Abstract
Pontin is a multifunctional protein having roles in various cellular processes including regulation of gene expression. Here, we addressed Pontin intracellular localization using two different monoclonal antibodies directed against different Pontin epitopes. For the first time, Pontin was directly visualized in nucleoli where it co-localizes with Upstream Binding Factor and RNA polymerase I. Nucleolar localization of Pontin was confirmed by its detection in nucleolar extracts and by electron microscopy, which revealed Pontin accumulation specifically in the nucleolar fibrillar centers. Pontin localization in the nucleolus was dynamic and Pontin accumulated in large nucleolar dots mainly during S-phase. Pontin concentration in the large nucleolar dots correlated with reduced transcriptional activity of nucleoli. In addition, Pontin was found to associate with RNA polymerase I and to interact in a complex with c-Myc with rDNA sequences indicating that Pontin is involved in the c-Myc-dependent regulation of rRNA synthesis.
Introduction
The Pontin protein was described as a putative mammalian DNA-helicase containing Walker A and Walker B motifs and belonging to the family of AAA+ ATPases. Pontin was independently described by several research groups that studied diverse cellular processes in different model organisms, which resulted in many Pontin synonyms—Tip49a, Tih2p, rp50, RUVBL1, NMP 238, ECP 54, p55. As part of several protein complexes, the major role of Pontin is apparently in the regulation of chromatin structure and gene expression and in DNA repair and stability. In this context, Pontin was identified as a component of different chromatin remodeling and modifying complexes including the Tip60, Ino80 and Uri complexes (for review see Gallant 2007). In addition, it has been shown that Pontin interacts with several transcription regulators like β-catenin, c-Myc and E2F1 and associates with the promoters targeted by these factors (Bauer et al. 2000; Bauer et al. 1998; Dugan et al. 2002; Frank et al. 2003; Taubert et al. 2004; Wood et al. 2000). Through its interaction with Hint1, Pontin modulates β-catenin mediated transcription in the canonical Wnt-signaling pathway and apoptosis (Weiske and Huber 2005; Weiske and Huber 2006). Besides transcriptional regulation Pontin has additional functions in maturation of small nucleolar ribonucleoprotein particles (snoRNPs; King et al. 2001; McKeegan et al. 2007; Watkins et al. 2002; 2004), in assembly of the telomerase holoenzyme complex (Venteicher et al. 2008) and through association with the mitotic spindle it appears that Pontin is involved in cell division (Gartner et al. 2003).
Several lines of evidence suggest that Pontin may act in the nucleolus: (1) Pontin interacts with c-Myc, which regulates the activity of all three RNA polymerases including RNA polymerase I (Pol I; Arabi et al. 2005; Grandori et al. 2005), (2) Pontin is a part of the Uri complex that includes Rpb5, a protein associated with Pol I (Zaros et al. 2007) and (3) Pontin is involved in the maturation of snoRNPs that mostly reside in the nucleolus. Indeed, Pontin was detected in nucleoli by extensive proteomic analysis (Leung et al. 2006). However, direct nucleolar localization of Pontin was never observed. Here, we focused on Pontin localization in more detail using immunofluorescence and electron microscopy and showed that Pontin is localized in the nucleolus, specifically in the nucleolar fibrillar centers. Moreover, we observed a dynamic, cell cycle-specific redistribution of Pontin within the nucleolus and found interactions between Pontin and the Pol I transcription machinery.
Materials and methods
Cell culture, synchronization
HeLa and HepG2 cells were cultured in Dulbecco’s Modified Eagle’s Medium with 10% (v/v) fetal bovine serum and supplemented with penicillin-streptomycin. For partial cell synchronization, cells were incubated for 16 h with 3 mM thymidine (Sigma). Thymidine was washed out to release cells into the S-phase as determined by 5-bromo-2′-deoxyuridine (BrdU) labeling 1 h after thymidine removal (Koberna et al. 2005).
siRNA
To reduce expression levels of the Pontin protein HeLa cells were transiently transfected with two siRNA duplexes (21-mers with 3′-dTdT overhangs) as described in Watkins et al. (2004). Following the protocol of Elbashir et al. (2002), siRNA oligonucleotides were annealed to produce siRNA duplexes. Annealing quality of siRNA duplexes was controlled by electrophoresis on a 4% (w/v) agarose gel. Transfection was performed with Oligofectamine (Invitrogen) according to the manufacturer’s protocol. Cells were analyzed 48 and 72 h post transfection.
Antibodies, immunofluorescence and light microscopy
Two monoclonal antibodies raised against Pontin were used in this study. Immunization and generation of antibodies has been reported previously (Weiske and Huber 2005). Clones 5G3-11 and 3A4-1 were isolated by ELISA screening and specificity was tested by Western blotting of cell lysates prepared as described previously (Meyer zum Buschenfelde et al. 2006). Epitope mapping was performed with purified recombinant MBP-Pontin fusion proteins with deletions either at the N- or C-terminus (Weiske and Huber 2005). Human autoantibodies anti-fibrillarin, anti-Upstream Binding Factor (UBF), anti-RNA polymerase I (kindly provided by Marvin Fritzler, Medical School, University of Calgary) and mouse anti-fibrillarin 17C12 (kindly provided by K.M. Pollard, The SCRIPPS Research Institute, CA; Yang et al. 2001) were used as nucleolar markers. Rabbit anti-SART3 antibodies (Stanek et al. 2003) were used as nucleoplasmic markers. The monoclonal anti-vimentin antibody VI-01 (Draberova et al. 1986) was kindly provided by Pavel Draber (Institute of Molecular Genetics, AS CR, Prague, Czech Republic). The anti-c-Myc (N-262 X) was obtained from Santa Cruz Biotechnology.
