Abstract
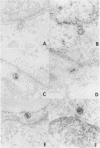
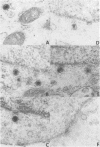
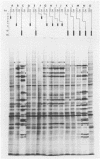
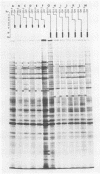
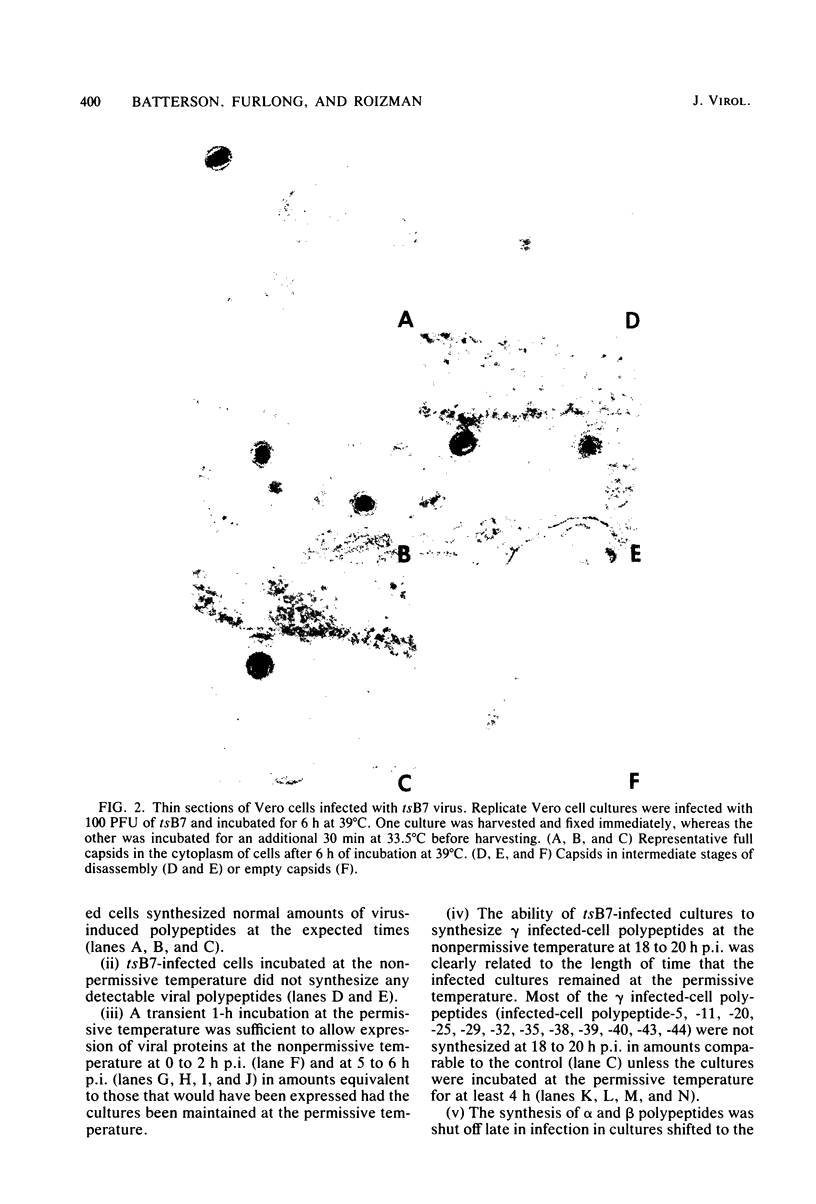
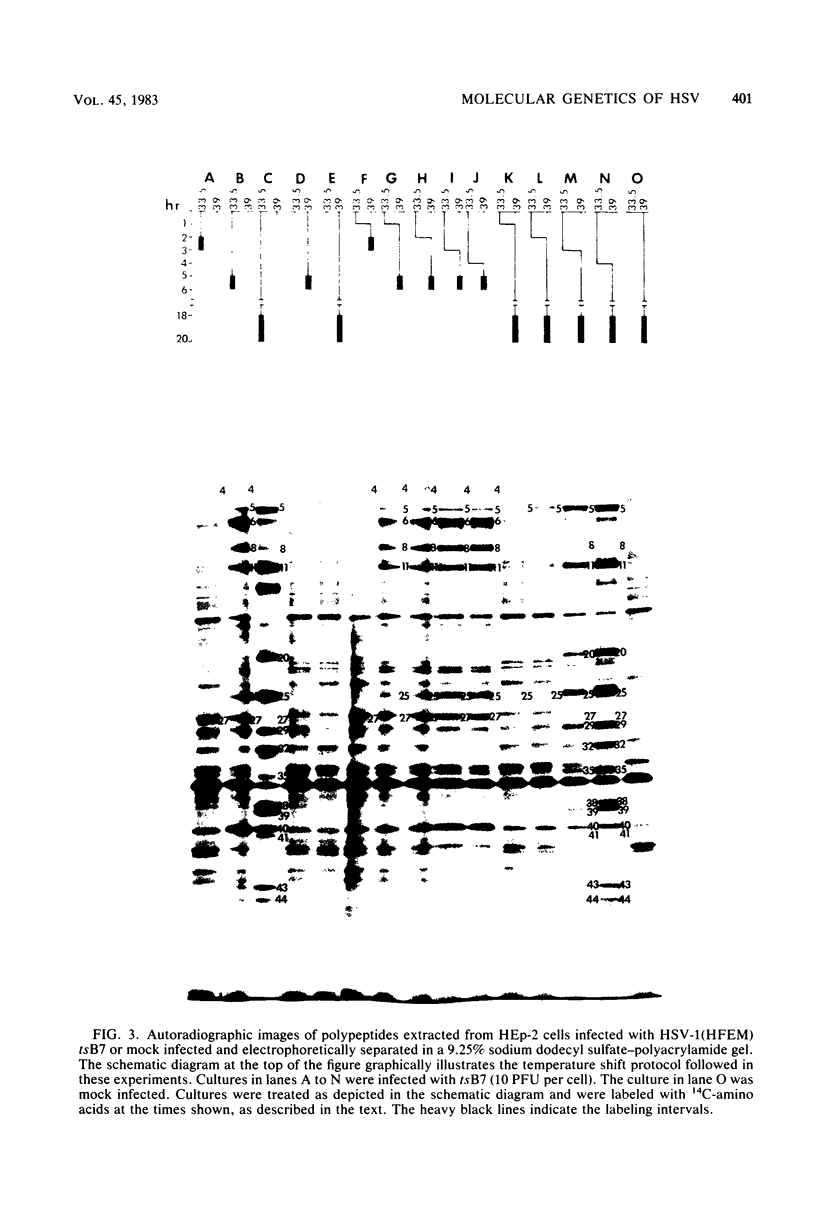
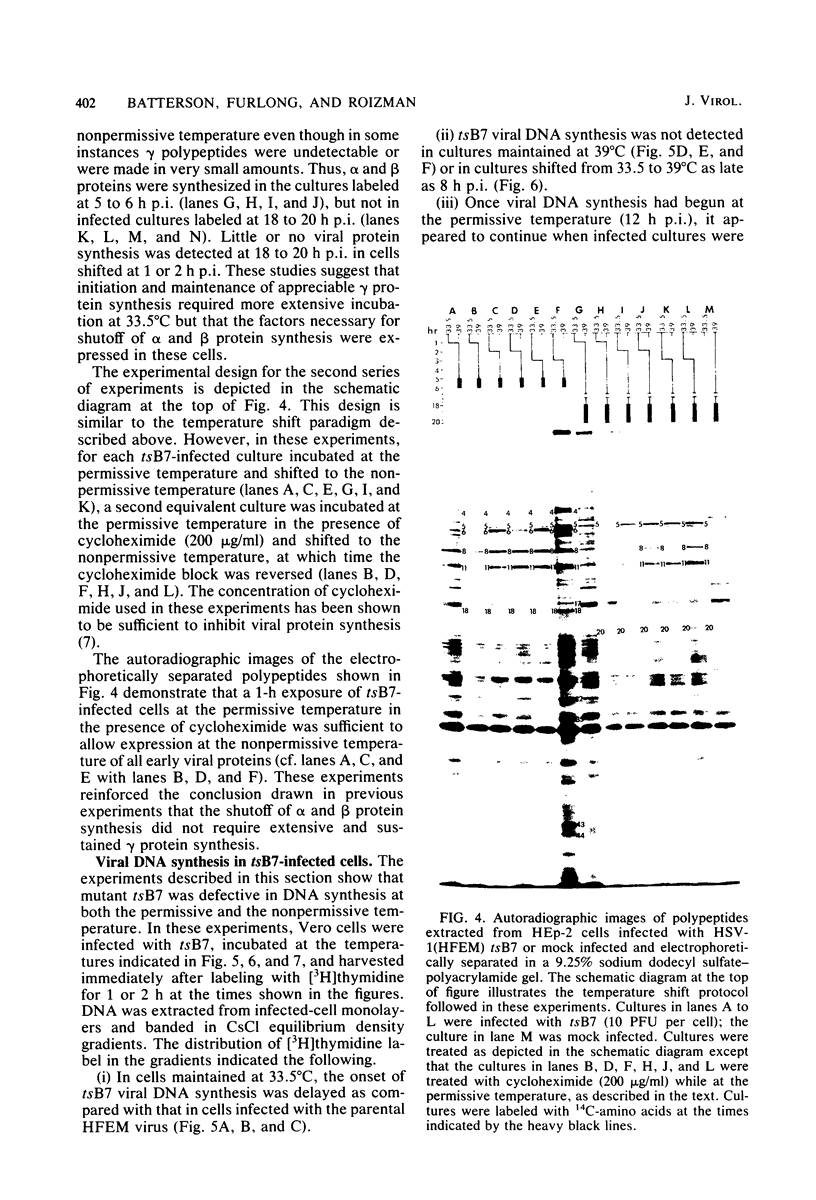
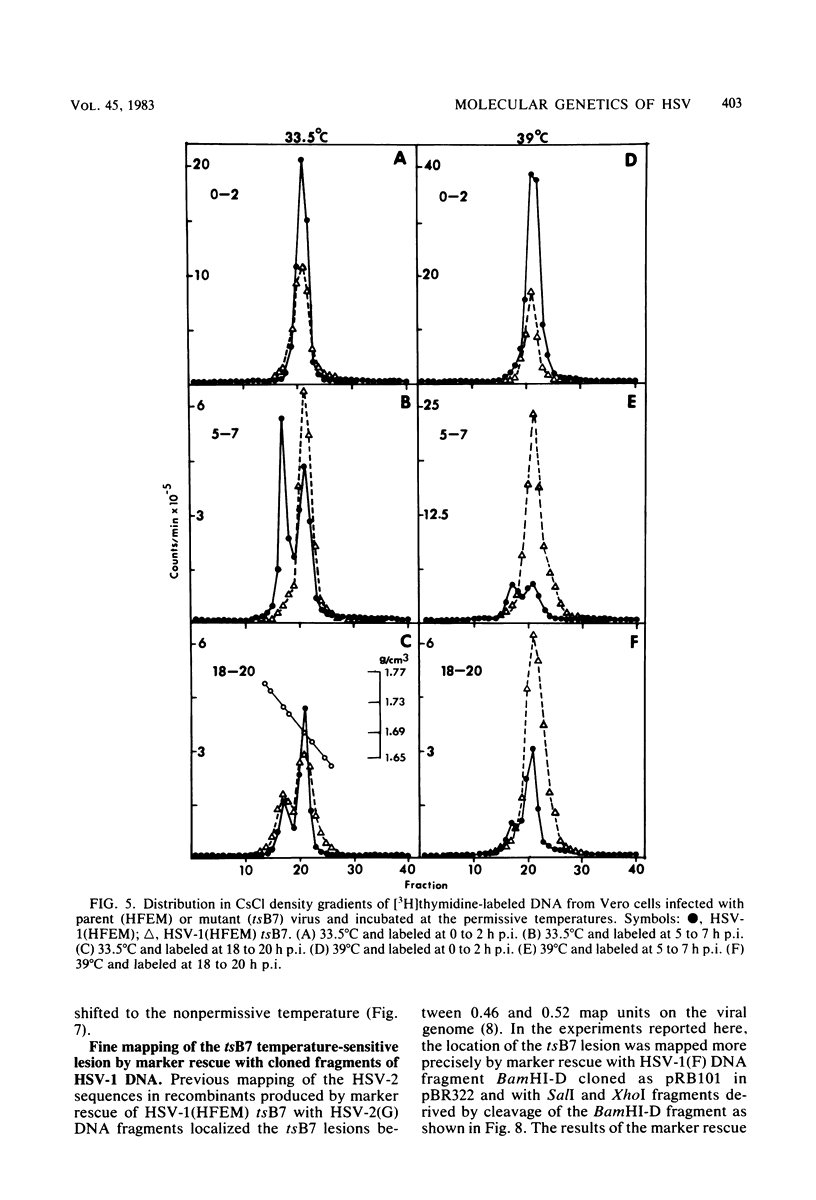
Previous studies have shown that cells infected with the herpes simplex virus 1(HFEM) mutant tsB7 and maintained at the nonpermissive temperature fail to accumulate viral polypeptides. Analyses of intertypic recombinants generated by marker rescue of tsB7 with herpes simplex virus 2 DNA fragments localized the mutation between 0.46 and 0.52 map units on the viral genome (Knipe et al., J. Virol. 38:539-547, 1981). In this paper we report that the mutation in tsB7 affects several aspects of the reproductive cycle of the virus at the nonpermissive temperature. Thus, (i) viral capsids accumulate at the nuclear pores and do not release viral DNA for at least 6 h postinfection at 39 degrees C. The DNA was released within 30 min after a shift to the permissive temperature. (ii) Experiments involving shifts from the permissive to the nonpermissive temperature indicated that viral protein synthesis was not sustained in cells