF receptor associated with Nck-2 in the absence of EGF stimulation (Figure 5B, lane 3). Stimulation with EGF dramatically increased the amount of EGF receptors that were associated with Nck-2 (Figure 5B, lane 2). In control experiments, EGF receptors also formed a complex with Nck-1, and the complex formation is significantly enhanced upon EGF stimulation (Figure 5B, lanes 6 and 7). Additionally, no EGF receptor was associated with GST, either with (Figure 5B, lane 4) or without (Figure 5B, lane 5) EGF stimulation. Taken together, these results demonstrate that Nck-2 is capable of associating with EGF receptors, preferentially the ligand-activated EGF receptor.
Figure 4.
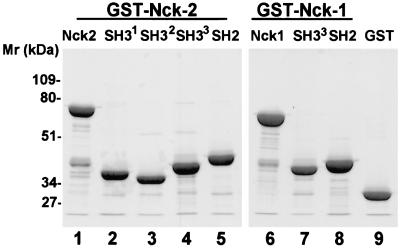
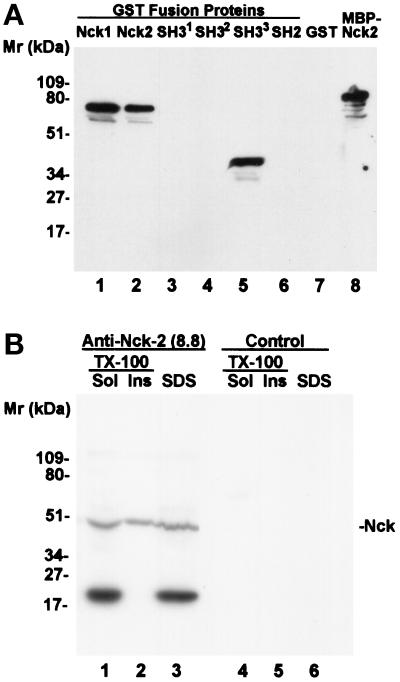
Expression of recombinant Nck-2 fusion proteins. The GST fusion proteins containing various Nck-2 and Nck-1 sequences were expressed in E. coli cells as described in MATERIALS AND METHODS. The fusion proteins were isolated with glutathione-Sepharose 4B beads, separated on 10% SDS-PAGE (reduced), and detected by staining with Coomassie Brilliant Blue R-250. Lanes were loaded with GST fusion proteins containing the full- length Nck-2 (residues 1–380) (lane 1), the first SH3 domain of Nck-2 (residues 1–76) (lane 2), the second SH3 domain of Nck-2 (residues 115–190) (lane 3), the third SH3 domain of Nck-2 (residues 176–274) (lane 4), the Nck-2 SH2 domain (residues 267–380) (lane 5), the full-length Nck-1 (residues 1–377)(lane 6), the third SH3 domain of Nck-1 (residues 171–270) (lane 7), the Nck-1 SH2 domain (residues 264–377), and GST (lane 9) (10 μg protein/lane).
Figure 5.
Association of Nck-2 with EGF receptors. Human A431 epidermoid carcinoma cell monolayers (∼80% confluent) were starved in DMEM medium containing 0.1% FBS for 18 h. At the end of serum starvation, the cells were either harvested (−EGF), or stimulated with 250 ng/ml EGF (+EGF) for 5 min and then lysed. (A) Tyrosine phosphorylation of EGF receptors. The cell lysates (1 ml of 2.8 mg protein/ml) were incubated with 1 μg of rabbit polyclonal anti-EGF receptor IgG (1005, Santa Cruz Biotechnology), and the EGF receptor immune complex was precipitated with 20 μl protein G-Sepharose 4B beads. The EGF receptor immunoprecipitates (EGFR, lanes 1, 2, 5, and 6, ¼ of the immunoprecipitates/lane) and cell lysates (Lysate, lanes 3 and 4, 21 μg protein/lane; lanes 7 and 8, 3 μg protein/lane) were analyzed by immunoblotting with mouse monoclonal anti-phosphotyrosine antibody (PY20, 0.2 μg/ml) (lanes 1–4) or rabbit anti-EGF receptor antibody (1005, 2 μg/ml) (lanes 5–8), appropriate horseradish peroxidase-conjugated secondary antibodies and the SuperSignal chemiluminescent substrate(Pierce Chemical). (B and C) Association of Nck-2 with EGF receptors. The cell lysates (0.9 mg) were incubated with equal amount (10 μg) of GST fusion proteins containing the full-length Nck-2 (panel B, lanes 2 and 3), the full-length Nck-1 (panel B, lanes 6 and 7), each of the four SH domains of Nck-2 (panel C), or GST (panel B, lanes 4 and 5) as indicated in the figure (final volume = 500 μl). The GST fusion proteins and associated proteins were precipitated with 23 μl glutathione-Sepharose 4B beads. The EGF receptors associated with the Nck-2 or Nck-1 fusion proteins were detected with the anti-EGF receptor (1005, 2 μg/ml) by immunoblotting. The exposure times for the x-ray films shown in panels B and C were identical. Lane 1 in panel B was loaded with 4 μg of the EGF-stimulated cell lysate.