For immunofluorescence detection, cells were grown on coverslips, washed with PBS, fixed in 2% (w/v) paraformaldehyde for 10 min and permeabilized with 0.2% (v/v) Triton X-100 in PBS for 5 min. Cells were then washed with PBS and incubated with monoclonal antibodies against Pontin diluted in 1% (w/v) BSA in PBS for 1 h, washed and incubated with secondary antibodies conjugated to TRITC or FITC (Jackson ImmunoResearch Laboratories) in 1% (w/v) BSA in PBS for 1 h. In the case of the double labeling procedure, cells were incubated for 1 h with mouse anti-Pontin antibodies (5G3-11 or 3A4-1) and one of the human autoantibodies mentioned above, both diluted in 1% (w/v) BSA in PBS, washed in PBS and incubated for another hour with a mixture of secondary anti-mouse and anti-human antibodies conjugated with TRITC and FITC, respectively. All steps were performed at room temperature.
To test the specificity of the monoclonal antibody staining, antibodies were pre-incubated in PBS containing 1% BSA for 1 h at room temperature with 5× excess of GST-Pontin fusion protein or with GST alone and subsequently used for immunofluorescence staining.
Coverslips were mounted to glycerol containing DAPI (for DNA staining) and DABCO (as an anti-fade agent). Images were taken with the Leica SP2 laser scanning confocal microscope.
Transcription labeling
Cell were incubated with 2 mM 5-fluorouridine (Sigma) for 10 min, fixed in 3.7% (w/v) paraformaldehyde for 5 min and permeabilized with 1% (v/v) Tritone X-100 in PBS for 10 min. Cells were then incubated with a mixture of mouse anti-Pontin (3A4-1) and rat anti-BrdU antibodies (Abcam) diluted in 1% (w/v) BSA in PBS for 1 h followed by incubation with secondary antibodies anti-mouse conjugated with TRITC and anti-rat conjugated with FITC (Jackson ImmunoResearch Laboratories).
Cell lysate preparation and Western blot analysis
For whole-cell lysate preparation, cells grown on Petri dishes were washed with ice-cold PBS and scraped off the dish in 2× sample buffer (2× SB consists of 20% (v/v) glycerol, 125 mM Tris-Cl, pH 6.8 and 4% (w/v) SDS). Lysates were homogenized by passing through a 22G needle. Nucleoplasmic and nucleolar fractions were prepared according to a previously published protocol (Andersen et al. 2002). Protein concentration was determined using the bicinchonic acid assay. Before electrophoresis, mercaptoethanol and bromphenol blue were added to final concentrations 5% (v/v) and 0.01 mg/ml, respectively, and samples were incubated for 5 min at 85°C. Whole-cell lysates were separated on 12% SDS-PAGE and transferred to nitrocellulose membranes (Protran). Membranes were blocked and incubated with mouse anti-Pontin antibodies diluted in PBST containing 5% (w/v) Blotto (Santa Cruz Biotechnology, Inc) and then with goat anti-mouse secondary antibodies conjugated with horseradish peroxidase (Biorad) diluted in 5% (w/v) Blotto in PBST for 1 h at room temperature. In control experiments anti-vimentin VI-01, anti-fibrillarin 17C12 and anti-SART3 antibodies were applied, each diluted in 5% (w/v) Blotto in PBST. Horseradish peroxidase activity was detected using an ECL chemiluminescence system (Pierce) and captured on X-ray film (Foma). Intensity of bands was quantified by ImageJ software.
Electron microscopy
HeLa cells were partially synchronized to S-phase by thymidine treatment (see above) and fixed in freshly prepared 8% (w/v) formaldehyde in 0.2 M PIPES buffer, pH 6.95 for 12 h, washed in PBS, scraped and centrifuged at 1,000 rpm for 5 min and mixed with 10% (w/v) gelatin in PBS at 37°C. Cells were again centrifuged for 5 minutes at 1,000 rpm and the excess of gelatin was removed. Cells in gelatin were chilled at 4°C, fixed with 8% (w/v) formaldehyde in 0.2 M PIPES, pH 6.95 for 20 min and washed in PBS. Cells embedded in gelatin were removed from centrifuge tube, and cut into small pieces (less than 1 mm3). Cells were cryo-preserved and frozen according to protocol described previously (Raska et al. 1995). Thin cryosections were cut on the ultracut (Leica) equipped with a cryochamber. Grids with cryosections were incubated with mouse anti-Pontin (5G3-11) antibody for 30 min, washed in PBS, incubated with 6 nm gold anti-mouse adduct (Jackson ImmunoResearch Laboratories) for 20 min, washed in PBS and water, embedded in mixture of 3% (w/v) polyvinyl alcohol (MW. 30-70 kDa, Sigma) and 0.1% (w/v) uranylacetate and dried. The sections were viewed using a Zeiss EM 900 electron microscope equipped with a KeenView CCD camera (Soft Imaging Systems).
Immunoprecipitation
Cells (7×106) were washed twice with ice-cold PBS containing Mg2+ and incubated with 500 μl of Lysis buffer A (10 mM imidazol pH 6.8, 100 mM KCl, 300 mM sucrose, 2 mM MgCl2, 10 mM EGTA, 2% (v/v) Triton X-100, protease inhibitor cocktail (Calbiochem)) for 10 min on ice. Cells were removed from the plate and centrifuged at 20,800 ×g for 10 min. A 450 μl volume of supernatant was immunoprecipitated with 2 μg of 3A4-1 antibody prebound to 30 μl of Protein-G beads under continual agitation for 4 h at 4°C. After five washings with Lysis buffer A, the immunoprecipitated proteins were resuspended in 30 μl of 2× sample buffer and separated by 12% SDS-PAGE.
