Abstract
Cockayne syndrome (CS) is a human genetic disorder characterized by UV sensitivity, developmental abnormalities, and premature aging. Two of the genes involved, CSA and CSB, are required for transcription-coupled repair (TCR), a subpathway of nucleotide excision repair that removes certain lesions rapidly and efficiently from the transcribed strand of active genes. CS proteins have also been implicated in the recovery of transcription after certain types of DNA damage such as those lesions induced by UV light. In this study, site-directed mutations have been introduced to the human CSB gene to investigate the functional significance of the conserved ATPase domain and of a highly acidic region of the protein. The CSB mutant alleles were tested for genetic complementation of UV-sensitive phenotypes in the human CS-B homologue of hamster UV61. In addition, the CSB mutant alleles were tested for their ability to complement the sensitivity of UV61 cells to the carcinogen 4-nitroquinoline-1-oxide (4-NQO), which introduces bulky DNA adducts repaired by global genome repair. Point mutation of a highly conserved glutamic acid residue in ATPase motif II abolished the ability of CSB protein to complement the UV-sensitive phenotypes of survival, RNA synthesis recovery, and gene-specific repair. These data indicate that the integrity of the ATPase domain is critical for CSB function in vivo. Likewise, the CSB ATPase point mutant failed to confer cellular resistance to 4-NQO, suggesting that ATP hydrolysis is required for CSB function in a TCR-independent pathway. On the contrary, a large deletion of the acidic region of CSB protein did not impair the genetic function in the processing of either UV- or 4-NQO-induced DNA damage. Thus the acidic region of CSB is likely to be dispensable for DNA repair, whereas the ATPase domain is essential for CSB function in both TCR-dependent and -independent pathways.
INTRODUCTION
Cockayne syndrome (CS) is an autosomal recessive human disorder with diverse clinical symptoms that include severe mental and physical growth retardation, microcephaly, progressive neurological and retinal degeneration, skeletal abnormalities, and a hypersensitivity to sunlight (Friedberg, 1996). Genetic analysis of fused heterodikaryons have identified two complementation groups involved in CS, designated CSA and CSB (Tanaka et al., 1981; Lehmann, 1982). CS cells demonstrate a reduced rate of nucleotide excision repair (NER) of active genes and, more specifically, of the transcribed strand of such genes (Venema et al., 1990; van Hoffen et al., 1993; Evans and Bohr, 1994). However, no defect in global NER is observed in CS. Defective transcription-coupled repair (TCR) in CS cells is found not only after UV exposure but also after exposure to certain forms of oxidative stress, suggesting that TCR is responsible for processing at least some types of oxidative DNA damage as well (Leadon and Cooper, 1993; Cooper and Leadon, 1994; Cooper et al., 1997).
A characteristic feature of CS cells is the lack of recovery of RNA synthesis after UV irradiation (Lehmann et al., 1979; Lehmann, 1982; Mayne and Lehmann, 1982). The CSB gene was originally cloned by its ability to complement the delay in RNA synthesis recovery displayed in the hamster mutant cell line UV61 isolated from the repair-proficient parental cell line AA8. Transfection of the human CSB gene into UV61 effectively restored RNA synthesis and UV resistance to normal levels (Troelstra et al., 1990). Gene-specific repair studies have demonstrated that the human CSB gene corrects the TCR defect in the hamster CS-B mutant UV61 (Orren et al., 1996). Thus the hamster system has been extremely useful to demonstrate that the human CSB gene is directly implicated in TCR. Subsequent studies confirmed that the CSB gene complements the UV sensitivity and the delay in RNA synthesis recovery of the human CS-B cell line CS1AN.S3.G2 (Troelstra et al., 1992). By sequence homology, the CSB gene product belongs to superfamily 2 and, more specifically, the SWI/SNF family of proteins, which have roles in transcription regulation, chromosome stability, and DNA repair (Eisen et al., 1995). All proteins belonging to the SWI/SNF family contain seven sequence motifs similar to those found in DNA and RNA helicase families. The CSB protein is a DNA-stimulated ATPase but fails to exhibit DNA unwinding activity as measured by the conventional strand displacement assay (Selby and Sancar, 1997a; Tantin et al., 1997). It is quite possible that CSB may play a role in chromatin remodeling as has been suggested for the SWI/SNF complex and related proteins in the gene family (Pazin and Kadonaga, 1997). However, some members of SWI/SNF subfamilies, such as MOT1, appear to function independently of chromatin (Pazin and Kadonaga, 1997). The mechanism of how ATP hydrolysis is coupled to the diverse roles of these proteins in chromatin regulation, stability, and repair remains to be determined.
The mechanism of TCR in mammalian cells and the precise role of CSB in this pathway remain to be elucidated. Using a biochemical approach to address the role of CSB protein in TCR, Tantin et al. (1997) have demonstrated that hydrolysis of the ATP β-γ phosphoanhydride bond is required for the formation of a stable RNA polymerase II (RNAPII)–CSB–DNA–RNA complex. However, in vitro studies indicate that CSB protein does not release RNAPII stalled at a T<>T lesion from the DNA (Selby and Sancar, 1997A). Furthermore, CSB protein has no effect on repair of a lesion protected by a stalled RNAPII (Selby et al., 1997). Most recently, it was shown that the RNAPII–CSB–DNA–RNA quaternary complex recruits a molecular complex containing the TFIIH core subunits p62 and XPB (Tantin, 1998). This finding suggests that CSB may facilitate repair of active genes by recruiting proteins involved in an early step of DNA damage recognition (Tantin, 1998). More studies are necessary to understand the role of CSB in the mechanism of TCR.
TCR may not be the only pathway defective in CS. There is some evidence for a defect in basal transcription. The transcription defect has been observed in human CS-B lymphoblastoid cells and fibroblasts without any exposure to stress such as UV light (Balajee et al., 1997). The reduced transcription in CS-B cells is complemented in chromatin by the addition of normal cell extract and in intact cells by transfection with the CSB gene (Balajee et al., 1997). In a reconstituted system, purified CSB protein enhances the rate of transcription by RNAPII, suggesting that CSB protein may indirectly stimulate TCR by facilitating the process of transcription (Selby and Sancar, 1997b). Conceivably CSB may serve dual roles as a transcription elongation factor and a repair coupling factor at the site of the RNAPII-blocking lesion in vivo. Direct involvement of CSB protein in transcription is supported by the finding that 10–15% of RNAPII is tightly associated with CSB protein in whole-cell extracts (van Gool et al., 1997). Thus, the CS phenotype may arise from a combined deficiency in repair and transcription. It is possible that the biological functions of CSB in different DNA metabolic pathways may be mediated by distinct functional domains of the protein.
Limited information is available addressing the functional significance of specific domains in the CSB protein. In vitro data suggest that ATP hydrolysis is required for the formation of a stable quaternary complex between CSB and a stalled RNAPII on a DNA template (Tantin et al., 1997). However, it was recently shown that a CSB mutant protein totally defective in ATP hydrolysis was able to partially rescue the delay in RNA synthesis recovery after exposure to UV light (Citterio et al., 1998). These data suggest that CSB-catalyzed ATP hydrolysis plays a role in TCR but may not be essential for the pathway to proceed.
In addition to the ATPase domain, CSB contains a negatively charged region located in the amino terminus before the ATPase and helicase motifs. Approximately 60% of the residues in a 39-amino-acid stretch are acidic. Acidic domains are found in a number of nuclear proteins that associate with chromatin or histones, suggesting that this domain may play a role in chromatin remodeling or transcriptional activation (Lapeyre et al., 1987; Ptashne, 1988; Sung et al., 1988; Wen et al., 1989). The fact that CSB belongs to a gene family containing members implicated in chromatin remodeling (for review, see Pazin and Kadonaga; 1997) suggests that the acidic region may be important to CSB function. In addition to a putative role in chromatin remodeling, acidic regions have been implicated in the function of transcriptional activator proteins (Ma and Ptashne, 1987; Hope et al., 1988). In fact, acidic transcriptional activation domains have been shown to directly interact with the RNAPII transcription machinery. For example, the acidic activation domain of VP16 directly binds to TATA box-binding factor TFIID (Stringer et al., 1990) as well as RNAPII initiation factor TFIIB (Gupta et al., 1996). The functional significance of the strikingly rich acidic region in CSB protein is presently unknown but it may play a role in the repair and/or transcription functions of CSB.