To identify Nck-2 domains that are involved in association with EGF receptors, we expressed each of the four SH domains of Nck-2 (Figure 4, lanes 2–5) and tested their ability to associate with EGF receptors (Figure 5C). The Nck-2 SH2 domain was able to form a complex with EGF receptors (Figure 5C, lane 1), although to a lesser extent than the full-length Nck-2 (Figure 5B, lane 2). The complex formation between the Nck-2 SH2 domain and EGF receptors was completely dependent on EGF activation (Figure 5C, compare lanes 1 and 2). Additionally, we have detected relatively weak associations between EGF receptors and the SH3 domains of Nck-2 (Figure 5C, lanes 3–8). The second SH3 domain of Nck-2 (Figure 5C, lanes 5 and 6), and to a lesser extent the third SH3 domain (Figure 5C, lanes 3 and 4) or the first SH3 domain (Figure 5C, lanes 7 and 8), was able to associate with EGF receptors. Furthermore, in contrast to the Nck-2 SH2 domain (Figure 5C, lanes 1 and 2), the associations of the Nck-2 SH3 domains with EGF receptors were independent of EGF stimulation (Figure 5C, lanes 3–8). Because the full-length Nck-2 (Figure 5B, lane 2) associated with the ligand-activated EGF receptors much more efficiently than each of the individual SH domains (Figure 5C, lanes 1, 3, 5, and 7), both the SH2 domain and the SH3 domains likely contributed to the association of Nck-2 with the ligand-activated EGF receptors. However, because the association of Nck-2 with EGF receptors is largely dependent on EGF stimulation, the Nck-2 SH2 domain most likely plays a more prominent role in mediating the complex formation between Nck-2 and the activated EGF receptors.
Nck-2 Associates with PDGF Receptor-β upon PDGF Stimulation
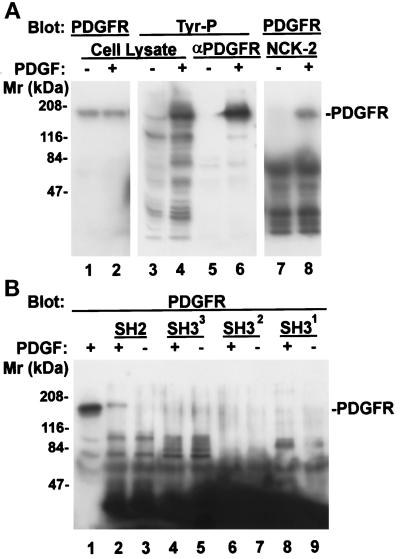
We next tested whether Nck-2 could associate with PDGF receptors either with or without PDGF activation. Stimulation of NIH3T3 cells with PDGF dramatically induced tyrosine phosphorylation of PDGF receptor-β (Figure 6A, lanes 5 and 6), whereas the overall protein level of PDGF receptor-β was not altered (Figure 6A, lanes 1 and 2). Coprecipitation experiments show that PDGF receptor-β associated with Nck-2 only upon PDGF stimulation (Figure 6A, lanes 7 and 8), suggesting that Nck-2 recognizes PDGF receptor-β in a ligand activation-dependent manner. To further analyze this, we tested the ability of each of the four SH domains of Nck-2 (Figure 4) to associate with PDGF receptor-β (Figure 6B). The Nck-2 SH2 domain was able to form a complex with PDGF receptor-β (Figure 6B, lane 2). Again, the complex formation was completely dependent on PDGF activation (Figure 6B, compare lanes 2 and 3). In contrast, none of the Nck-2 SH3 domains associated with PDGF receptor-β, either with or without PDGF stimulation (Figure 6B, lanes 4–9). In control experiments, no PDGF receptor-β was associated with GST either in the presence or absence of PDGF stimulation (our unpublished results). We conclude from these experiments that Nck-2 is capable of associating with PDGF receptor-β upon PDGF activation, and the SH2 domain of Nck-2 mediates the association with the activated PDGF receptor-β.
Figure 6.
Association of Nck-2 with PDGF receptor-β. NIH3T3 cell monolayers (∼80% confluent) were starved in DMEM containing 0.2% FBS for 18 h. At the end of serum starvation, the cells were either harvested (−PDGF), or stimulated with 50 ng/ml PDGF (+PDGF) for 5 min and then lysed. The PDGF receptor-β was precipitated from the cell lysates (1 ml of 2.8 mg protein/ml) with 1 μg of rabbit polyclonal anti-PDGF receptor-β antibody (P-20, Santa Cruz Biotechnology) and protein G-Sepharose 4B beads (panel A, lanes 5 and 6, one half of the immunoprecipitates/lane). The protein tyrosine phosphorylation was determined by immunoblotting with a mouse monoclonal anti-phosphotyrosine antibody (PY20, 0.2 μg/ml) (panel A, lanes 3–6). For GST fusion protein pull-down experiments, the cell lysates (0.37 mg protein) were incubated with 10 μg of the GST fusion protein containing the full-length Nck-2 (panel A, lanes 7 and 8) or its individual SH domains (panel B, lanes 2–9) as indicated in the figure (final volume = 220 μl). The GST fusion proteins were precipitated with glutathione-Sepharose beads. The PDGF receptor-β that was associated with the Nck-2 proteins (panel A, lanes 7 and 8; panel B, lanes 1–9) or those in the total cell lysates (panel A, lanes 1 and 2) were detected with a rabbit anti-PDGF receptor-β antibody (P-20, 1 μg/ml). Lanes 1–4 in panel A and lane 1 in panel B were loaded with cell lysates (21 μg protein/lane).