Chromatin immunoprecipitation (ChIP)
HeLa cells were grown to confluency and fixed with 1% formaldehyde for 10 min at room temperature. Subsequently, the cells were washed twice with PBS and the cross-linking reaction was stopped by incubation with glycine at a final concentration of 125 mM for 10 min, followed by washing twice with PBS. Cells were lysed by incubation in 1 ml cell-lysis buffer (5 mM Pipes pH 8, 85 mM KCl, 0.5% (v/v) NP40, 1 mM PMSF, protease inhibitor cocktail (Roche)) for 10 min at 4°C. Subsequently, cells were scraped from the cell culture dish and nuclei were pelleted by centrifugation (2,700 ×g, 5′, 4°C). The pellet was resuspended in 500 μl RIPA-buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 1% (w/v) Nonidet-P40, 0.5% (w/v) Na-Deoxicholate, 0.1% (w/v) SDS, 1 mM PMSF and Protease inhibitor cocktail (Roche)). Nuclei were disrupted by sonication with three 20 s pulses in a UP 50H sonicator (Hielscher Ultraschall Technologie) at a setting of cycle 0.5 and 30% amplitude, yielding genomic DNA fragments with a bulk size of 200-1,000 bp. For ChIP 50 μg of DNA diluted in RIPA-buffer were incubated with 2 μg anti-Pontin (5G3-11) or anti-c-Myc (N-262 X) antibodies overnight at 4°C in an orbital shaker. Immune complexes were precipitated by the addition of 35 μl Protein A-Sepharose CL4B beads for 1 h at 4°C. Precipitates were serially washed three times with RIPA-buffer, three times with high salt-buffer (100 mM Tris-HCl pH 8, 500 mM LiCl, 1% (v/v) Nonidet-P40, 0.5% (w/v) Na-deoxycholate, Protease inhibitor cocktail (Roche)) and twice with TE-buffer (10 mM Tris-HCl pH 8, 1 mM EDTA). Precipitates were then treated with 50 ng/μl RNase A in 200 μl TE-buffer at 37°C for 1 h. Subsequently proteins were digested by addition of 50 μl 5× Proteinase K-buffer (50 mM Tris-HCl pH 7.5, 25 mM EDTA, 1.25% (w/v) SDS) to the RNase A-reaction and the addition of 50 ng/ml Proteinase K. Samples were incubated at 65°C overnight under constant agitation. DNA was purified by phenol/chloroform extraction and subsequent ethanol precipitation. For PCR analysis 2 μl of the extracted DNA (total volume 50 μl) was used as a template for 25-35 cycles of amplification. For the two-step ChIP assay, components were eluted from the first immunoprecipitation reaction by incubation with 10 mM dithiothreitol at 37°C for 30 min and diluted 1:40 in RIPA-buffer followed by immunoprecipitation with the second antibodies. ReChIP was essentially performed as the first ChIP. For the detection of rDNA initiator- and terminator-regions primer-sets H1 and H13 were used as described previously (Grandori et al. 2005). The following primers were used for detection of the GAPDH promoter: Forward—5′-TACTAGCGGTTTTACGGGCG-3′ and reverse—5′-TCGAACAGGAGGAGCAGAGAGCGA-3′ giving a 166-bp PCR product.
Results
Characterization of anti-Pontin monoclonal antibodies
Monoclonal antibodies were raised against the whole Pontin protein to obtain tools for studying Pontin intracellular localization and function. Individual clones were tested for their ability to immunostain Pontin and specificity of the two selected monoclonal antibodies were further tested using whole-cell lysates and immunoblotting (Fig. 1a). Both, the 5G3-11 and the 3A4-1 antibodies recognized a protein band with an apparent molecular weight of ∼50 kD that corresponds to Pontin. To further test antibody specificity, protein extracts were prepared from cells treated with siRNA against Pontin (Fig. 1b). A significant decrease of signal was observed in cells treated with specific siRNAs but not with negative control siRNAs. In addition, both antibodies specifically immunoprecipitated Myc-tagged Pontin from cell extracts (Fig. 1b). To define the regions in the Pontin protein containing the epitopes recognized by the antibodies, different Pontin deletion constructs were bacterially expressed, purified and used for epitope mapping (Fig. 1d). These data demonstrate that 5G3-11 and 3A4-1 antibodies interact with different regions of the Pontin protein and specifically recognize Pontin in cellular extracts.
Fig. 1.
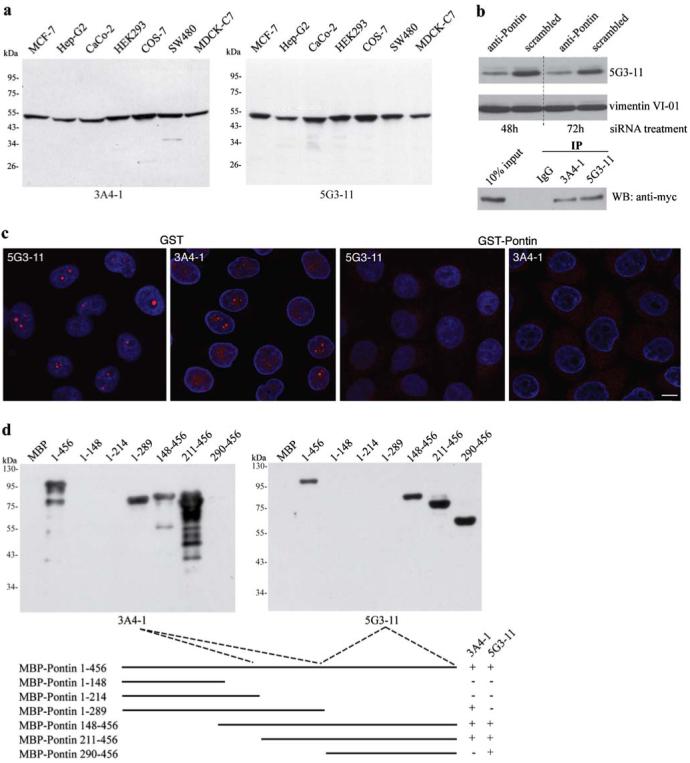
Characterization of monoclonal antibodies against Pontin. a Monoclonal antibodies were raised against the full-length recombinant protein and their specificity tested on whole-cell extracts from different cell lines. Only one band of an apparent molecular weight of 50 kD is detected by either one of the antibodies. b To further confirm the antibody specificity, Pontin was detected by the anti-Pontin (5G3-11) antibody in extracts made from cells treated for 48 h or 72 h with anti-Pontin siRNA. A ∼50% reduction at the protein level was observed after the siRNA treatment. In addition, both anti-Pontin antibodies specifically immunoprecipitated Myc-tagged Pontin from cell extracts. c Immunofluorescence labeling of HeLa cells with mouse anti-Pontin monoclonal antibodies (red). DNA counter-stained with DAPI (blue). Prominent dots within the nucleolus were observed in a subset of cells. Staining in the nucleus disappeared after pre-incubation of antibodies with GST-Pontin but not with GST alone showing the specificity of the staining. Scale bar represents 10 μm. d Different regions of Pontin were expressed in E. coli as MBP fusion proteins, purified and used for epitope mapping. The anti-Pontin (3A4-1) antibody interacts with an epitope between the Walker A and B motifs of the protein (amino acids 214-289) while anti-Pontin (5G3-11) antibody binds to an epitope within the C-terminal amino acids 290-456
Intracellular localization of Pontin
Pontin localization has been elusive and different groups referred to diverse localization based on the antibody and/or fixation conditions used (Bauer et al. 1998; Holzmann et al. 1998; Makino et al. 1998; Salzer et al. 1999). Moreover, a GFP-tag on either end of the protein prevents Pontin localization to the nucleus (data not shown) and thus impairs studying the intranuclear distribution. Here we used two different monoclonal antibodies to analyze Pontin localization in situ (Fig. 1c). Both antibodies revealed a similar staining pattern and detected Pontin in the nucleoplasm. Surprisingly, a signal in nucleoli was also observed and a significant number of cells exhibited bright dots. Similar results were obtained with the human hepatocellular carcinoma cell line HepG2 (data not shown). Specificity of the staining was further confirmed by pre-incubation of the antibodies with GST-Pontin. Nuclear and nucleolar staining disappeared after antibody pre-incubation with GST-Pontin but not with GST alone (Fig. 1c).