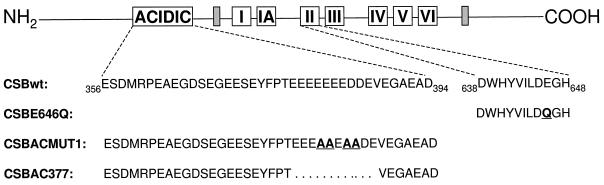
To gain a better understanding of the functional importance of different domains in the CSB protein in DNA repair, site-specific mutations were introduced in motif II of the ATPase domain and the acidic region of CSB protein (Figure 1). We have characterized the sensitivity of the CS-B mutant cell lines to two DNA-damaging agents, UV light and the chemical carcinogen 4-nitroquinoline-1-oxide (4-NQO). The lesions induced by these agents are processed by different repair pathways. The most abundant UV-induced lesions, cyclobutane pyrimidine dimers (CPDs), are repaired by TCR (Mellon et al., 1986). In contrast, 4-NQO induces both alkali-labile single-strand DNA breaks and UvrABC excinuclease-cleavable bulky adducts, which are repaired without any strand bias in mammalian cells (Snyderwine and Bohr, 1992). Genetic characterization of the ATPase mutant indicates that replacement of the highly conserved glutamic acid in motif II with glutamine abolishes the genetic function of the CSB protein in survival, RNA synthesis recovery, and gene-specific repair upon treatment with UV light. In contrast, mutations in the acidic region of CSB do not impair the ability of CSB protein to repair UV-induced damage. Similarly, the ATPase mutant failed to complement the sensitivity of the CS-B mutant cell line to 4-NQO, whereas the acidic mutants confer resistance to 4-NQO. These results provide genetic evidence that the ATPase activity of CSB protein is essential in TCR as well as in a TCR-independent pathway.
Figure 1.
Site-directed mutations introduced in the ATPase motif II and the acidic region of CSB. The CSB protein contains the ATPase and helicase motifs conserved in superfamily 2 as well as a negatively charged domain in which 22 of 39 residues are acidic (56%). The E646Q mutation in CSB changes the highly conserved glutamic acid in ATPase motif II to glutamine (CSBE646Q). The ACMUT1 mu-tation replaces acidic residues at positions 381, 382, 384, and 385 with alanines (CSBACMUT1). The AC377 mutation deletes 10 consecutive acidic residues (378–387) from the CSB protein (CSBAC377).
MATERIALS AND METHODS
Cell Lines and Culture Conditions
AA8 and UV61, the hamster cell lines used in this study, and their repair characteristics have been previously described (Mitchell and Nairn, 1989). AA8 is a repair-proficient wild-type cell line. UV61 cells, derived from AA8, belong to rodent complementation group 6 and are homologous to human CS-B cells based on genetic complementation by the human CSB gene (Troelstra et al., 1990; Orren et al., 1996). UV61 cell lines transfected with the mammalian expression vector pcDNA3.1 (Invitrogen, San Diego, CA; abbreviated pc3.1) or pc3.1 containing the wild-type human CSB gene are designated UV61/pc3.1 and UV61/pc3.1-CSBwt, respectively. UV61 cells transfected with pc3.1-CSB containing various mutations in the CSB gene were designated UV61/pc3.1-CSBE646Q, UV61/pc3.1-CSBAC377, and UV61/pc3.1-CSBACMUT1, respectively. All the cell lines were routinely grown in Ham’s F-10 and Dulbecco’s minimal essential medium (1:1) supplemented with 10% fetal bovine serum and antibiotics. UV61 cell lines transfected with pc3.1, which contains the neomycin resistance gene, or pc3.1 derivatives were grown in media containing 400 μg/ml geneticin (Life Technologies, Gaithersburg, MD).
DNA, Nucleotides, and Enzymes
pcBLsSE6, a plasmid containing the entire human CSB cDNA, was kindly provided by Dr. Jan Hoeijmakers (Rotterdam University, Rotterdam, The Netherlands). Nucleotides were purchased from Boehringer Mannheim (Indianapolis, IN). Primers used for site-directed mutagenesis, sequencing, reverse transcription, and PCR were purchased from Life Technologies. Restriction endonucleases SalI, XhoI, and PvuI, phage T7 DNA polymerase, calf intestinal alkaline phosphatase, phage T4 polynucleotide kinase, and DNA ligase were purchased from New England Biolabs (Beverly, MA). Restriction endonuclease KpnI was from Boehringer Mannheim. T4 Endo V was purified from an overproducing strain. Ready to Go T4 DNA ligase was from Amersham Pharmacia Biotech (Piscataway, NJ). The reaction conditions were essentially the same as suggested by the supplier.
Site-directed Mutagenesis and DNA Constructions
pcBLsSE6, a bacterial phagemid containing the f1 origin of replication and the entire human CSB gene, was used for site-directed mutagenesis by published procedures (Kunkel et al., 1991). A description of the mutations introduced in the CSB gene is shown in Figure 1. The oligonucleotide 5′-TTGTGTCCTTGGTCCAAGATC-3′ was used to replace the negatively charged Glu646 in the CSB protein to a neutral glutamine (E646Q). The oligonucleotide 5′-CTCTGCCCCCTCCACTGTGGGGAAATACTC-3′ was used to precisely delete 10 consecutive acidic residues, 377–386, in the CSB protein (AC377). The oligonucleotide 5′-CACCTCGTCAGCTGCCTCCGCTGCCTCCTCCTC-3′ was used to replace Glu381, Glu382, Glu384, and Asp385 with neutral alanine residues (ACMUT1).
The CSB gene, containing the mutant or wild-type sequence, was cloned into the mammalian expression vector pc3.1 to yield pc3.1-CSB. Briefly, pcBLsSE6 was digested to completion with SalI and a 4.7-kb linear DNA fragment containing the entire CSB gene was excised from a 0.8% agarose gel and purified using Gene-Clean II kit (BIO 101, La Jolla, CA). pc3.1 was digested with XhoI to completion, and the 5.4-kb linear DNA fragment was excised from a 0.8% agarose gel and purified using Gene-Clean II. The 5′ phosphates were removed from the linear 5.4-kb DNA fragments using calf intestinal alkaline phosphatase, and the 4.7-kb DNA fragment containing the CSB gene was ligated into pc3.1 using Ready to Go T4 DNA ligase. Site-directed mutations in the CSB gene in each construct were verified by sequencing. The entire CSB coding sequence of pc3.1-CSBwt and pc3.1-CSBE646Q was sequenced to verify that the coding sequences of the two plasmids were identical with the exception of the site-specific mutation.
Construction and Selection of UV61 Stable Transfectant Cell Lines
CsCl-purified pc3.1-CSB plasmids (100 μg) were linearized with PvuI (200 U) at 37°C for 16 h. The DNA sample was treated with proteinase K (200 μg/ml) in 1 M lithium acetate and 0.1% SDS at 50°C for 2 h, phenol extracted, and ethanol precipitated. UV61 cells were transfected with pc3.1 and pc3.1-CSB plasmids using liposomes (N-[1-(2,3-dioleoyloxyl)propyl]-N,N,N-trimethylammoniummethyl sulfate; Boehringer Mannheim) according to manufacturer’s procedure. Briefly, 250,000 cells were seeded in a 3-cm2 dish and allowed to grow until 50% confluent. To these cells were added 40 μg of N-[1-(2,3-dioleoyloxyl)propyl]-N,N,N-trimethylammoniummethyl sulfate and 10 μg linearized plasmid in 1 ml of media. The cells were incubated with the liposome-DNA mixture for 6 h at 37°C. The media were then replaced, and cells were grown for an additional 48 h at 37°C. The cells were trypsinized and transferred to a 10-cm2 dish, and geneticin was added to a final concentration of 400 μg/ml for selection of antibiotic-resistant cells. After 10 d of selection, the surviving cells were trypsinized and seeded for isolation of clones (50 cells per 10-cm2 dish). Individual colonies were isolated and reseeded for UV and 4-NQO experiments.
Analysis of CSB Expression
Whole-cell lysate and cytoplasmic and nuclear extracts of hamster CS-B transfectant cell lines were tested by immunoblot to detect CSB protein using an affinity-purified rabbit polyclonal antibody against a recombinant fragment of the human CSB protein (amino acids 528-1222) graciously provided by Drs. C. Selby and A. Sancar (University of North Carolina, Chapel Hill, NC) (Selby and Sancar, 1997a). Although the CSB antibody effectively reacted with native CSB protein expressed in human cell lines GM0637D and HeLa by immunoblot analysis (our unpublished results), we were unable to detect CSB protein in the UV-resistant hamster cell line UV61/pc3.1-CSBwt. The lack of detection of CSB protein in the hamster transfectant cell line may be accounted for by several explanations. The level of CSB protein may be quite low in the hamster CS-B transfectant cell line but at a sufficient level to complement the TCR deficiency by the wild-type CSB protein. Alternatively, the CSB protein may be poorly recognized by the CSB antibody when the human protein is present in a preparation (whole-cell lysate or cytoplasmic or nuclear fraction) in a background of hamster proteins.