Nck-2 Associates with Insulin Receptor Substrate (IRS)-1
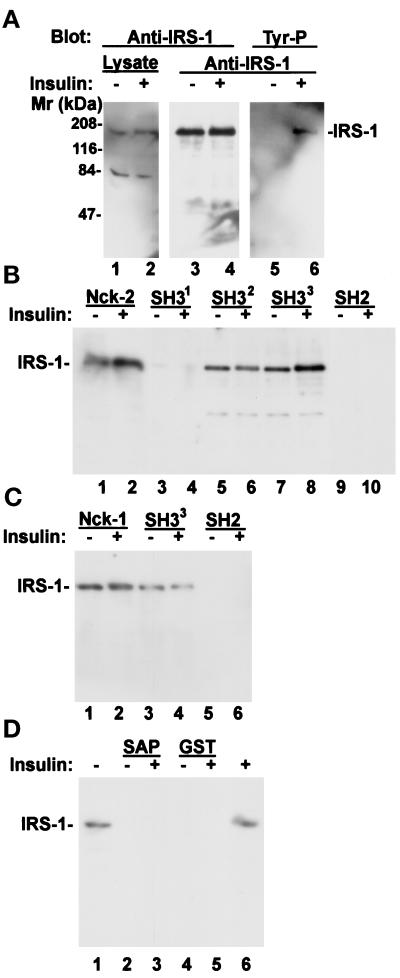
We have also analyzed the ability of Nck-2 to associate with IRS-1 from human 293 cells that were either serum starved or stimulated with insulin. IRS-1 was not tyrosine phosphorylated in the serum-starved 293 cells (Figure 7A, lane 5). Stimulation of the serum-starved 293 cells with insulin resulted in significant tyrosine phosphorylation on IRS-1 (Figure 7A, lane 6). Coprecipitation experiments showed that the recombinant GST-Nck-2 protein (Figure 7B, lane 2), but not GST (Figure 7D, lane 5), readily associated with IRS-1 upon insulin stimulation. However, unexpectedly, abundant IRS-1 was also coprecipitated with the recombinant Nck-2 protein in the absence of insulin stimulation (Figure 7B, lane 1). In additional experiments, recombinant GST-Nck-1, like GST-Nck-2, was also able to form a complex with IRS-1, either in the presence (Figure 7C, lane 2) or absence (Figure 7C, lane 1) of insulin stimulation. To further analyze this, we examined the ability of the Nck-2 SH2 domain, which was able to form a complex with ligand-activated EGF or PDGF receptors (Figure 5C, lane 1 and Figure 6B, lane 2), to associate with IRS-1. Under the conditions used (either with or without insulin stimulation), no association between the Nck-2 SH2 domain and IRS-1 was detected (Figure 7B, lanes 9 and 10). By contrast, the second or the third SH3 domain of Nck-2 was able to associate with IRS-1 (Figure 7B, lanes 5–8). Like the full-length Nck-2, the second or the third SH3 domain of Nck-2 associated with IRS-1 either in the presence (Figure 7B, lanes 6 and 8) or absence (Figure 7B, lanes 5 and 7) of insulin stimulation. On the other hand, no association between the first SH3 domain of Nck-2 and IRS-1 was detected under either experimental conditions (Figure 7B, lanes 3 and 4). In additional experiments, the third SH3 domain of Nck-1 (Figure 7C, lanes 3 and 4), but neither the SH2 domain of Nck-1 (Figure 7C, lanes 5 and 6) nor the SH3 domain of an unrelated protein (SAP97) (Figure 7D, lanes 2 and 3), was able to form a complex with IRS-1 either with or without insulin stimulation. Taken together, these experiments demonstrate that Nck-2, like Nck-1, is capable of associating with IRS-1. Moreover, they reveal that at least under certain conditions, the association of Nck-2 with IRS-1 could be mediated primarily via its second and third SH3 domains in a manner that is independent of insulin stimulation.
Figure 7.
Association of Nck-2 with IRS-1. Human 293 cell monolayers (∼70% confluent) were starved in serum-free medium (Eagle’s MEM) for 20 h. At the end of serum starvation, the cells were either harvested (−insulin) or stimulated with 6 μg/ml insulin (+insulin) for 5 min and then lysed. (A) Tyrosine phosphorylation of IRS-1. The cell lysates (0.8 mg) were incubated with 1 μg of rabbit polyclonal anti-IRS-1 receptor IgG (C-20, Santa Cruz Biotechnology), and the IRS-1 immune complex was precipitated with protein G-Sepharose 4B beads. The cell lysates (Lysate, lanes 1 and 2; 2 μg protein/lane) and the IRS-1 immunoprecipitates (anti-IRS-1, lanes 3–6, ¼ of the immunoprecipitates/lane) were analyzed by immunoblotting with rabbit anti-IRS-1 antibody (C-20, 1 μg/ml) (lanes 1–4) or mouse monoclonal anti-phosphotyrosine antibody (PY20, 0.2 μg/ml) (lanes 5 and 6), appropriate horseradish peroxidase-conjugated secondary antibodies and the SuperSignal chemiluminescent substrate (Pierce Chemical). (B and C) Association of Nck-2 with IRS-1. The cell lysates (100 μg) were incubated with equalamounts (10 μg) of GST fusion proteins containing the full-length Nck-2 or its individual SH domains (panel B), the full-length Nck-1 or its SH domains (panel C), the SH3 domain of SAP97 (panel D, lanes 2 and 3), or GST alone (panel D, lanes 4 and 5) as indicated in the figure (final volume = 170 μl). The GST fusion proteins and associated proteins were precipitated with glutathione-Sepharose 4B beads. The IRS-1 associated with the Nck-2 or Nck-1 fusion proteins were detected with the anti-IRS-1 antibody (C-20, 1 μg/ml) by immunoblotting. The exposure time for the x-ray films shown in panels B–D was identical. Lanes 1 and 6 in panel D were loaded with cell lysates (6 μg protein/lane).