Pontin dots are localized in the nucleolus
Based on DAPI staining and phase contrast, the Pontin dots appeared preferentially in nucleoli (Fig. 1c). To confirm this nucleolar localization, cells were double labeled with antibodies against Pontin and chosen nucleolar markers (fibrillarin, Upstream Binding Factor or RNA polymerase I; Fig. 2a,b). Fibrillarin associates with C/D box snoRNPs that function in site-specific 2′-O-methylation of pre-ribosomal RNA and is present in nucleoli and Cajal bodies. We observed partial co-localization of Pontin dots with fibrillarin inside nucleoli, but not in Cajal bodies. Fibrillarin staining often surrounded the Pontin signal. Upstream Binding Factor (UBF) and Pol I represent important factors of ribosomal DNA (rDNA) transcription machinery localized in many dots inside the nucleolus. We found that Pontin-positive nucleolar dots overlapped with the UBF/Pol I signal (Fig 2a,b).
Fig. 2.
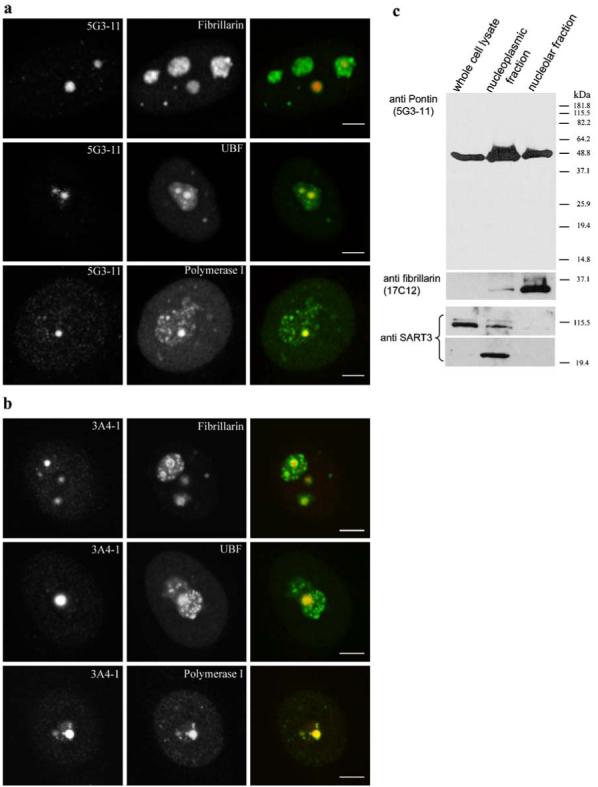
Pontin is localized in nucleoli. Double-labeling of HeLa cells with anti-Pontin antibodies 5G3-11 a or 3A4-1 b and anti-fibrillarin, anti-UBF or anti-RNA polymerase I antibodies. The large Pontin dots co-localize with UBF and RNA polymerase I containing dots. In contrast, Pontin revealed only partial co-localization with fibrillarin, which often formed a rim around Pontin dots. Note that in contrast to the three standard nucleolar proteins, Pontin dots appear only in a subset of nucleoli. Scale bar represents 5 μm. c Whole-cell lysate, nucleoplasmic and nucleolar fractions were prepared and used for immunoblotting with the anti-Pontin (5G3-11) antibody. Purity of the nucleolar fraction was verified by detection of a nucleolar marker fibrillarin and absence of the SART3 protein, which is present only in the nucleoplasm and devoid of the nucleolus. SART3 was partially cleaved during the nucleolar preparation and a lower molecular weight band appeared in the nucleoplasmic fraction
To further confirm Pontin nucleolar localization, nucleoplasmic and nucleolar fractions were prepared and Pontin was detected by immunoblotting using the 5G3-11 antibody (Fig. 2c). Purity of the nucleolar fraction was verified by the detection of fibrillarin, which accumulates in the nucleolus and SART3, a nucleoplasmic protein associated with spliceosomal snRNPs that is devoid of nucleoli. Fibrillarin was not detected in the whole-cell extract probably due to lower concentration of this protein in the total cell lysate. Taken together these data show that a significant fraction of Pontin is localized in nucleoli.