Two approaches were used to examine the integrity and relative amounts of CSB transcripts from UV61 transfectant cell lines. To evaluate the expression of intact CSB transcript in isolated clones of UV61 transfectants, RNA was isolated and evaluated by RT-PCR using CSB-specific primers. The first-strand synthesis was performed according to the method of Mallery et al. (1998) with the following modifications. RNA was extracted from 107 cells using RNA STAT-60 (Tel-Test, Friendswood, TX) according to manufacturer’s protocol. cDNA synthesis was performed by incubating 5 μg of total RNA, 1.5 μg oligo(dT)12–18 primer (Life Technologies), and 500 U of Superscript II (Life Technologies) at 44°C for 60 min. Samples were subsequently processed using the manufacturer’s recommended procedure for Superscript II. For PCR amplification of cDNA products, 5 U of AmpliTaq Gold (Perkin Elmer, Norwalk, CT) were used according to the manufacturer’s procedures. The 4.7-kb cDNA product was amplified as six overlapping fragments ranging in size from ∼0.6 to 1.5 kb using primers and annealing conditions previously described (Mallery et al., 1998). DNA products were verified by electrophoresis on a 1.2% agarose-1× Tris acetate-EDTA gel and analysis by ethidium bromide staining (our unpublished results). For sequencing, PCR products were purified using a QIAquick PCR purification kit (Qiagen, Hilden, Germany) and analyzed using an Applied Biosystems (Foster City, CA) automatic sequencer. Comparison of sequence analysis of cDNA obtained from UV61/pc3.1-CSBE646Q, UV61/pc3.1-CSBAC377, or UV61/pc3.1-CSBACMUT1 with UV61/pc3.1-CSBwt verified the presence of the engineered mutation at the defined position (our unpublished results). The entire cDNAs of UV61/pc3.1-CSBwt and UV61/pc3.1-CSBE646Q were sequenced to verify that the only difference between the wild-type and E646Q mutant CSB genes was the engineered mutation (our unpublished results).
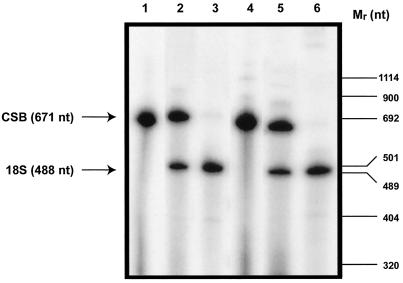
To quantitate the relative levels of CSB expression in UV61/pc3.1-CSBwt and UV61/pc3.1-CSBE646Q, we performed relative quantitative RT-PCR experiments (QuantumRNA module; Ambion, Austin, TX). The QuantumRNA module provides a method for comparing relative transcript abundances standardized by coamplification of a highly conserved fragment of 18S rRNA as an invariant internal standard. The CSB transcript and 18S rRNA were analyzed during the exponential phase of amplification to determine an accurate level of relative expression. Initial experiments were performed to determine the optimal conditions for the linear range of amplification of the CSB fragment. Once this was established, the amplification efficiency of the 18S rRNA standard was optimized to the efficiency of the amplification of the CSB transcript by adjusting the ratio of 18S:competimers. Competimers are modified 18S rRNA primers blocked at their 3′ ends to prevent extension by DNA polymerase (QuantumRNA module). PCR cycling of the CSB fragment and 18S RNA standard was performed in a single tube. For CSB cDNA amplification, the primers 5′-GGTGTTAGGTGGCTGTGGGAATT-3′ (3F) and 5′-GTATCTCGTAAGACACACATGCACAC-3′ (3R) were used, which produce a 671-bp product from the coding sequence of CSB mRNA (Mallery et al., 1998). The RNA used for cDNA synthesis was isolated from UV61 transfectant cell lines by the procedure described above. cDNA synthesis was performed by incubating 2.5 μg of total RNA, 250 ng of random decamer primer (Ambion), and 500 U of Superscript II at 42°C for 50 min. For PCR amplification of cDNA products, 5 U of AmpliTaq Gold (Perkin-Elmer) were used according to the manufacturer’s procedures. In addition to the standard reaction components, PCR mixtures contained 20 pmol of CSB primers 3F and 3R, 20 pmol of 18S RNA primer:competimer (9:1), and [α-32P]dCTP (0.1 μCi/μl) in a total volume of 50 μl. Reaction products were amplified by 31 cycles (94°C, 1 min; 67°C, 1.5 min; and 72°C, 3 min). Ten microliters of the reaction products were mixed with 10 μl of 2× denaturing gel loading buffer (95% formamide) and denatured at 95°C for 3 min. The samples were loaded onto an 8 M urea-6% polyacrylamide gel. Radiolabeled DNA species in polyacrylamide gels were visualized using a PhosphorImager and quantitated using the ImageQuant software (Molecular Dynamics, Sunnyvale, CA). The ratio of the counts of the upper CSB band (671 nucleotides [nt]) and the lower 18S RNA standard (488 nt) band was determined.
The results of the relative RT-PCR experiments are shown in Figure 2. The ratios of CSB transcript to 18S RNA standard (671-nt CSB fragment:488-nt 18S RNA internal standard) for UV61/pc3.1-CSBwt and UV61/pc3.1-CSBE646Q were determined to be 1.68 ± 0.05 and 1.37 ± 0.15, respectively. The results demonstrate that the wild-type and mutant CSB transcripts are expressed with nearly the same efficiency in the UV61 transfectant cell lines.
Figure 2.
Relative quantitative RT-PCR of CSB transcripts isolated from UV61/pc3.1-CSBwt and UV61/pc3.1-CSBE646Q. CSB expression in hamster transfectant cell lines was determined relative to the internal 18S rRNA standard as described in MATERIALS AND METHODS. RT-PCR products were electrophoresed on an 8 M urea-6% polyacrylamide gel. The RT-PCR products from the CSB mRNA and the 18S rRNA using site-specific CSB and 18S primers respectively are indicated. Reaction products from RT-PCR amplifications are as follows: lanes 1–3, UV61/pc3.1-CSBwt using CSB primers (lane 1), CSB primers plus 18S RNA primers (lane 2), and 18S RNA primers (lane 3); lanes 4–6, UV61/pc3.1-CSBE646Q using CSB primers (lane 4), CSB primers plus 18S RNA primers (lane 5), and 18S RNA primers (lane 6).
UV and 4-NQO Survival Assays
Chinese hamster cell AA8 and UV61 transfectants were trypsinized, and 300 cells were seeded per 10-cm2 dish and allowed to grow overnight. For UV treatment, the cells were washed once with PBS and then irradiated at the indicated doses of UV light (254 nm). The cells were grown for 6 d, washed once in PBS, and fixed with methanol for 10 min. The fixed cells were then stained with methylene blue and washed once in PBS, and blue colonies were counted to determine the clonogenic survival of cells. For 4-NQO treatment, the cells were washed once with PBS and incubated in media lacking serum but containing 4-NQO at the indicated concentrations for 1 h at 37°C. Cells were washed once with PBS, and the media were replaced with media supplemented with 10% fetal bovine serum and antibiotics. The cells were then allowed to grow for 6 d and treated as described above to determine colony-forming units.
RNA Synthesis Recovery
UV61 transfectant cell lines were grown in the presence of [14C]thymidine (0.02 μCi/ml) for 3 d to uniformly label the DNA. For UV experiments, the cells were washed with PBS and irradiated with UV light (254 nm) at the indicated doses. For 4-NQO experiments, the cells were washed with PBS and exposed to the indicated concentrations of 4-NQO in serum-free media for 60 min. Cells were restored to complete media and incubated for 16 h. Cells were subsequently pulse labeled with 5 μCi/ml [3H]uridine for 60 min at 37°C, washed once with PBS, and lysed in 10 mM Tris, pH 8.0, 1 mM EDTA buffer containing 0.5% SDS and 100 μg/ml proteinase K for 2 h at 37°C. Trichloroacetic acid (TCA) was added to the cell lysate at a final concentration of 10%, and the samples were then spotted onto glass fiber discs (Whatman, Maidstone, United Kingdom). The filters were sequentially washed in 5% TCA, 70% ethanol, and acetone. The TCA-precipitable radioactivity was then scintillation counted.
Gene-specific Repair
Treatment of cells in culture with UV light and subsequent isolation of genomic DNA was essentially performed according to previously published techniques (Bohr and Okumoto, 1988). In brief, cells were either untreated or irradiated with UV light (254 nm) at a dose of 20 J/m2. The cells were harvested (0 h) or incubated in fresh medium containing bromodeoxyuridine and fluorodeoxyuridine and harvested after 8 or 24 h. Cells were lysed by a proteinase K-SDS treatment, and total genomic DNA was extracted after an NaCl salting out protocol (Miller et al., 1988). Genomic DNA was treated with RNase A (100 g/ml) and KpnI restriction enzyme as described previously (Bohr and Okumoto, 1988). The resulting restriction fragment (14 kb) resides entirely within the 5′ end of the DHFR gene (May et al., 1993). Unreplicated DNA was isolated from replicated DNA (containing bromodeoxyuridine) in CsCl density gradients, dialyzed against Tris, pH 8.0, 1 mM EDTA buffer, and ethanol precipitated. All DNA concentrations were measured spectrophotometrically at 260 nm.