Generation and Characterization of a Monoclonal Antibody Recognizing Nck-2
We have generated a monoclonal antibody that recognizes Nck-2 protein. Screening of monoclonal antibodies raised against a GST fusion protein containing the C-terminal region of Nck-2 (residues 115–380) with a MBP fusion protein containing the full-length Nck-2 in ELISA (our unpublished results) and immunoblotting assays (Figure 8A, lane 8) identified a positive monoclonal antibody (clone 8.8). The monoclonal antibody 8.8 recognized GST-Nck-2 (Figure 8A, lane 2) but not GST (Figure 8A, lane 7), confirming the specificity of the monoclonal antibody. Additionally, the monoclonal antibody 8.8 also recognized GST-Nck-1 (Figure 8A, lane 1), suggesting that the Nck-2 protein shares certain epitopes with the Nck-1 protein. To map the epitope recognized by the monoclonal antibody 8.8, we test its ability to interact with each of the four SH domains of Nck-2. The results showed that the monoclonal antibody 8.8 specifically recognized an epitope located within the residues 176–274 (Figure 8A, lane 5).
Figure 8.
Immunoblot detection of Nck proteins with monoclonal antibody 8.8. (A) Lanes 1–8 were loaded with GST fusion proteins containing the full-length Nck-1 (lane 1), the full-length Nck-2 (lane 2), the first SH3 domain of Nck-2 (residues 1–76) (lane 3), the second SH3 domain of Nck-2 (residues 115–190) (lane 4), the third SH3 domain of Nck-2 (residues 176–274) (lane 5), the Nck-2 SH2 domain (residues 267–380) (lane 6), GST (lane 7), or an MBP fusion protein containing the full-length Nck-2 (lane 8)(5 ng protein/lane). (B) Equal amount of human 293 cells was extracted with 0.4 ml of SDS sample buffer (2% [wt/vol] SDS, 5% [vol/vol] 2-mercaptoethanol, 10% [vol/vol] glycerol, 0.05% [wt/vol] bromophenol blue in 62.5 mM Tris-HCl, pH 6.8) (lanes 3 and 6), or 0.2 ml of 0.5% Triton X-100 in 10 mM Tris-HCl buffer, pH 7.1, containing 50 mM NaCl, 30 mM sodium pyrophosphate, 50 mM sodium fluoride, 200 μm sodium orthovanadate, 1 mg/ml BSA, 0.2 mM 4-(2-aminoethyl)benzenesulfonylfluoride, HCl, 10 μg/ml aprotinin, 1 μg/ml pepstatin A and 5 μg/ml leupeptin. The Triton X-100 insoluble fraction was pelleted by centrifugation, and then mixed with 0.4 ml (insoluble fraction) of the SDS sample buffer (lanes 2 and 5). The Triton X-100 soluble fraction (total volume = 0.2 ml) was mixed with 0.2 ml of the SDS sample buffer (lanes 1 and 4). Each lane was loaded with 20 μl of the samples as indicated in the figure. The membranes were probed with monoclonal antibody 8.8-conditioned culture supernatant (lanes 1–3) and the unconditioned plain medium (lanes 4–6), respectively.
A Fraction of the Nck Proteins Are Present in the Triton-X 100 Insoluble Cytoskeleton Fraction
We utilized the monoclonal anti-Nck antibody 8.8 to analyze the subcellular distribution of the Nck proteins. Immunofluorescence staining of cultured human 293 cells with the monoclonal antibody 8.8 indicated that the Nck proteins were present predominantly in the cytoplasm (our unpublished results). Immunoblotting analyses of SDS extracts of human cellular proteins showed that the monoclonal antibody 8.8 recognized a band with apparent molecular mass equivalent to that of Nck-1 (∼47 kDa) (Figure 8B, lane 3). Because the anti-Nck antibody is capable of recognizing both Nck-1 and Nck-2 proteins (Figure 8A, lanes 1 and 2) and the calculated molecular weight of Nck-2 (42,889.0) is very close to that of Nck-1 (42,864.4), the 47-kDa band likely represented human Nck-1 and Nck-2 proteins. Interestingly, the anti-Nck antibody recognized an additional band with apparent molecular mass of ∼20 kDa (Figure 8B, lane 3), which likely represented a protein sharing a common epitope with the third SH3 domain of the Nck proteins. Extraction of the human cells with Triton-X 100 revealed that a significant portion of the Nck proteins were insoluble in Triton-X 100 (Figure 8B, lane 2), indicating that a fraction of the Nck proteins are associated with the cytoskeleton. By contrast, the 20-kDa protein that is immunologically related to the Nck proteins was completely extractable with the Triton-X 100 (Figure 8B, lanes 1 and 2).
DISCUSSION
In this study, we have identified, cloned, and characterized Nck-2, a novel SH2- and SH3-domain–containing protein that is widely expressed in human tissues. Structurally, Nck-2 is closely related to Nck-1, both in overall domain organization and in amino acid sequence within each domain. Functionally, Nck-2 also possesses several activities that are similar to those of Nck-1. We have found in this study that Nck-2 is capable of recognizing several key components of the growth factor receptor kinase-signaling pathways including EGF receptors, PDGF receptor-β, and IRS-1, which are known protein targets of Nck-1. However, while Nck-2 and Nck-1 are structurally related and share several common protein targets, they are clearly not redundant. A marked difference between the two members of the Nck family is that Nck-2, but not Nck-1, interacts with the LIM-only adaptor protein PINCH.