Pontin accumulates in nucleolar fibrillar centers
The nucleolus consists of three different regions: fibrillar centers, dense fibrillar components, and granular components. rDNA was mainly mapped to the fibrillar centers that also contain the rDNA transcription machinery (e.g. Pol I, UBF). In contrast, newly synthesized pre-ribosomal RNA (pre-rRNA) and factors involved in pre-rRNA processing (e.g. snoRNPs, fibrillarin) are localized in the dense fibrillar components (Raska 2003; Raska et al. 2006a, b). Co-localization of Pontin with Pol I and UBF indicates that Pontin is specifically localized to the fibrillar centers. However, light microscopy does not posses enough resolution to specifically address sub-nucleolar localization (Koberna et al. 2002; Malinsky et al. 2002). Therefore, we used electron microscopy to locate Pontin within the nucleolus. Thin sections from synchronized HeLa cells were incubated with anti-Pontin (5G3-11) antibody and subsequently visualized by antibodies bound to gold particles (Fig. 3). Labeling within the nucleolus was specifically enriched in the fibrillar centers.
Fig. 3.
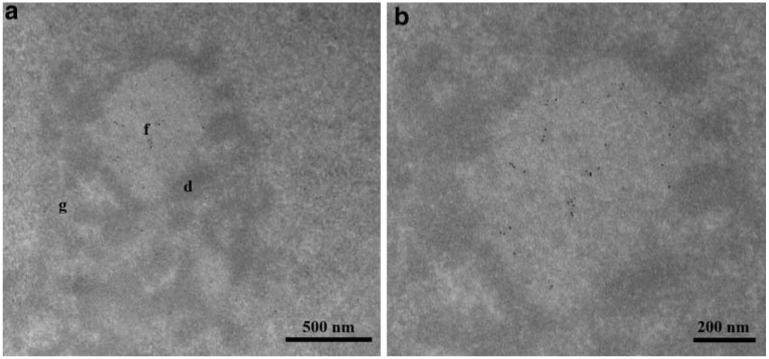
Ultrastructural localization of Pontin. a HeLa cells were synchronized in S-phase and Pontin was detected by the anti-Pontin (5G3-11) monoclonal antibody by means of electron microscopy. In this nucleolar section, Pontin is specifically enriched in a fibrillar center. b Detail of picture a. f fibrillar centers, d dense fibrillar components, g granular components
Nucleolar localization of Pontin during S-phase
The Pontin-labeling pattern differs between individual cells within an unsynchronized cell population (Fig. 1c). While the Pontin nucleolar signal was presented in all cells, bright dots appeared only in a fraction of cells. To test whether accumulation of Pontin in the nucleolar dots is cell cycle dependent, we partially synchronized HeLa cells in S-phase by a thymidine block and stained them with monoclonal antibodies 1 h after release from the thymidine block. The number of cells in S-phase was controlled in parallel experiments by labeling of newly synthesized DNA with BrdU. Almost all of the cells were in S-phase 1 h after release from the thymidine block (data not shown). In the S-phase, the number of cells containing Pontin nucleolar dots increased to 96% while only 42% of cells in the unsynchronized culture exhibited nucleolar accumulation of Pontin (Fig. 4a,b).
Fig. 4.
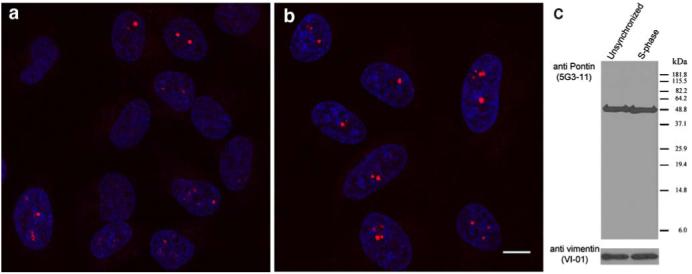
Pontin accumulates in the nucleolar dots during S-phase. Pontin was immunodetected by the anti-Pontin (5G3-11) antibody in a unsynchronized cell population or b cells synchronized to S-phase of the cell cycle. Ninety six percent of cells in S-phase accumulated Pontin in the nucleolus. Scale bar represents 10 μm. c Equivalent amounts of proteins from S-phase cells and from unsynchronized cells were loaded in each line (vimentin antibody used as a loading control). No significant differences in protein levels of Pontin were observed between the unsynchronized and synchronized cell populations
To test whether Pontin accumulation in the nucleolar dots corresponded to increased expression of the Pontin protein in S-phase, the amount of Pontin protein was analyzed in S-phase cells and in unsynchronized cell populations (Fig. 4c). Although the number of cells with Pontin dots significantly increased in S-phase, the amount of protein did not change. These data indicate that accumulation of Pontin in big dots is rather due to its relocalization within the nucleolus than to a higher expression of the Pontin protein during S-phase.
Pontin interacts with the nucleolar transcription machinery
To get further inside into Pontin function in the nucleolus, immunoprecipitation with the anti-Pontin (3A4-1) antibody was performed and the association of nucleolar proteins with Pontin was analyzed (Fig. 5a). The anti-Pontin antibody co-precipitated Pol I but not fibrillarin or UBF. These data suggest that Pontin in the nucleolus may be involved in the regulation of rRNA synthesis. In this respect, Pontin was identified as a c-Myc co-factor that regulates activity of several genes. c-Myc was recently identified as an activator of rRNA transcription that interacts with specific regions of rDNA genes (Grandori et al. 2005). To test whether Pontin associates with c-Myc at these rDNA loci, we performed chromatin immunoprecipitation (ChIP) with anti-Pontin and anti-c-Myc antibodies. Cis-elements H1 (close to the transcriptional start) and H13 (close to the termination site), which were previously identified as major c-Myc binding targets were analyzed (Fig. 5b). These experiments confirmed the earlier findings showing that c-Myc binds to these sites in the rDNA gene cluster (Grandori et al. 2005). In addition, our data show that Pontin is also associated with these rDNA loci. Since Pontin has been shown to interact with c-Myc and to modulate c-Myc transcriptional activity, the formation of Pontin/c-Myc complexes at these rDNA sites was analyzed by two-step ChIP. The first ChIP was performed with the anti-Pontin (5G3-11) antibody followed by a second ChIP using an anti-c-Myc antibody or vice versa (Fig. 5b). Both combinations gave the same result indicating that Pontin binds to rDNA cis-elements in a complex with c-Myc.