The presence of CPDs in DNA samples was measured in an enzymatic assay as described previously (Bohr et al., 1985). In brief, unreplicated DNA samples (10 μg) from the different time points were incubated with T4 endonuclease V in a reaction buffer containing 10 mM Tris, pH 8.0, 100 mM NaCl, and 10 mM EDTA for 20 min at 37°C, and the reactions were stopped by the addition of alkaline loading dye containing 10 mM EDTA and 500 mM NaOH. In parallel, comparable reactions minus T4 endonuclease V were performed to determine the original distribution of restriction fragments.
T4 endonuclease V-treated and untreated DNA samples were electrophoresed under alkaline conditions on a 0.5% agarose gel. The DNA was transferred to a Hybond N+ membrane (Amersham Pharmacia Biotech) by Posiblot (Stratagene, La Jolla, CA) using standard protocols (Bohr and Okumoto, 1988). The membrane was baked at 80°C for 2 h before treatment with prehybridization buffer (0.342 M Na2HPO4 + 0.088 M NaH2PO4, pH 7.2, 7% SDS, and 2 mM EDTA) for a minimum of 2 h. Double-stranded DNA probes were synthesized with a random-primed labeling kit (Amersham Pharmacia Biotech) using a 3.4-kb DHFR gene template as described (May et al., 1993). Membranes were hybridized with the probe at 68°C overnight and washed to remove nonhybridized probe. The blots were visualized using a PhosphorImager and quantitated using the ImageQuant software. By comparison of untreated and T4 endonuclease V-treated DNA samples, the average number of CPDs per fragment was determined from the zero class of the Poisson distribution. Percent repair is calculated as the frequency of lesions remaining at a given time point compared with the CPD frequency at time zero. In some experiments, negative values for repair were obtained as observed previously (Orren et al., 1996), indicating no detectable repair.
RESULTS
To evaluate the relative biological importance of the ATPase and acidic domains of CSB protein, site-specific mutations were introduced into these motifs, and the CSB mutant alleles were transfected into the hamster CS-B homologue UV61. Specifically, a point mutation in the ATPase domain of CSB, designated CSBE646Q, replaced the highly conserved glutamic acid found in a large number of ATPases and helicases with a neutral glutamine (Figure 1). In addition, two mutations were engineered in a stretch of 10 consecutive acidic residues in a negatively charged region of CSB located in the amino terminus before the helicase motifs. CSBAC377 is a CSB deletion mutant in which the entire 10-acidic-residue stretch is removed from the protein (Figure 1). CSBMUT1 is a mutant in which four acidic residues in the acidic core are replaced with neutral alanines (Figure 1). The CSB mutant alleles were cloned behind a cytomegalovirus strong promotor in the mammalian expression vector pc3.1. The CSB expression plasmids were transfected into the hamster CS-B homologue UV61, and isogenic stable transfectants were obtained for studies. Quantitation of relative amounts of CSB transcript from the cell lines UV61/pc3.1-CSBwt and UV61/pc3.1-CSBE646Q demonstrated very similar levels of expression (Figure 2). However, the inability to detect CSB protein in the hamster transfectant cell lines implies that we cannot dismiss the possibility that CSB protein levels may differ between the cell lines because of translation and degradation effects. We examined repair and transcription of CS-B transfectants by genetic complementation assays after treatment with the DNA-damaging agents UV light and 4-NQO. The phenotypes measured to assess DNA damage sensitivity included 1) viability, 2) RNA synthesis recovery, and 3) gene-specific DNA repair.
UV Sensitivity of CS-B Transfectants
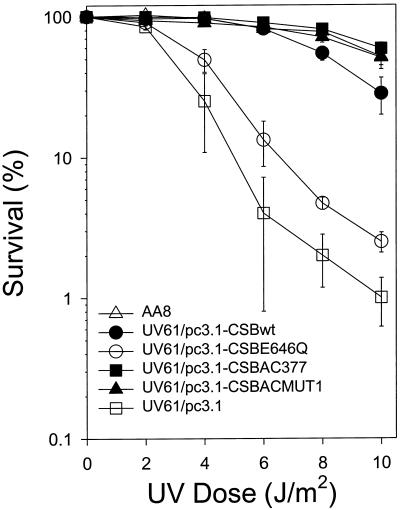
We determined the relative UV sensitivity of various CS-B-transfected clonal cell lines compared with AA8 by clonogenic survival assays. As shown in Figure 3, CSBE646Q, containing a point mutation in motif II of the ATPase domain, was severely compromised in its ability to complement the UV-sensitive phenotype exhibited by the vector alone (UV61/pc3.1), whereas the wild-type CSB allele restored UV resistance. In contrast to CSBE646Q, the acidic mutants CSBACMUT1 and CSBAC377 fully complemented the UV sensitivity to a level comparable with UV61/pc3.1-CSBwt.
Figure 3.
UV sensitivity of AA8 and isogenic clonal populations of UV61 transfectant cell lines. Cells were irradiated with UV light as described in Materials and Methods. Data (percent survival) are expressed as the number of UV-irradiated cells forming colonies as a fraction of the colonies formed by unirradiated cells and represent the average of at least three independent experiments.
The viability studies clearly show that replacement of the highly conserved glutamic acid in the ATPase domain of CSB with a neutral glutamine dramatically impairs the function of the CSB protein. Thus the integrity of the ATPase domain of CSB is critical for cellular resistance to UV light. On the contrary, deletion of 10 consecutive acidic residues in the highly negatively charged motif of CSB does not impair the genetic pathway conferring UV resistance. These data suggest that the acidic region, which resides outside the ATPase and helicase motifs, is dispensable, whereas the ATPase domain of CSB is important for cell survival after UV-induced DNA damage.
RNA Synthesis Recovery after UV Treatment
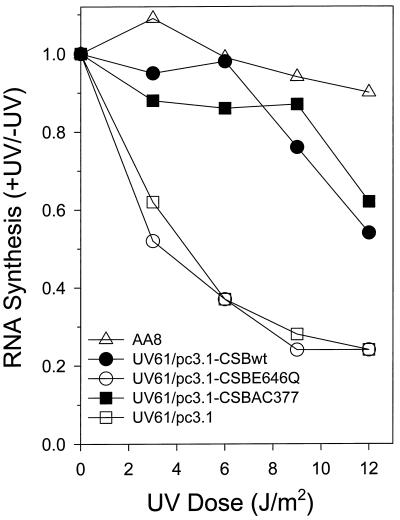
To further characterize the molecular defects giving rise to the increased cell killing of CS-B mutant cell lines by UV light, we examined RNA synthesis recovery in clonal populations of UV61 cell lines transfected with wild-type or site-specific mutant CSB genes (Figure 4). Recovery of RNA synthesis was an early hallmark of the CS phenotype (Mayne and Lehmann, 1982). This assay is generally thought to reflect TCR. RNA synthesis was measured by [3H]uridine incorporation during a 60-min pulse 16 h after exposure of cells to UV light. Cells were either unirradiated or irradiated with 3, 6, 9, or 12 J/m2. After a dose of 3 J/m2, transcription levels in UV61/pc3.1 and UV61/pc3.1-CSBE646Q were reduced to approximately one-half the level of the parental cell line AA8. In contrast, UV61 cell lines transfected with either pc3.1-CSBwt or pc3.1-CSBAC377 showed a level of transcription only slightly reduced compared with AA8. At a UV dose of 6 J/m2, the RNA synthesis of UV61/pc3.1-CSBE646Q and UV61/pc3.1 was reduced to 0.37, but UV61/pc3.1-CSBwt remained close to 1 (0.98). UV61/pc3.1- CSBAC377 also retained a high level of RNA synthesis (0.86), only slightly reduced compared with UV61/pc3.1-CSBwt. Similar results were obtained at a UV dose of 9 J/m2. At 12 J/m2, the recovery of RNA synthesis was compromised for UV61/pc3.1-CSBwt and UV61/pc3.1-CSBAC377 but still nearly threefold greater than either UV61/pc3.1 or UV61/pc3.1-CSBE646Q. These results correlate well with the viability studies. The failure of the ATPase point mutant to complement the RNA synthesis inhibition in UV61 cells strongly suggests that ATPase activity is instrumental for CSB protein to genetically function in the TCR pathway. In contrast, deletion of 10 consecutive acidic residues in the acidic region of CSB does not impair RNA synthesis recovery, indicating that the acidic domain of CSB is not likely to be important for its repair function.
Figure 4.
RNA synthesis recovery of UV61 transfectant cell lines after UV irradiation with increasing dose. Cells unirradiated or UV irradiated with the indicated dose were pulse labeled with [3H]uridine 16 h after irradiation, and acid-insoluble radioactivity was determined.