It is well established that a number of cellular processes, including cell proliferation, survival, differentiation, and migration, are coordinately controlled by both cell adhesion- and growth factor-signaling pathways. In a recent study, Delcommenne et al. (1998) have shown that the kinase activity of ILK can be regulated not only by cell adhesion to fibronectin but also by insulin in a phosphoinositide-3-OH kinase-dependent manner. Furthermore, ILK can directly phosphorylate PKB/AKT on serine-473, one of the two phosphorylation sites involved in the activation of PKB/AKT, and regulate GSK-3 activity (Delcommenne et al., 1998), suggesting that ILK is involved in the regulation of both growth factor- and integrin-signaling pathways. We recently have demonstrated that ILK interacts with PINCH via its N-terminal ANK repeats-containing domain and mapped a major ILK-binding site to the N-terminal-most LIM domain (LIM1) of PINCH. Thus, the Nck-2-PINCH interaction, which is mediated by the fourth LIM domain of PINCH (LIM4), could potentially provide an important physical connection between the growth factor receptor-signaling pathways and the cell adhesion receptor integrin-mediated signaling pathways. It has been shown in a number of studies that integrin receptors could colocalize and physically associate with components of growth factor-signaling pathways (Bartfeld et al., 1993; Vuori and Ruoslahti, 1994; Plopper et al., 1995; Miyamoto et al., 1996; Schneller et al., 1997). For example, the αvβ3-integrin, which associates with ILK in vivo (Dedhar and Hannigan, 1996; Hannigan et al., 1996), could also form a complex with activated PDGF receptors (Bartfeld et al., 1993; Schneller et al., 1997), IRS-1 (Vuori and Ruoslahti, 1994), or the insulin receptor (Schneller et al., 1997). The molecular basis leading to the multiprotein complex formation between the growth factor receptors and the integrins, however, was not clear. The modular structures of Nck-2 and PINCH, and the protein binding activities associated with their SH and LIM domains, raise an interesting possibility that Nck-2 and PINCH could be involved in connecting ILK and the integrins with PDGF receptors, IRS-1, or other components of the growth factor-signaling pathways. One of the simplest models would be that the integrins associate with the ligand-activated growth factor receptors (e.g., PDGF receptors) via ILK, PINCH, and Nck-2. The identification of Nck-2 and the biochemical activities associated with it described in this study should allow us to determine in the future whether the interactions between the integrins, ILK, PINCH, Nck-2, and the PDGF receptors (or other proteins such as EGF receptors or IRS-1) are involved in promoting or regulating the formation of multiprotein complexes that integrate the signals from integrins and the growth factor receptors.
In addition to demonstrating that Nck-2 is capable of associating with PINCH, EGF receptors, PDGF receptor-β, and IRS-1, we have determined in this study the domains of Nck-2 that are involved in each of the molecular associations. One of the important findings is that the PINCH LIM4 domain specifically recognizes the third SH3 region of Nck-2. Several lines of evidence suggest that this PINCH/Nck-2 recognition system is distinctive from the conventional proline-rech motif/SH3 recognition system. First, the PINCH LIM4 domain (residues 192–249) recognized by the Nck-2 SH3 region lacks a characteristic PxxP motif. Second, deletion of 19 amino acid residues (176–194) that are N-terminal to the SH3 conserved sequence in this region reduced, although did not eliminate, the PINCH binding activity (Figure 3A, Nck-2(SH33)195-274), suggesting that residues outside the conserved SH3 sequence could either directly contribute to the binding or indirectly influence the interaction between the LIM and SH3 domains. Finally, substitution of a highly conserved tryptophan (residues 234) with lysine in the third SH3 domain of Nck-2, which eliminates the recognition of the proline-rich sequences by several other SH3 domains (Tanaka et al., 1995), did not affect the PINCH binding activity. These results suggest a novel protein–protein recognition system involving both LIM and SH3 domains, two well described protein-binding structures. Additionally they indicate that a region containing a single SH3 domain (e.g., the third SH3 region of Nck-2) could be potentially involved in mediating multiple protein–protein interactions.
Additionally, we have found that the SH2 domain is primarily responsible for the association of Nck-2 with the ligand-activated PDGF receptor-β or EGF receptors. The involvement of the SH2 domain in connecting Nck-2 to the growth factor receptors is consistent with a major role of tyrosine phosphorylation in signal transduction. Indeed, the associations of Nck-2 with PDGF receptor-β and the EGF receptors were regulated by ligand-induced tyrosine phosphorylation. A finding that we found intriguing was that the SH3 domains of Nck-2 also contributed to the associations of Nck-2 with certain specific components of the growth factor-signaling pathways. For example, there appear to exist weak associations between the Nck-2 SH3 domains and the EGF receptor. More prominently, using IRS-1 derived from human 293 cells, we have detected stable associations of IRS-1 with specific Nck-2 SH3 domains (the second and third SH3 domains). Furthermore, Nck-1, the other member of the Nck family, could also utilize the SH3-dependent mechanism to associate with IRS-1. In a previous study, Lee et al. (1993) have shown that the SH2 domain of Nck-1 could associate with tyrosine-phosphorylated IRS-1 proteins and identified a sequence containing phosphotyrosine 147 on rat IRS-1 as a major recognition site of the Nck-1 SH2 domain. Thus, there exist at least two distinct mechanisms by which Nck-1, and possibly Nck-2, could form a complex with IRS-1. The first one involves the SH2 domain and the second one involves the SH3 domains. The capacity of Nck proteins to associate with IRS-1 via two different types of protein-binding motifs provides cells with greater versatility in controlling the complex formation between IRS-1 and the Nck proteins. Under the experimental conditions used in this study, the associations between the human IRS-1 and the Nck proteins that we observed were mediated primarily by their SH3 domains. This could reflect the fact that the tyrosine 147-containing motif is not conserved in human IRS-1. However, although we did not detect an association between human IRS-1 and the SH2 domain of Nck-2 (or that of Nck-1), we could not rule out the possibility that there exists a potential binding site in human IRS-1 for the SH2 domain of Nck-2 (or that of Nck-1) that could be activated (tyrosine-phosphorylated) upon proper stimulation. This site, if it exists, was clearly not activated under the experimental conditions we used.