Fig. 5.
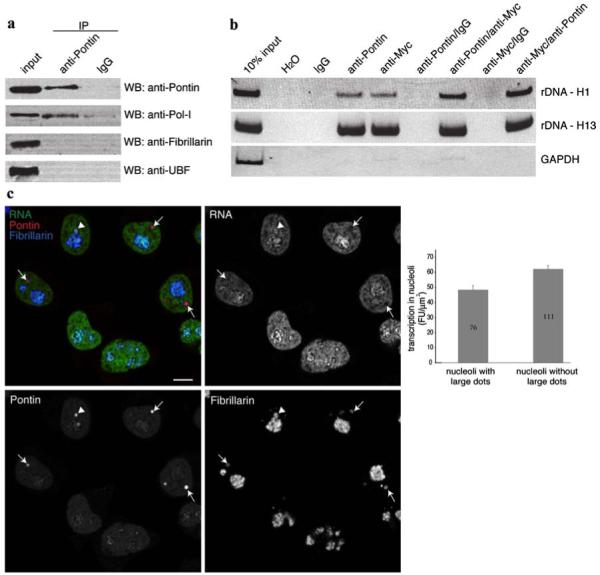
Pontin is involved in the regulation of rRNA transcription. a Pontin was immunoprecipitated using the anti-Pontin (3A4-1) antibody and nuclear proteins detected by immunoblotting. Pol I was specifically precipitated with anti-Pontin antibody but not with a control antibody. b rDNA loci H1 close to the transcription initiation site (952-1,030 bp) and H13 close to the termination site (12,855-12,970 bp) were enriched after chromatin immunoprecipitation with anti-Pontin (5G3-11) or anti-Myc antibodies. Moreover, both regions were specifically amplified after two-step immunoprecipitation with anti-Pontin/anti-Myc or anti-Myc/anti-Pontin antibodies but not with control IgG antibodies showing that Pontin and c-Myc binds to the same regions of rDNA in a complex. Constitutively active GAPDH promoter served as a negative control. c Transcriptional activity of nucleoli containing large Pontin dots is reduced. Newly synthesized RNA (green) was labeled by 10 min incubation with 5-fluorouridine and visualized together with Pontin (red) and fibrillarin (blue) as a marker of nucleoli. In transcriptionally active nucleoli, rRNA signal surrounded Pontin labelling (arrowhead). However, most nucleoli that contained Pontin dots exhibited reduce transcription activity (arrows). Quantification of nucleolar transcription (fibrillarin signal was used as a mask) revealed that transcription activity was reduced by ∼20% in nucleoli where Pontin is sequestered in large dots (average nucleolar fluorescence of transcription signal with standard error of the mean is shown together with number of nucleoli analyzed)
The observation that Pontin interacts with Pol I, c-Myc and rDNA indicate that Pontin is involved in the regulation of rRNA transcription. In order to analyze the relationship between nucleolar Pontin and transcription, cells were incubated for 10 min with 5-fluorouridine and newly synthesized RNA immunostained together with Pontin and fibrillarin (Fig. 5c). In a few nucleoli, newly synthesized rRNA that had accumulated in the dense fibrillar components surrounded the Pontin dots, which is consistent with Pontin localization in the fibrillar centers. However, transcription was reduced in many nucleoli containing Pontin dots. Quantification of the transcription signal revealed that the accumulation of Pontin in the large nucleolar dots correlate with ∼20% reduction of nucleolar transcriptional activity.
Discussion
Pontin is a protein with many faces and its proposed functions range from chromatin remodeling and transcriptional regulation, the processing of small nucleolar RNAs, mitotic spindle function (for review see Gallant 2007) to assembly of telomerase (Venteicher et al. 2008). Despite a large number of reports analyzing Pontin functions, a relatively few groups have analyzed Pontin cellular distribution (Bauer et al. 1998; Holzmann et al. 1998; Makino et al. 1998; Salzer et al. 1999). However, outcomes of these studies have been inconsistent. Considering the multitude of potential Pontin interactions and the number of protein complexes Pontin participates in, various localizations are likely. These discrepant results could be explained by the combination of different epitope availabilities in different complexes and antibody specificities towards these epitopes. An additional factor that may play a role in Pontin recognition is a fixation procedure that can significantly influence epitope recognition (data not shown). Depending on the combination of antibodies and fixation conditions, each study might have detected different functional pools of Pontin and their results are rather complementary.
Here, we used two different monoclonal antibodies recognizing different epitopes in Pontin to determine its sub-cellular distribution. In accordance with previous studies, we detected Pontin predominantly in the cell nucleus. Surprisingly, Pontin was also found in the nucleolus. In a large number of cells, Pontin concentrated in big nucleolar dots and this localization was cell cycle dependent. Presence of Pontin in the nucleolus was further confirmed by its detection in nucleolar extracts. To determine Pontin sub-nucleolar distribution, Pontin was detected by means of electron microscopy and was found to be localized specifically to fibrillar centers. Our data are in agreement with multiple mass spectrometric analyses that found Pontin in isolated nucleoli (Leung et al. 2006) and provide the first direct visualization of Pontin in the nucleolus.
Nucleolar fibrillar centers contain proteins involved in rDNA transcription but there is overwhelming evidence that they are mostly transcriptionally inactive. It is widely believed that rDNA transcription occurs at the border of the fibrillar centers and dense fibrillar components and within the dense fibrillar components themselves (Raska et al. 2006a,b). Pontin in the fibrillar centers may interact with the rDNA transcription machinery and regulate its activity during the cell cycle. In this respect, we found that Pontin specifically interacts with Pol I but not UBF even though both proteins are concentrated in the fibrillar centers.