Gene-specific Repair of UV-induced DNA Damage
To directly measure the ability of CSB mutants to function in preferential repair of UV-induced DNA damage, gene-specific repair experiments were performed. We measured the induction and removal of CPDs in the DHFR housekeeping gene in AA8 and UV61 transfectant cell lines. Quantitative Southern blots were used to measure the amount of CPDs in the DHFR gene 8 and 24 h after irradiation (Figure 5). Seventy-one percent of the CPDs in the DHFR gene in AA8 were repaired by 24 h compared with 43% repair in UV61/pc3.1-CSBwt (Table 1). Both acidic mutant cell lines UV61/pc3.1-CSBAC377 and UV61/pc3.1-CSBMUT1 demonstrated repair to the level of the wild-type CS-B transfectant, indicating that these CSB mutant proteins are competent in TCR. In contrast, no repair of CPDs was detected in UV61/pc3.1-CSBE646Q or UV61/pc3.1 at 8 or 24 h, indicating that the E646Q mutant CSB protein was completely nonfunctional in TCR (Table 1). The gene-specific repair analysis demonstrates that the ATPase point mutation has rendered CSB protein defective in removing CPDs. The repair deficiency observed in UV61/pc3.1-CSBE646Q is consistent with the increased UV sensitivity and prolonged transcription inhibition after UV damage. In contrast, both the AC377 and ACMUT1 acidic mutants retained the ability to preferentially repair UV-induced DNA damage in an active gene. We thus do not observe any phenotypic defect in TCR with the acidic mutations.
Figure 5.
Formation and removal of CPDs in the DHFR gene in AA8 and UV61 transfectant cell lines. Genomic DNA (10 μg) was isolated from either unirradiated (−UV) or UV irradiated (20 J/m2) cells at 0, 8, and 24 h after exposure as described in MATERIALS AND METHODS. The DNA was subsequently digested with KpnI and either treated with T4 endonuclease V (+) or untreated (−). DNA was electrophoresed through an alkaline agarose gel (0.5%) and quantitatively transferred to a nylon membrane. The membrane was hybridized to 32P-labeled denatured double-stranded DNA probe for the DHFR gene. Hybridization of gene-specific 32P-labeled DHFR probes to membranes was detected by PhosphorImager analysis.
Table 1.
Induction and removal of CPD in the DHFR gene
| Cell line | Initial CPD | Repair (%)
|
|
|---|---|---|---|
| 8 h | 24 h | ||
| AA8 | 1.55 ± 0.05 | 35.5 ± 4.95 | 71.5 ± 10.6 |
| UV61/pc3.1-CSBwt | 1.53 ± 0.06 | 20.2 ± 10.7 | 46.3 ± 15.28 |
| UV61/pc3.1-CSBAC377 | 1.72 ± 0.13 | 26.1 ± 7.64 | 51.9 ± 8.46 |
| UV61/pc3.1-CSBACMUT1 | 1.45 ± 0.19 | 27.1 ± 10.14 | 37.9 ± 12.10 |
| UV61/pc3.1-CSBE646Q | 1.35 ± 0.12 | −17.1 ± 6.42 | −16.4 ± 9.04 |
| UV61/pc3.1 | 1.39 ± 0.06 | 5.7 ± 5.41 | −4.6 ± 6.8 |
Mean ± SD are calculated from three independent experiments.
4-NQO Survival Studies
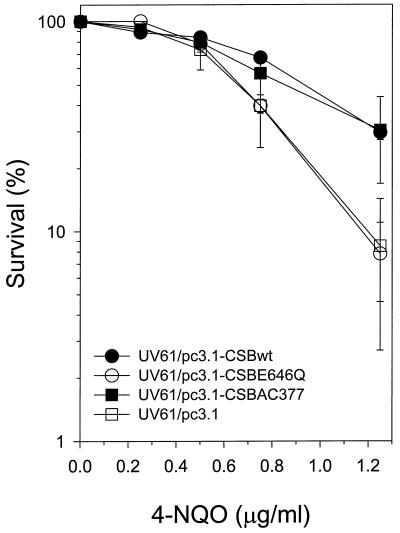
The genotoxin 4-NQO introduces a number of types of DNA damage, including bulky lesions, oxidative damage such as 8-hydroxyguanine, and strand breaks (Friedberg et al., 1995). However, unlike UV-induced CPDs, the 4-NQO-induced bulky adducts recognized by UvrABC excinuclease are repaired without strand bias (Snyderwine and Bohr, 1992) and thus without transcription coupling. To characterize the functional significance of the ATPase and acidic domains of CSB in this TCR-independent pathway, the respective CSB mutant alleles were examined for their abilities to complement the 4-NQO-sensitive phenotype of CS-B mutant cell line UV61 as previously demonstrated by clonogenic survival assays (Wade and Chu, 1979). Clonal populations of the CS-B transfectant cell lines were evaluated for survival as a function of 4-NQO dose (Figure 6). The ATPase point mutant CSBE646Q failed to complement UV61 in resistance to 4-NQO, whereas the wild-type CSB allele, introduced by the same plasmid during transfection, exhibited complementation. The acidic mutant CSB allele CSBAC377 complemented the 4-NQO sensitivity to a level similar to that of the wild-type CSB allele. We conclude that the CSBE646Q allele fails to complement the 4-NQO-sensitive phenotype, reflecting a phenotype similar to that observed with UV light.
Figure 6.
4-NQO sensitivity of AA8 and isogenic clonal populations of UV61 transfectant cell lines. Cells were treated with 4-NQO as described in MATERIALS AND METHODS. Data (percent survival) are expressed as the number of 4-NQO-treated cells forming colonies as a fraction of the colonies formed by untreated cells and represent the average of at least three independent experiments.
RNA Synthesis Recovery after 4-NQO Treatment
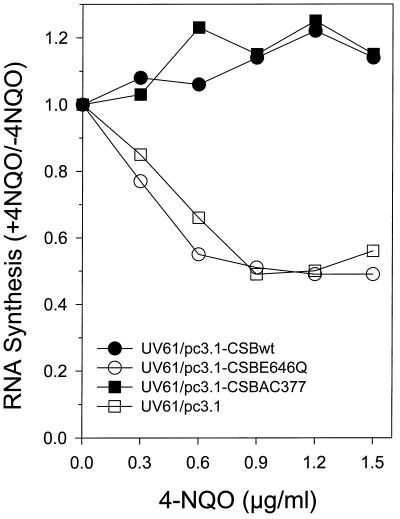
To further characterize the sensitivity of CS-B cell lines to the DNA-damaging agent 4-NQO, recovery of RNA synthesis after exposure of cells to 4-NQO was measured (Figure 7). At all doses tested, UV61/pc3.1 and UV61/pc3.1-CSBE646Q exhibited closely matched reduced levels of [3H]uridine incorporation compared with the 4-NQO-resistant lines UV61/pc3.1-CSBwt and UV61/pc3.1-CSBAC377. The peak reduction in RNA synthesis of UV61/pc3.1-CSBE646Q, ∼50% of the control, was attained at a minimal 4-NQO concentration of 0.6 μg/ml. At this concentration, both UV61/pc3.1-CSBwt and UV61/pc3.1-CSBAC377 exhibited levels of RNA synthesis ≥1. These results demonstrate that the E646Q replacement inactivated the genetic function of CSB to restore transcription after 4-NQO-induced DNA damage. On the contrary, the CSBAC377 mutant protein with a deletion of the core 10 acidic residues in the largely negatively charged region of CSB facilitated RNA synthesis recovery, suggesting that the acidic domain is not important for processing DNA damage adducts induced by either 4-NQO or UV.
Figure 7.
RNA synthesis recovery of UV61 transfectant cell lines after 4-NQO treatment with increasing dose. Cells unexposed or exposed to 4-NQO at the indicated dose were pulse labeled with [3H]uridine 16 h after treatment, and acid-insoluble radioactivity was determined.
DISCUSSION
The results in this study demonstrate that the single amino acid substitution of glutamine for an invariant glutamic acid in motif II of the ATPase domain severely reduced the function of CSB protein in cellular pathways conferring resistance to UV light. Specifically, the CSBE646Q mutant was seriously compromised in its ability to complement the UV sensitivity of the hamster CSB homologue UV61 by a colony-forming survival assay. Moreover, the CSB ATPase point mutant failed to function in RNA synthesis recovery and gene-specific repair. The genetic data suggest that ATP hydrolysis catalyzed by CSB protein is an important step in TCR of UV-induced DNA damage in vivo. Abrogation of CSB function in the TCR pathway by the ATPase point mutation resulted in UV-induced cell killing and failure to resume RNA synthesis.
The genetic results presented here suggest that CSB functions in TCR by a step involving ATP hydrolysis. This notion is consistent with biochemical data that demonstrate that hydrolysis of the ATP β-γ phosphoanhydride bond is required for the interaction of CSB with a stalled RNAPII on a DNA template (Tantin et al., 1997). The ability of the RNAPII–CSB–DNA–RNA quaternary complex to recruit a molecular complex containing the TFIIH core subunits p62 and XPB provides a biochemical basis to facilitate repair of active genes (Tantin, 1998). However, the precise role of CSB-catalyzed ATP hydrolysis in TCR remains to be elucidated. It is quite likely that the enzymatic turnover of ATP may be important in the role of CSB protein as a repair-coupling factor. Alternatively, ATP hydrolysis may be essential to the function of CSB as a transcription elongation factor. Stimulation of the efficiency and rate of RNAPII transcription would boost the number of encounters of RNAPII with sites of DNA damage, which may serve as the damage-signaling function in mammalian TCR. Thus CSB would stimulate the rate of lesion removal from the transcribed strand by increasing the efficiency and rate of RNAPII transcription. Either possibility would explain the genetic deficiency of the CSBE646Q mutant in TCR.