Recently, Myers et al. (1996) have demonstrated that an IRS-1 mutant in which all 18 potential tyrosine phosphorylation sites were replaced by phenylalanine could mediate, although to a less extent than the wild-type IRS-1, insulin-stimulated mitogenesis. Thus, in addition to serving as a docking protein for SH2 domains, IRS-1 may engage other protein-binding domains that could transduce phosphotyrosine-independent signals leading to mitogenesis. The modular structure and the ability of the Nck proteins to associate with IRS-1 via the SH3-domains suggest that Nck-2 and Nck-1 likely play a role in transducing the phosphotyrosine-independent signals, in addition to the SH2/phosphotyrosine-dependent signals, from IRS-1.
ACKNOWLEDGMENTS
We thank Dr. Oliver Hobert (Massachusetts General Hospital) for sharing unpublished results, Drs. Louise T. Chow, Stuart J. Frank, Craig C. Garner, and Jeffrey E. Kudlow for valuable discussions and reagents, and the Hybridoma Core Facility of University of Alabama at Birmingham for technical assistance in the production of the mouse monoclonal anti-Nck antibodies. This work was supported in part by National Institutes of Health grant DK-54639, Research Project grant 98-220-01-CSM from the American Cancer Society, and research grants from the American Heart Association, the American Lung Association, and the Francis Families Foundation (to C.W.). F.L. was supported by the Cell Adhesion and Matrix Research Center of University of Alabama at Birmingham.
REFERENCES
- Anton IM, Lu W, Mayer BJ, Ramesh N, Geha RS. The Wiskott-Aldrich Syndrome Protein-interacting Protein (WIP) binds to the adaptor protein Nck. J Biol Chem. 1998;273:20992–20995. doi: 10.1074/jbc.273.33.20992. [DOI] [PubMed] [Google Scholar]
- Bartfeld NS, Pasquale EB, Geltosky JE, Languino LR. The alpha v beta 3 integrin associates with a 190-kDa protein that is phosphorylated on tyrosine in response to platelet-derived growth factor. J Biol Chem. 1993;268:17270–17276. [PubMed] [Google Scholar]
- Birge RB, Knudsen BS, Besser D, Hanafusa H. SH2 and SH3-containing adaptor proteins: redundant or independent mediators of intracellular signal transduction. Genes Cells. 1996;1:595–613. doi: 10.1046/j.1365-2443.1996.00258.x. [DOI] [PubMed] [Google Scholar]
- Bokoch GM, Wang Y, Bohl BP, Sells MA, Quilliam LA, Knaus UG. Interaction of the Nck adapter protein with p21-activated kinase (PAK1) J Biol Chem. 1996;271:25746–25749. doi: 10.1074/jbc.271.42.25746. [DOI] [PubMed] [Google Scholar]
- Chou MM, Fajardo JE, Hanafusa H. The SH2- and SH3-containing Nck protein transforms mammalian fibroblasts in the absence of elevated phosphotyrosine levels. Mol Cell Biol. 1992;12:5834–5842. doi: 10.1128/mcb.12.12.5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou MM, Hanafusa H. A novel ligand for SH3 domains. The Nck adaptor protein binds to a serine/threonine kinase via an SH3 domain. J Biol Chem. 1995;270:7359–7364. doi: 10.1074/jbc.270.13.7359. [DOI] [PubMed] [Google Scholar]
- Choudhury GG, Marra F, Abboud HE. Thrombin stimulates association of src homology domain containing adaptor protein Nck with Pp 125FAK. Am J Physiol. 1996;270:F295–F300. doi: 10.1152/ajprenal.1996.270.2.F295. [DOI] [PubMed] [Google Scholar]
- Dawid IB, Toyama R, Taira M. LIM domain proteins. C R Acad Sci Ser III Sci Vie. 1995;318:295–306. [PubMed] [Google Scholar]
- Dedhar S, Hannigan GE. Integrin cytoplasmic interactions and bidirectional transmembrane signal transduction. Curr Opin Cell Biol. 1996;8:657–669. doi: 10.1016/s0955-0674(96)80107-4. [DOI] [PubMed] [Google Scholar]
- Delcommenne M, Tan C, Gray V, Rue L, Woodgett J, Dedhar S. Phosphoinositide-3-OH kinase-depedent regulation of glycogen synthase kinase 3 and protein kinase B/AKT by the integrin-linked kinase. Proc Natl Acad Sci USA. 1998;95:11211–11216. doi: 10.1073/pnas.95.19.11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galisteo ML, Chernoff J, Su YC, Skolnik EY, Schlessinger J. The adaptor protein Nck links receptor tyrosine kinases with the serine-threonine kinase Pak1. J Biol Chem. 1996;271:20997–21000. doi: 10.1074/jbc.271.35.20997. [DOI] [PubMed] [Google Scholar]
- Garrity PA, Rao Y, Salecker I, McGlade J, Pawson T, Zipursky SL. Drosophila photoreceptor axon guidance and targeting requires the dreadlocks SH2/SH3 adapter protein. Cell. 1996;85:639–650. doi: 10.1016/s0092-8674(00)81231-3. [DOI] [PubMed] [Google Scholar]
- Gill GN. The enigma of LIM domains. Structure. 1995;3:1285–1289. doi: 10.1016/s0969-2126(01)00265-9. [DOI] [PubMed] [Google Scholar]