It was previously shown that Pontin interacts with c-Myc and functions as a c-Myc co-activator (Bellosta et al. 2005; Etard et al. 2005; Wood et al. 2000). Recently, c-Myc was localized to nucleoli during cell serum stimulation where it stimulated the acetylation of rDNA chromatin and rRNA synthesis (Arabi et al. 2005; Grandori et al. 2005). We demonstrated that Pontin interacts with the same rDNA sequences as c-Myc and moreover, two-step ChIP experiments indicated that Pontin associated with rDNA in a complex with c-Myc (Fig. 5). Thus, Pontin is likely an important c-Myc co-factor regulating rDNA transcription. Based on our finding that the accumulation of Pontin in large nucleolar dots correlates with the reduction of rRNA synthesis, we speculate that Pontin is a positive regulator of Pol I activity and its sequestering to transcriptionally inactive fibrillar centers might represent a regulatory mechanism of rRNA transcription during the cell cycle.
Acknowledgments
We would like to thank Pavel Draber, Marvin Fritzer and Kenneth M. Pollard for providing us with antibodies, Klaus Holzmann (University of Vienna, Vienna, Austria) for Pontin-GFP constructs and Jasper Manning for comments on the manuscript. This work was supported by grants from the Grant Agency of the Czech Republic 301/05/0601 (to D.S.), 304/05/0374 (to K.K.), the Czech Ministry of Education (MSM0021620806, 1K05009 and LC535 to I.R.), the Wellcome trust grant 075834/04/Z (to I.R.) and the institutional projects AV0Z50520514, AV0Z50110509 and AV0Z50390512 awarded by the Academy of Sciences of the Czech Republic. The work of O.H. was supported by grants SFB366/C12 and HU881/5-1 of the Deutsche Forschungsgemeinschaft, LOM grants of the Charité—Universitätsmedizin Berlin and the Sonnenfeld Stiftung. Z.C. was supported by a grant from the Grant Agency of the Czech Republic 303/03/H065 and A.L. by 204/07/0133.
Contributor Information
Zuzana Cvǎková, Institute of Cellular Biology and Pathology, 1st Faculty of Medicine, Charles University in Prague, Albertov 4, 128 00 Prague 2, Czech Republic; Department of Cell Biology, Institute of Physiology AS CR v.v.i., Albertov 4, 128 00 Prague 2, Czech Republic.
Kai F. Albring, Institute of Laboratory Medicine and Pathobiochemistry, Charité-Universitätsmedizin Berlin, Hindenburgdamm 30, 12200 Berlin, Germany
Karel Koberna, Institute of Cellular Biology and Pathology, 1st Faculty of Medicine, Charles University in Prague, Albertov 4, 128 00 Prague 2, Czech Republic; Department of Cell Biology, Institute of Physiology AS CR v.v.i., Albertov 4, 128 00 Prague 2, Czech Republic; Laboratory of Cell Biology, Institute of Experimental Medicine AS CR v.v.i., Vídeňská 1083, 142 20 Prague 4, Czech Republic.
Anna Ligasová, Institute of Cellular Biology and Pathology, 1st Faculty of Medicine, Charles University in Prague, Albertov 4, 128 00 Prague 2, Czech Republic; Department of Cell Biology, Institute of Physiology AS CR v.v.i., Albertov 4, 128 00 Prague 2, Czech Republic; Laboratory of Cell Biology, Institute of Experimental Medicine AS CR v.v.i., Vídeňská 1083,142 20 Prague 4, Czech Republic.
Otmar Huber, Institute of Laboratory Medicine and Pathobiochemistry, Charité-Universitätsmedizin Berlin, Hindenburgdamm 30, 12200 Berlin, Germany.
Ivan Raška, Institute of Cellular Biology and Pathology, 1st Faculty of Medicine, Charles University in Prague, Albertov 4, 128 00 Prague 2, Czech Republic; Department of Cell Biology, Institute of Physiology AS CR v.v.i., Albertov 4, 128 00 Prague 2, Czech Republic.
David Staněk, Department of RNA Biology, Institute of Molecular Genetics AS CR v.v.i., Vídeňská 1083, 142 20 Prague 4, Czech Republic, e-mail: stanek@img.cas.cz.
References
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, Larsson LG, Wright AP. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7:303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- Bauer A, Huber O, Kemler R. Pontin52, an interaction partner of beta-catenin, binds to the TATA box binding protein. Proc Natl Acad Sci USA. 1998;95:14787–14792. doi: 10.1073/pnas.95.25.14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A, Chauvet S, Huber O, Usseglio F, Rothbacher U, Aragnol D, Kemler R, Pradel J. Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. Embo J. 2000;19:6121–6130. doi: 10.1093/emboj/19.22.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellosta P, Hulf T, Balla Diop S, Usseglio F, Pradel J, Aragnol D, Gallant P. Myc interacts genetically with Tip48/Reptin and Tip49/Pontin to control growth and proliferation during Drosophila development. Proc Natl Acad Sci USA. 2005;102:11799–11804. doi: 10.1073/pnas.0408945102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draberova E, Draber P, Havlicek F, Viklicky V. A common antigenic determinant of vimentin and desmin defined by monoclonal antibody. Folia Biol (Praha) 1986;32:295–303. [PubMed] [Google Scholar]
- Dugan KA, Wood MA, Cole MD. TIP49, but not TRRAP, modulates c-Myc and E2F1 dependent apoptosis. Oncogene. 2002;21:5835–5843. doi: 10.1038/sj.onc.1205763. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Weber K, Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- Etard C, Gradl D, Kunz M, Eilers M, Wedlich D. Pontin and Reptin regulate cell proliferation in early Xenopus embryos in collaboration with c-Myc and Miz-1. Mech Dev. 2005;122:545–556. doi: 10.1016/j.mod.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B. MYC recruits the TIP60 histon acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant P. Control of transcription by Pontin and Reptin. Trends Cell Biol. 2007;17:187–192. doi: 10.1016/j.tcb.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Gartner W, Rossbacher J, Zierhut B, Daneva T, Base W, Weissel M, Waldhausl W, Pasternack MS, Wagner L. The ATP-dependent helicase RUVBL1/TIP49a associates with tubulin during mitosis. Cell Motil Cytoskeleton. 2003;56:79–93. doi: 10.1002/cm.10136. [DOI] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- Holzmann K, Gerner C, Korosec T, Poltl A, Grimm R, Sauermann G. Identification and characterization of the ubiquitously occurring nuclear matrix protein NMP 238. Biochem Biophys Res Commun. 1998;252:39–45. doi: 10.1006/bbrc.1998.9604. [DOI] [PubMed] [Google Scholar]