The functional significance of ATP hydrolysis by CSB protein has also been examined in the immortalized human CS-B fibroblast cell line CS1AN (Citterio et al., 1998). In these studies, replacement of the invariant lysine in motif I of the ATPase domain with an arginine (K538R) abolished the ATPase activity of the protein. Purified mutant and wild-type CSB proteins were microinjected into the cytoplasm of CS1AN fibroblasts, and recovery of RNA synthesis was measured 16–20 h after exposure to UV light (15 J/m2). The authors observed 90% recovery of RNA synthesis by cells injected with the wild-type CSB protein compared with basal transcription of unirrradiated cells (Citterio et al., 1998). In contrast, the level of RNA synthesis of cells injected with the K538R mutant protein was restored to 38% of that of the control. CS1AN cells that were not injected with any CSB protein, mutant or wild-type, exhibited only 22% of the level of RNA synthesis of unirradiated cells. The 1.7-fold difference in RNA synthesis recovery after UV exposure between CS1AN injected with K538R mutant protein and CS1AN not injected with CSB protein suggested that the catalytically inactive mutant CSB protein retained partial function in vivo. Citterio et al. (1998) proposed that the enzymatically inactive mutant CSB-K538R protein possibly functions to stabilize a CSB-containing protein complex (involving RNAPII?), enabling the complex to retain residual activity. A previous demonstration of a stable association of a fraction of RNAPII (10–15%) with wild-type CSB in cell-free extracts (van Gool et al., 1997) is consistent with this proposal.
The results in this study clearly show that the E646Q substitution in motif II of the ATPase domain severely reduced the ability of the CSB protein to complement the UV-sensitive phenotypes of survival, RNA synthesis recovery, and gene-specific repair of the hamster cell line UV61. The slightly higher levels of cell survival of UV61/pc3.1-CSBE646Q compared with UV61/pc3.1 may suggest that the ATPase point mutant CSB protein may weakly function to confer UV resistance; however, the difference between UV61/pc3.1-CSBwt and UV61/pc3.1-CSBE646Q is an order of magnitude at the higher doses of UV light (8 and 10 J/m2), suggesting that the E646Q mutation has greatly diminished the biological function of CSB protein. In addition, the CSBE646Q mutant completely failed to complement the UV-sensitive phenotypes of RNA synthesis recovery and gene-specific repair of the UV61 cell line, consistent with the notion that the mutant CSB protein fails to function in TCR.
Based on the UV survival data (Figure 3), it is possible that the CSBE646Q mutant protein retains residual biological activity. It was previously shown that mutations in swi2 that reduce ATP hydrolysis impair cellular transcription of SWI/SNF-dependent genes and abolish chromatin remodeling in vitro by the reconstituted complex (Laurent et al., 1993; Richmond and Peterson, 1996). A double point mutation of acidic residues D894 and E895 of SWI2, which are conserved in motif II of the ERCC6 and SWI/SNF subfamilies, resulted in a complete loss of transcription activity for yeast SWI2/SNF2 (Richmond and Peterson, 1996). The SWI2 D894AE895A mutant protein could be incorporated into a multiprotein complex in vivo, although the complex was less stable in vitro. Richmond and Peterson (1996) concluded that swi2 mutations in ATPase motif II do not grossly impair SWI/SNF complex assembly. More recently, it was reported that a yeast swi2K798A allele containing a point mutation in motif I of the ATPase domain supported a wild-type growth rate in contrast to the reduced viability of the swi2 deletion mutant allele (Biggar and Crabtree, 1999). These genetic data suggest that there may be residual ATP-independent activity of the SWI2 protein. Likewise, it is possible that the ATPase mutant CSB protein is able to weakly complement the UV-sensitive phenotype.
The dramatic loss of RNA synthesis recovery after UV treatment of the E646Q mutant versus the significantly compromised but residual function of the K538R mutant (Citterio et al., 1998) may be explained by a number of reasons, some of which will be considered here. First, we used a hamster cell line that was originally used to identify the human CSB gene by cross-complementation, whereas Citterio et al. (1998) used an SV40-transformed human CS-B cell line. It is certainly possible that the difference in genetic background may influence the amount, stability, or activity of the CSB protein. Second, the site-directed mutations introduced to the CSB protein are directed at two spatially separated sequence motifs and may differentially impact CSB function in vivo. Crystal structure data (Subramanya et al., 1996; Korolev et al., 1997) and mutational analysis (Sung et al., 1988; George et al., 1994; Brosh and Matson, 1995) have implicated both motifs I and II in nucleotide binding and/or hydrolysis. Motif I, which contains the signature NTP-binding sequence GSGKS, is responsible for binding the triphosphate tail of the ATP. Motif II, which contains the DEXH sequence, is involved in binding Mg+2 via the acidic residues and is required for hydrolysis of ATP in these enzymes. Thus the conserved residues of both domains play an important role in ATP hydrolysis. It should be pointed out, however, that the phenotypic difference between the CSBK538R and CSBE646Q mutant cell lines may reflect the differential effect of the point mutations on the CSB protein. In both studies the recovery of RNA synthesis of CS-B cells with the ATPase mutant CSB protein present was significantly reduced compared with cells containing the wild-type CSB protein, indicating that mutations in either domain seriously affect the function of CSB in vivo. These data are consistent with the notion that CSB exerts its effect on the TCR pathway by a step involving ATP hydrolysis.
A third consideration to explain the lack of genetic complementation by the CSBE646Q allele is that the mutant protein is rendered unstable by the amino acid replacement. Contrary to this notion, a number of studies in which similar motif II mutations have been made to ATPases and helicases have shown that the stability of the mutant proteins did not change (Pause and Sonenberg, 1992; Jindal et al., 1994; Brosh and Matson, 1995; Richmond and Peterson, 1996; Washington et al., 1996). In addition, biochemical analysis of the mutant proteins has demonstrated that properties other than ATP hydrolysis or unwinding activity have remained intact (Pause and Sonenberg, 1992; Jindal et al., 1994; Brosh and Matson, 1995; Richmond and Peterson, 1996; Washington et al., 1996). For example, motif II point mutants retain nucleotide binding (UvrD, eIF-4A, and NS-1), DNA binding (UvrD and T7 primase and helicase), and protein interaction (T7 primase and helicase and Swi2). These findings indicate that replacement of the highly conserved acidic residues in motif II with neutral amino acids does not grossly impair the overall structure of these proteins. These results suggest that the lack of genetic function of the CSBE646Q allele is not likely to be a consequence of protein destabilization because of the single amino acid substitution.
In contrast to the critical importance of the ATPase domain of CSB, genetic characterization of UV61/pc3.1-CSBAC377 suggests that the acidic region is not important in TCR. The CSBAC377 mutant is characterized by a deletion of 10 consecutive acidic residues in the “core” of a negatively charged domain in the CSB protein. The possibility exists that the remaining 11 acidic residues of the region fulfill the functional requirement of CSB in TCR. Similarly, the replacement of four acidic residues in the core of the acidic region with neutral alanine residues did not impact the function of CSB protein. Perhaps the acidic domain of CSB has some other specialized function not detected by the genetic assays used in this study. An interesting area to explore is the transcriptional status of various CS-B mutant cell lines. The presence of an acidic region in the domain responsible for activation of the transcriptional machinery in a large class of transcriptional activators suggests an important role in transcription (Ma and Ptashne, 1987; Hope et al., 1988). In addition, functional domains of the CSB protein, such as the acidic region, may be important in processing of DNA damage other than UV-induced photoproducts. For example, the molecular functions required of CSB protein to process oxidative DNA damage have not been addressed in this study. Protein–protein interactions, mediated by the acidic domain of CSB, may be essential in one of these pathways. Further studies using a structure–function approach should prove useful in understanding the requirements of CSB protein in DNA repair of various types of DNA damage as well as the process of transcription. In a recent study, Mallery et al. (1998) have analyzed the location of mutations in the CSB gene in CS patients. Their findings demonstrate that the site or nature of the mutation (truncation vs. amino acid substitution) did not correlate well with the severity of the clinical features. These data suggest fairly complex relationships among genotype, molecular phenotype, and clinical phenotype of CS.