- Hannigan GE, Leung-Hagesteijn C, Fitz-Gibbon L, Coppolino MG, Radeva G, Filmus J, Bell JC, Dedhar S. Regulation of cell adhesion and anchorage-dependent growth by a new beta 1-integrin-linked protein kinase. Nature. 1996;379:91–96. doi: 10.1038/379091a0. [DOI] [PubMed] [Google Scholar]
- Holland SJ, Gale NW, Gish GD, Roth RA, Songyang Z, Cantley LC, Henkemeyer M, Yancopoulos GD, Pawson T. Juxtamembrane tyrosine residues couple the Eph family receptor EphB2/Nuk to specific SH2 domain proteins in neuronal cells. EMBO J. 1997;16:3877–3888. doi: 10.1093/emboj/16.13.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Milfay D, Williams LT. Binding of NCK to SOS and activation of ras-dependent gene expression. Mol Cell Biol. 1995;15:1169–1174. doi: 10.1128/mcb.15.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Kitamura Y, Yonezawa K, Totty NF, Gout I, Hara K, Waterfield MD, Sakaue M, Ogawa W, Kasuga M. Molecular cloning of p125Nap1, a protein that associates with an SH3 domain of Nck. Biochem Biophys Res Commun. 1996;219:509–514. doi: 10.1006/bbrc.1996.0264. [DOI] [PubMed] [Google Scholar]
- Lee CH, Li W, Nishimura R, Zhou M, Batzer AG, Myers MG, Jr, White MF, Schlessinger J, Skolnik EY. Nck associates with the SH2 domain-docking protein IRS-1 in insulin-stimulated cells. Proc Natl Acad Sci USA. 1993;90:11713–11717. doi: 10.1073/pnas.90.24.11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann JM, Riethmuller G, Johnson JP. Nck, a melanoma cDNA encoding a cytoplasmic protein consisting of the src homology units SH2 and SH3. Nucleic Acids Res. 1990;18:1048. doi: 10.1093/nar/18.4.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Hu P, Skolnik EY, Ullrich A, Schlessinger J. The SH2 and SH3 domain-containing Nck protein is oncogenic and a common target for phosphorylation by different surface receptors. Mol Cell Biol. 1992;12:5824–5833. doi: 10.1128/mcb.12.12.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Katz S, Gupta R, Mayer BJ. Activation of Pak by membrane localization mediated by an SH3 domain from the adaptor protein Nck. Curr Biol. 1997;7:85–94. doi: 10.1016/s0960-9822(06)00052-2. [DOI] [PubMed] [Google Scholar]
- Lussier G, Larose L. A casein kinase I activity is constitutively associated with Nck. J Biol Chem. 1997;272:2688–2694. doi: 10.1074/jbc.272.5.2688. [DOI] [PubMed] [Google Scholar]
- Margolis B, Silvennoinen O, Comoglio F, Roonprapunt C, Skolnik E, Ullrich A, Schlessinger J. High-efficiency expression/cloning of epidermal growth factor-receptor-binding proteins with Src homology 2 domains. Proc Natl Acad Sci USA. 1992;89:8894–8898. doi: 10.1073/pnas.89.19.8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisenhelder J, Hunter T. The SH2/SH3 domain-containing protein Nck is recognized by certain anti-phospholipase C-gamma 1 monoclonal antibodies, and its phosphorylation on tyrosine is stimulated by platelet-derived growth factor and epidermal growth factor treatment. Mol Cell Biol. 1992;12:5843–5856. doi: 10.1128/mcb.12.12.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S, Teramoto H, Gutkind JS, Yamada KM. Integrins can collaborate with growth factors for phosphorylation of receptor tyrosine kinases and MAP kinase activation: roles of integrin aggregation and occupancy of receptors. J Cell Biol. 1996;135:1633–1642. doi: 10.1083/jcb.135.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller BM, Kistner U, Veh RW, Cases-Langhoff C, Becker B, Gundelfinger ED, Garner CC. Molecular characterization and spatial distribution of SAP97, a novel presynaptic protein homologous to SAP90 and the Drosophila discs-large tumor suppressor protein. J Neurosci. 1995;15:2354–2366. doi: 10.1523/JNEUROSCI.15-03-02354.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MG, Jr, et al. YMXM motifs and signaling by an insulin receptor substrate 1 molecule without tyrosine phosphorylation sites. Mol Cell Biol. 1996;16:4147–4155. doi: 10.1128/mcb.16.8.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura R, Li W, Kashishian A, Mondino A, Zhou M, Cooper J, Schlessinger J. Two signaling molecules share a phosphotyrosine-containing binding site in the platelet-derived growth factor receptor. Mol Cell Biol. 1993;13:6889–6896. doi: 10.1128/mcb.13.11.6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak A, Hsu SC, Leunghagesteijn C, Radeva G, Papkoff J, Montesano R, Roskelley C, Grosschedl R, Dedhar S. Cell adhesion and the integrin-linked kinase regulate the Lef-1 and beta-catenin signaling pathways. Proc Natl Acad Sci USA. 1998;95:4374–4379. doi: 10.1073/pnas.95.8.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D. Cloning, sequencing, and overexpression of SH2/SH3 adaptor protein Nck from mouse thymus. Mol Cells. 1997;7:231–236. [PubMed] [Google Scholar]