- King TH, Decatur WA, Bertrand E, Maxwell ES, Fournier MJ. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol Cell Biol. 2001;21:7731–7746. doi: 10.1128/MCB.21.22.7731-7746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koberna K, Malinsky J, Pliss A, Masata M, Vecerova J, Fialova M, Bednar J, Raska I. Ribosomal genes in focus: new transcripts label the dense fibrillar components and form clusters indicative of “Christmas trees” in situ. J Cell Biol. 2002;157:743–748. doi: 10.1083/jcb.200202007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koberna K, Ligasova A, Malinsky J, Pliss A, Siegel AJ, Cvackova Z, Fidlerova H, Masata M, Fialova M, Raska I, Berezney R. Electron microscopy of DNA replication in 3-D: evidence for similar-sized replication foci throughout S-phase. J Cell Biochem. 2005;94:126–138. doi: 10.1002/jcb.20300. [DOI] [PubMed] [Google Scholar]
- Leung AK, Trinkle-Mulcahy L, Lam YW, Andersen JS, Mann M, Lamond AI. NOPdb: Nucleolar Proteome Database. Nucleic Acids Res. 2006;34:D218–D220. doi: 10.1093/nar/gkj004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino Y, Mimori T, Koike C, Kanemaki M, Kurokawa Y, Inoue S, Kishimoto T, Tamura T. TIP49, homologous to the bacterial DNA helicase RuvB, acts as an autoantigen in human. Biochem Biophys Res Commun. 1998;245:819–823. doi: 10.1006/bbrc.1998.8504. [DOI] [PubMed] [Google Scholar]
- Malinsky J, Koberna K, Bednar J, Stulik J, Raska I. Searching for active ribosomal genes in situ: light microscopy in light of the electron beam. J Struct Biol. 2002;140:227–231. doi: 10.1016/s1047-8477(02)00574-9. [DOI] [PubMed] [Google Scholar]
- McKeegan KS, Debieux CM, Boulon S, Bertrand E, Watkins NJ. A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol Cell Biol. 2007;27:6782–6793. doi: 10.1128/MCB.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer zum Buschenfelde D, Tauber R, Huber O. TFF3-peptide increases transepithelial resistance in epithelial cells by modulating claudin-1 and -2 expression. Peptides. 2006;27:3383–3390. doi: 10.1016/j.peptides.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Raska I. Oldies but goldies: searching for Christmas trees within the nucleolar architecture. Trends Cell Biol. 2003;13:517–525. doi: 10.1016/j.tcb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Raska I, Dundr M, Koberna K, Melcak I, Risueno MC, Torok I. Does the synthesis of ribosomal RNA take place within nucleolar fibrillar centers or dense fibrillar components? A critical appraisal. J Struct Biol. 1995;114:1–22. doi: 10.1006/jsbi.1995.1001. [DOI] [PubMed] [Google Scholar]
- Raska I, Shaw PJ, Cmarko D. New insights into nucleolar architecture and activity. Int Rev Cytol. 2006a;255:177–235. doi: 10.1016/S0074-7696(06)55004-1. [DOI] [PubMed] [Google Scholar]
- Raska I, Shaw PJ, Cmarko D. Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol. 2006b;18:325–334. doi: 10.1016/j.ceb.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Salzer U, Kubicek M, Prohaska R. Isolation, molecular characterization, and tissue-specific expression of ECP-51 and ECP-54 (TIP49), two homologous, interacting erythroid cytosolic proteins. Biochim Biophys Acta. 1999;1446:365–370. doi: 10.1016/s0167-4781(99)00104-9. [DOI] [PubMed] [Google Scholar]
- Stanek D, Rader SD, Klingauf M, Neugebauer KM. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J Cell Biol. 2003;160:505–516. doi: 10.1083/jcb.200210087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan HM, Livingston DM, Amati B. E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol Cell Biol. 2004;24:4546–4556. doi: 10.1128/MCB.24.10.4546-4556.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Dickmanns A, Luhrmann R. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol Cell Biol. 2002;22:8342–8352. doi: 10.1128/MCB.22.23.8342-8352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, Luhrmann R. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell. 2004;16:789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Weiske J, Huber O. The histidine triad protein Hint1 interacts with Pontin and Reptin and inhibits TCF-beta-catenin-mediated transcription. J Cell Sci. 2005;118:3117–3129. doi: 10.1242/jcs.02437. [DOI] [PubMed] [Google Scholar]
- Weiske J, Huber O. The histidine triad protein Hint1 triggers apoptosis independent of its enzymatic activity. J Biol Chem. 2006;281:27356–27366. doi: 10.1074/jbc.M513452200. [DOI] [PubMed] [Google Scholar]
- Wood MA, McMahon SB, Cole MD. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol Cell. 2000;5:321–330. doi: 10.1016/s1097-2765(00)80427-x. [DOI] [PubMed] [Google Scholar]
- Yang JM, Baserga SJ, Turley SJ, Pollard KM. Fibrillarin and other snoRNP proteins are targets of autoantibodies in xenobiotic-induced autoimmunity. Clin Immunol. 2001;101:38–50. doi: 10.1006/clim.2001.5099. [DOI] [PubMed] [Google Scholar]
- Zaros C, Briand JF, Boulard Y, Labarre-Mariotte S, Garcia-Lopez MC, Thuriaux P, Navarro F. Functional organization of the Rpb5 subunit shared by the three yeast RNA polymerases. Nucleic Acids Res. 2007;35:634–647. doi: 10.1093/nar/gkl686. [DOI] [PMC free article] [PubMed] [Google Scholar]