In this study we examined some repair characteristics of UV61 transfectant cell lines exposed to the chemical carcinogen 4-NQO. 4-NQO has been referred to as a “UV-mimetic” agent because it elicits similar repair mechanisms to correct DNA damage induced by the compound (Kondo and Kato, 1968; Ikenaga et al., 1975) and because XP cells are sensitive to it (Takebe et al., 1972). NER is responsible for repairing bulky 4-NQO adducts in a manner similar to UV-induced pyrimidine dimers. CS and XP cells have been previously shown to exhibit a hypersensitivity to 4-NQO compared with normal cells, as demonstrated by clonogenic survival assays (Wade and Chu, 1979). However, Snyderwine and Bohr (1992) demonstrated that the gene-specific repair characteristics operating for UV dimers and 4-NQO adducts are different from one another. UV dimers are repaired faster in actively transcribing genes than inactive regions. In contrast, 4-NQO adducts, recognized by UvrABC excinuclease, are repaired without any bias toward the transcriptionally active genes. In addition, there is no preferential repair of these 4-NQO adducts in the transcribed strand over the nontranscribed strand. Collectively these data indicate that 4-NQO adducts are processed by a pathway that is different from TCR of UV-induced DNA damage. Aside from the bulky adducts, several types of oxidative lesions are introduced to DNA in cells exposed to 4-NQO (Galiegue-Zouitina et al., 1985; Kohda et al., 1987). Defective TCR of oxidative base damage has been shown in CS cells (CS-B) (Leadon and Cooper, 1993) and XP-G/CS (Cooper and Leadon, 1994; Cooper et al., 1997), suggesting that oxidative damage may also play a role in the hypersensitivity of CS cell lines to 4-NQO (Wade and Chu, 1979). Therefore, it was interesting to determine what effect mutations in the CSB gene would have on the sensitivity of CS-B transfectant cell lines to 4-NQO. Our results demonstrate that the E646Q (ATPase) point mutation inactivates CSB function as determined by clonogenic survival assays and RNA synthesis recovery after 4-NQO treatment. The requirement for an intact ATPase domain of CSB to complement the deficiencies in survival and RNA synthesis after treatment with either UV light or 4-NQO suggests that ATP hydrolysis is important in a TCR-dependent pathway (operational for UV-induced CPDs) as well as a TCR-independent pathway. We propose that the molecular function of CSB to confer resistance to 4-NQO is likely to be a mechanism that allows the cell to overcome transcription inhibition by DNA adducts.
It was originally suggested by this laboratory that specific alterations to DNA structure, such as lesion size or degree of distortion caused by the adduct, may explain the lack of TCR of some adducts (Snyderwine and Bohr, 1992). 4-NQO-induced adducts recognized by UvrABC excinuclease, such as UV-induced pyrimidine 6-4 pyrimidone photoproducts, are efficiently removed by general global repair. Other forms of DNA damage, such as UV-induced CPDs, are predominantly repaired by TCR. Despite the existence of a global repair pathway for bulky 4-NQO adducts, RNA synthesis of CS-B cells fails to resume after 4-NQO treatment. Recent studies have also demonstrated a lack of TCR repair of N-acetoxy-2-acetylaminofluorene (N-AAF) adducts, yet the drug is extremely cytoxic and inhibits transcription (van Oosterwijk et al., 1996, 1998). Similar repair kinetics of N- (deoxyguanosine-8-yl)-2-acetylaminofluorene in CS cells compared with normal cells suggested that the elevated drug sensitivity and RNA synthesis inhibition of CS cells by NA-AAF are not consequences of defective TCR. It is conceivable that the lesions induced by 4-NQO or NA-AAF titrate out a critical factor required for transcription initiation and/or elongation. Recent work has suggested that proteins involved in RNAPII transcription may be titrated or hijacked by DNA damage (Vichi et al., 1997; You et al., 1998; Cullinane et al., 1999). Also, a model was proposed in which CS proteins act as repair–transcription uncoupling factors allowing the basal transcription factor TFIIH to convert from a repair mode to a transcription initiation mode (van Oosterwijk et al., 1996, 1998). Presumably, an ATPase-defective CSB protein fails to recycle TFIIH from global genome repair to transcription. Further molecular studies will be necessary to clearly address the functions of CS proteins in TCR-dependent and -independent pathways.
These studies demonstrate that the integrity of the ATPase domain of CSB is essential for the biological function of the protein in UV-resistant phenotypes of survival, RNA synthesis recovery, and TCR. Likewise, the ATPase domain plays a pivotal role in the genetic pathway responsible for the cellular resistance to DNA damage introduced by the chemical carcinogen 4-NQO. Further structure–function studies should provide insight to the molecular functions of CSB in pathways relating to DNA damage, repair, and transcription.
ACKNOWLEDGMENTS
We appreciate the comments by A. Majumdar and the technical help from A. May. We thank Drs. Hoeijmakers and Citterio for help with the Western blot. M.S. was supported by the Danish Center for Molecular Gerontology.
Abbreviations used:
- CPD
cyclobutane pyrimidine dimer
- CS
Cockayne syndrome
- NER
nucleotide excision repair
- 4-NQO
4-nitroquinoline-1-oxide
- nt
nucleotide
- RNAPII
RNA polymerase II
- TCA
trichloroacetic acid
- TCR
transcription-coupled repair
REFERENCES
- Balajee AS, May A, Dianov GL, Friedberg EC, Bohr VA. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc Natl Acad Sci USA. 1997;94:4306–4311. doi: 10.1073/pnas.94.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggar SR, Crabtree GR. Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J. 1999;18:2254–2264. doi: 10.1093/emboj/18.8.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr VA, Okumoto DS. Analysis of pyrmidine dimers in defined genes. In: Friedberg EC, Hanawalt PC, editors. A Laboratory Manual of Research Procedures. New York: Marcel Dekker; 1988. pp. 347–366. [Google Scholar]
- Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- Brosh RMJ, Matson SW. Mutations in motif II of Escherichia coli DNA helicase II render the enzyme nonfunctional in both mismatch repair and excision repair with differential effects on the unwinding reaction. J Bacteriol. 1995;177:5612–5621. doi: 10.1128/jb.177.19.5612-5621.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citterio E, Rademakers S, van der Horst GT, van Gool AJ, Hoeijmakers JH, Vermeulen W. Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J Biol Chem. 1998;273:11844–11851. doi: 10.1074/jbc.273.19.11844. [DOI] [PubMed] [Google Scholar]
- Cooper PK, Leadon SA. Defective repair of ionizing radiation damage in Cockayne’s syndrome and xeroderma pigmentosum group G. Ann NY Acad Sci. 1994;726:330–332. doi: 10.1111/j.1749-6632.1994.tb52842.x. [DOI] [PubMed] [Google Scholar]
- Cooper PK, Nouspikel T, Clarkson SG, Leadon SA. Defective transcription-coupled., repair of oxidative base damage in Cockayne syndrome patients from XP group G. Science. 1997;275:990–993. doi: 10.1126/science.275.5302.990. [DOI] [PubMed] [Google Scholar]
- Cullinane C, Mazur SJ, Essigmann JM, Phillips DR, Bohr VA. Inhibition of RNA polymerase II transcription in human cell extracts by cisplatin DNA damage. Biochemistry. 1999;38:6204–6212. doi: 10.1021/bi982685+. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MK, Bohr VA. Gene-specific DNA repair of UV-induced cyclobutane pyrimidine dimers in some cancer-prone and premature-aging human syndromes. Mutat Res. 1994;314:221–231. doi: 10.1016/0921-8777(94)90067-1. [DOI] [PubMed] [Google Scholar]
- Friedberg EC. Cockayne syndrome—a primary defect in DNA repair, transcription, both or neither? Bioessays. 1996;18:731–738. doi: 10.1002/bies.950180908. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 1995. pp. 42–43. [Google Scholar]
- Galiegue-Zouitina S, Bailleul B, Loucheux-Lefebvre MH. Adducts from in vivo action of the carcinogen 4-hydroxyaminoquinoline 1-oxide in rats and from in vitro reaction of 4-acetoxyaminoquinoline 1-oxide with DNA and polynucleotides. Cancer Res. 1985;45:520–525. [PubMed] [Google Scholar]
- George JW, Brosh RMJ, Matson SW. A dominant negative allele of the Escherichia coli uvrD gene encoding DNA helicase II. A biochemical and genetic characterization. J Mol Biol. 1994;235:424–435. doi: 10.1006/jmbi.1994.1003. [DOI] [PubMed] [Google Scholar]
- Gupta R, Emili A, Pan G, Xiao H, Shales M, Greenblatt J, Ingles CJ. Characterization of the interaction between the acidic activation domain of VP16 and the RNA polymerase II initiation factor TFIIB. Nucleic Acids Res. 1996;24:2324–2330. doi: 10.1093/nar/24.12.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope IA, Mahadevan S, Struhl K. Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature. 1988;333:635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- Ikenaga M, Ichikawa-Ryo H, Kondo S. The major cause of inactivation and mutation by 4-nitroquinoline 1-oxide in Escherichia coli: excisable 4NQO-purine adducts. J Mol Biol. 1975;92:341–356. doi: 10.1016/0022-2836(75)90233-8. [DOI] [PubMed] [Google Scholar]