- Park D, Rhee SG. Phosphorylation of Nck in response to a variety of receptors, phorbol myristate acetate, and cyclic AMP. Mol Cell Biol. 1992;12:5816–5823. doi: 10.1128/mcb.12.12.5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plopper GE, McNamee HP, Dike LE, Bojanowski K, Ingber DE. Convergence of integrin and growth factor receptor signaling pathways within the focal adhesion complex. Mol Biol Cell. 1995;6:1349–1365. doi: 10.1091/mbc.6.10.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilliam LA, Lambert QT, Mickelson-Young LA, Westwick JK, Sparks AB, Kay BK, Jenkins NA, Gilbert DJ, Copeland NG, Der CJ. Isolation of a NCK-associated kinase, PRK2, an SH3-binding protein and potential effector of Rho protein signaling. J Biol Chem. 1996;271:28772–28776. doi: 10.1074/jbc.271.46.28772. [DOI] [PubMed] [Google Scholar]
- Rao Y, Zipursky SL. Domain requirements For the Dock adapter protein in growth-cone signaling. Proc Natl Acad Sci USA. 1998;95:2077–2082. doi: 10.1073/pnas.95.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radeva G, Petrocelli T, Behrend E, Leunghagesteijn C, Filmus J, Slingerland J, Dedhar S. Overexpression of the integrin-linked kinase promotes anchorage-independent cell cycle progression. J Biol Chem. 1997;272:13937–13944. doi: 10.1074/jbc.272.21.13937. [DOI] [PubMed] [Google Scholar]
- Rearden A. A new Lim protein containing an autoepitope homologous to “senescent cell antigen.”. Biochem Biophys Res Commun. 1994;201:1124–1131. doi: 10.1006/bbrc.1994.1822. [DOI] [PubMed] [Google Scholar]
- Rivero-Lezcano OM, Marcilla A, Sameshima JH, Robbins KC. Wiskott-Aldrich syndrome protein physically associates with Nck through Src homology 3 domains. Mol Cell Biol. 1995;15:5725–5731. doi: 10.1128/mcb.15.10.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaepfer DD, Broome MA, Hunter T. Fibronectin-stimulated signaling from a focal adhesion kinase-c-Src complex: involvement of the Grb2, p130cas, and Nck adaptor proteins. Mol Cell Biol. 1997;17:1702–1713. doi: 10.1128/mcb.17.3.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeichel KL, Beckerle MC. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Schneller M, Vuori K, Ruoslahti E. Alphavbeta3 integrin associates with activated insulin and PDGFbeta receptors and potentiates the biological activity of PDGF. EMBO J. 1997;16:5600–5607. doi: 10.1093/emboj/16.18.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein E, Huynh-Do U, Lane AA, Cerretti DP, Daniel TO. Nck recruitment to Eph receptor, EphB1/ELK, couples ligand activation to c-Jun kinase. J Biol Chem. 1998;273:1303–1308. doi: 10.1074/jbc.273.3.1303. [DOI] [PubMed] [Google Scholar]
- Su YC, Han J, Xu S, Cobb M, Skolnik EY. NIK is a new Ste20-related kinase that binds NCK and MEKK1 and activates the SAPK/JNK cascade via a conserved regulatory domain. EMBO J. 1997;16:1279–1290. doi: 10.1093/emboj/16.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XJ, Rothenberg P, Kahn CR, Backer JM, Araki E, Wilden PA, Cahill DA, Goldstein BJ, White MF. Structure of the insulin receptor substrate IRS-1 defines a unique signal transduction protein. Nature. 1991;352:73–77. doi: 10.1038/352073a0. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Gupta R, Mayer BJ. Differential inhibition of signaling pathways by dominant-negative SH2/SH3 adapter proteins. Mol Cell Biol. 1995;15:6829–6837. doi: 10.1128/mcb.15.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Lu W, Gupta R, Mayer BJ. Expression of mutated Nck SH2/SH3 adaptor respecifies mesodermal cell fate in Xenopus laevis development. Proc Natl Acad Sci USA. 1997;94:4493–4498. doi: 10.1073/pnas.94.9.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Feng GS, Li W. Induced direct binding of the adapter protein Nck to the GTPase-activating protein-associated protein p62 by epidermal growth factor. Oncogene. 1997;15:1823–1832. doi: 10.1038/sj.onc.1201351. [DOI] [PubMed] [Google Scholar]
- Vuori K, Ruoslahti E. Association of insulin receptor substrate-1 with integrins. Science. 1994;266:1576–1578. doi: 10.1126/science.7527156. [DOI] [PubMed] [Google Scholar]
- Wu C, Keightley SY, Leung-Hagesteijn C, Radeva G, Coppolino M, Goicoechea S, McDonald JA, Dedhar S. Integrin-linked protein kinase (ILK) regulates fibronectin matrix assembly, E-cadherin expression and tumorigenicity. J Biol Chem. 1998;273:528–536. doi: 10.1074/jbc.273.1.528. [DOI] [PubMed] [Google Scholar]
- Xie W, Li F, Kudlow JE, Wu C. Expression of the Integrin-linked kinase (ILK) in mouse skins: loss of expression in suprabasal layers of the epidermis and up-regulation by erbB-2. Am J Pathol. 1998;153:367–372. doi: 10.1016/S0002-9440(10)65580-0. [DOI] [PMC free article] [PubMed] [Google Scholar]