maintained at the permissive temperature for less than 4 h. (iii) Viral DNA synthesis was delayed at the permissive temperature for as long as 8 h. Once initiated, it continued at 39 degrees C. (iv) Marker rescue of tsB7 by transfection with herpes simplex virus 1(F) DNA fragments localized the mutation to between 0.501 and 0.503 map units on the viral genome. These results are consistent with the tsB7 lesion being in a gene coding for a virion component which affects release of viral DNA from capsids and onset of viral DNA synthesis.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 1970 Oct 27;9(22):4428–4440. doi: 10.1021/bi00824a026. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney R. J., Powell K. L. Immunological and biochemical characterization of polypeptides induced by herpes simplex virus types 1 and 2. IARC Sci Publ. 1975;(11 Pt 1):63–73. [PubMed] [Google Scholar]
- Feldman L., Blankenship M. L., Ben-Porat T. Isolation and characterization of a temperature-sensitive uncoating mutant of pseudorabies virus. J Gen Virol. 1981 Jun;54(Pt 2):333–342. doi: 10.1099/0022-1317-54-2-333. [DOI] [PubMed] [Google Scholar]
- Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA VIII. Properties of the replicating DNA. J Virol. 1977 Aug;23(2):394–411. doi: 10.1128/jvi.23.2.394-411.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. C., Roizman B. Regulation of herpesvirus macromolecular synthesis. VIII. The transcription program consists of three phases during which both extent of transcription and accumulation of RNA in the cytoplasm are regulated. J Virol. 1979 Aug;31(2):299–314. doi: 10.1128/jvi.31.2.299-314.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe D. M., Batterson W., Nosal C., Roizman B., Buchan A. Molecular genetics of herpes simplex virus. VI. Characterization of a temperature-sensitive mutant defective in the expression of all early viral gene products. J Virol. 1981 May;38(2):539–547. doi: 10.1128/jvi.38.2.539-547.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipe D. M., Ruyechan W. T., Roizman B., Halliburton I. W. Molecular genetics of herpes simplex virus: demonstration of regions of obligatory and nonobligatory identity within diploid regions of the genome by sequence replacement and insertion. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3896–3900. doi: 10.1073/pnas.75.8.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse L. S., Buchman T. G., Roizman B., Schaffer P. A. Anatomy of herpes simplex virus DNA. IX. Apparent exclusion of some parental DNA arrangements in the generation of intertypic (HSV-1 X HSV-2) recombinants. J Virol. 1977 Oct;24(1):231–248. doi: 10.1128/jvi.24.1.231-248.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post L. E., Conley A. J., Mocarski E. S., Roizman B. Cloning of reiterated and nonreiterated herpes simplex virus 1 sequences as BamHI fragments. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4201–4205. doi: 10.1073/pnas.77.7.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B., Spear P. G. Preparation of herpes simplex virus of high titer. J Virol. 1968 Jan;2(1):83–84. doi: 10.1128/jvi.2.1.83-84.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyechan W. T., Morse L. S., Knipe D. M., Roizman B. Molecular genetics of herpes simplex virus. II. Mapping of the major viral glycoproteins and of the genetic loci specifying the social behavior of infected cells. J Virol. 1979 Feb;29(2):677–697. doi: 10.1128/jvi.29.2.677-697.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer P. A., Carter V. C., Timbury M. C. Collaborative complementation study of temperature-sensitive mutants of herpes simplex virus types 1 and 2. J Virol. 1978 Sep;27(3):490–504. doi: 10.1128/jvi.27.3.490-504.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]