- Jindal HK, Yong CB, Wilson GM, Tam P, Astell CR. Mutations in the NTP-binding motif of minute virus of mice (MVM) NS-1 protein uncouple ATPase and DNA helicase functions. J Biol Chem. 1994;269:3283–3289. [PubMed] [Google Scholar]
- Kohda K, Tada M, Hakura A, Kasai H, Kawazoe Y. Formation of 8-hydroxyguanine residues in DNA treated with 4-hydroxyaminoquinoline 1-oxide and its related compounds in the presence of seryl-AMP. Biochem Biophys Res Commun. 1987;149:1141–1148. doi: 10.1016/0006-291x(87)90527-4. [DOI] [PubMed] [Google Scholar]
- Kondo S, Kato T. Photoreactivation of mutation and killing in Escherichia coli. Adv Biol Med Phys. 1968;12:283–298. doi: 10.1016/b978-1-4831-9928-3.50015-1. [DOI] [PubMed] [Google Scholar]
- Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- Lapeyre B, Bourbon H, Amalric F. Nucleolin, the major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by the nucleotide sequence. Proc Natl Acad Sci USA. 1987;84:1472–1476. doi: 10.1073/pnas.84.6.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent BC, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes & Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- Leadon SA, Cooper PK. Preferential repair of ionizing radiation-induced damage in the transcribed strand of an active human gene is defective in Cockayne syndrome. Proc Natl Acad Sci USA. 1993;90:10499–10503. doi: 10.1073/pnas.90.22.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR. Three complementation groups in Cockayne syndrome. Mutat Res. 1982;106:347–356. doi: 10.1016/0027-5107(82)90115-4. [DOI] [PubMed] [Google Scholar]
- Lehmann AR, Kirk-Bell S, Mayne L. Abnormal kinetics of DNA synthesis in UV light-irradiated cells from patients with Cockayne’s syndrome. Cancer Res. 1979;39:4237–4241. [PubMed] [Google Scholar]
- Ma J, Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987;48:847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Mallery DL, Tanganelli B, Colella S, Steingrimsdottir H, van Gool AJ, Troelstra C, Stefanini M, Lehmann AR. Molecular analysis of mutations in the CSB (ERCC6) gene in patients with Cockayne syndrome. Am J Hum Genet. 1998;62:77–85. doi: 10.1086/301686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May A, Nairn RS, Okumoto DS, Wassermann K, Stevnsner T, Jones JC, Bohr VA. Repair of individual DNA strands in the hamster dihydrofolate reductase gene after treatment with UV light, alkylating agents, and cisplatin. J Biol Chem. 1993;268:1650–1657. [PubMed] [Google Scholar]
- Mayne LV, Lehmann AR. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne’s syndrome and xeroderma pigmentosum. Cancer Res. 1982;42:1473–1478. [PubMed] [Google Scholar]
- Mellon I, Bohr VA, Smith CA, Hanawalt PC. Preferential DNA repair of an active gene in human cells. Proc Natl Acad Sci USA. 1986;83:8878–8882. doi: 10.1073/pnas.83.23.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DL, Nairn RS. The biology of the (6–4) photoproduct. Photochem Photobiol. 1989;49:805–819. doi: 10.1111/j.1751-1097.1989.tb05578.x. [DOI] [PubMed] [Google Scholar]
- Orren DK, Dianov GL, Bohr VA. The human CSB (ERCC6) gene corrects the transcription-coupled repair defect in the CHO cell mutant UV61. Nucleic Acids Res. 1996;24:3317–3322. doi: 10.1093/nar/24.17.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazin MJ, Kadonaga JT. SWI2/SNF2 and related proteins: ATP-driven motors that disrupt protein-DNA interactions? Cell. 1997;88:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Richmond E, Peterson CL. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 1996;24:3685–3692. doi: 10.1093/nar/24.19.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Drapkin R, Reinberg D, Sancar A. RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Nucleic Acids Res. 1997;25:787–793. doi: 10.1093/nar/25.4.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Cockayne syndrome group B protein enhances by polymerase II. Proc Natl Acad Sci USA. 1997a;94:11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci USA. 1997b;94:11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyderwine EG, Bohr VA. Gene- and strand-specific damage and repair in Chinese hamster ovary cells treated with 4-nitroquinoline 1-oxide. Cancer Res. 1992;52:4183–4189. [PubMed] [Google Scholar]
- Stringer KF, Ingles CJ, Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- Subramanya HS, Bird LE, Brannigan JA, Wigley DB. Crystal structure of a DExx box DNA helicase. Nature. 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- Sung P, Prakash S, Prakash L. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes & Dev. 1988;2:1476–1485. doi: 10.1101/gad.2.11.1476. [DOI] [PubMed] [Google Scholar]
- Takebe H, Furuyama JI, Miki Y, Kondo S. High sensitivity of xeroderma pigmentosum cells to the carcinogen 4-nitroguinoline-1-oxide. Mutat Res. 1972;15:98–100. doi: 10.1016/0027-5107(72)90099-1. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Kawai K, Kumahara Y, Ikenaga M, Okada Y. Genetic complementation groups in cockayne syndrome. Somatic Cell Genet. 1981;7:445–455. doi: 10.1007/BF01542989. [DOI] [PubMed] [Google Scholar]
- Tantin D. RNA polymerase II elongation complexes containing the Cockayne syndrome group B protein interact with a molecular complex containing the transcription factor IIH components xeroderma pigmentosum B and p62. J Biol Chem. 1998;273:27794–27799. doi: 10.1074/jbc.273.43.27794. [DOI] [PubMed] [Google Scholar]
- Tantin D, Kansal A, Carey M. Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol Cell Biol. 1997;17:6803–6814. doi: 10.1128/mcb.17.12.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troelstra C, Odijk H, de Wit J, Westerveld A, Thompson LH, Bootsma D, Hoeijmakers JH. Molecular cloning of the human DNA excision repair gene ERCC-6. Mol Cell Biol. 1990;10:5806–5813. doi: 10.1128/mcb.10.11.5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers JH. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- van Gool AJ, Citterio E, Rademakers S, van Os R, Vermeulen W, Constantinou A, Egly JM, Bootsma D, Hoeijmakers JH. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 1997;16:5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoffen A, Natarajan AT, Mayne LV, van Zeeland AA, Mullenders LH, Venema J. Deficient repair of the transcribed strand of active genes in Cockayne’s syndrome cells. Nucleic Acids Res. 1993;21:5890–5895. doi: 10.1093/nar/21.25.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oosterwijk MF, Filon R, de Groot AJ, van Zeeland AA, Mullenders LH. Lack of transcription-coupled repair of acetylaminofluorene DNA adducts in human fibroblasts contrasts their efficient inhibition of transcription. J Biol Chem. 1998;273:13599–13604. doi: 10.1074/jbc.273.22.13599. [DOI] [PubMed] [Google Scholar]
- van Oosterwijk MF, Versteeg A, Filon R, van Zeeland AA, Mullenders LH. The sensitivity of Cockayne’s syndrome cells to DNA-damaging agents is not due to defective transcription-coupled repair of active genes. Mol Cell Biol. 1996;16:4436–4444. doi: 10.1128/mcb.16.8.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, Mullenders LH, Natarajan AT, van Zeeland AA, Mayne LV. The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc Natl Acad Sci USA. 1990;87:4707–4711. doi: 10.1073/pnas.87.12.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vichi P, Coin F, Renaud JP, Vermeulen W, Hoeijmakers JH, Moras D, Egly JM. Cisplatin- and UV-damaged DNA lure the basal transcription factor TFIID/TBP. EMBO J. 1997;16:7444–7456. doi: 10.1093/emboj/16.24.7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade MH, Chu EH. Effects of DNA damaging agents on cultured fibroblasts derived from patients with Cockayne syndrome. Mutat Res. 1979;59:49–60. doi: 10.1016/0027-5107(79)90194-5. [DOI] [PubMed] [Google Scholar]
- Washington MT, Rosenberg AH, Griffin K, Studier FW, Patel SS. Biochemical analysis of mutant T7 primase/helicase proteins defective in DNA binding, nucleotide hydrolysis, and the coupling of hydrolysis with DNA unwinding. J Biol Chem. 1996;271:26825–26834. doi: 10.1074/jbc.271.43.26825. [DOI] [PubMed] [Google Scholar]
- Wen L, Huang JK, Johnson BH, Reeck GR. A human placental cDNA clone that encodes nonhistone chromosomal protein HMG-1. Nucleic Acids Res. 1989;17:1197–1214. doi: 10.1093/nar/17.3.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Feaver WJ, Friedberg EC. Yeast RNA polymerase II transcription in vitro is inhibited in the presence of nucleotide excision repair: complementation of inhibition by Holo-TFIIH and requirement for RAD26. Mol Cell Biol. 1998;18:2668–2676. doi: 10.1128/mcb.18.5.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]