SUMMARY
V(D)J recombination is one of the most complex DNA transactions in biology. The RAG complex makes double-strand breaks adjacent to signal sequences and creates hairpin coding ends. Here we find that the kinase activity of the Artemis:DNA-PKcs complex can be activated by hairpin DNA ends in cis, thereby allowing the hairpins to be nicked and then to undergo processing and joining by nonhomologous DNA end joining. Based on these insights, we have reconstituted many aspects of the antigen receptor diversification of V(D)J recombination using 13 highly purified polypeptides, thereby permitting variable domain exon assembly using this fully defined system in accord with the 12/23 rule for this process. The features of the recombination sites created by this system include all of the features observed in vivo (nucleolytic resection, P nucleotides, and N nucleotide addition), indicating that most, if not all, of the end modification enzymes have been identified.
INTRODUCTION
V(D)J recombination is the vertebrate gene rearrangement process for assembling the antigen receptor genes, immunoglobulins and T cell receptors (Gellert, 1992; Jung et al., 2006). The V, D and J coding segments must be assembled to form the variable domain exon, which encodes the binding pocket for antigens. This process is essential for acquired immunity, and without it, humans and other mammals have severe combined immune deficiency (SCID) (Revy et al., 2005; Schwarz et al., 1996).
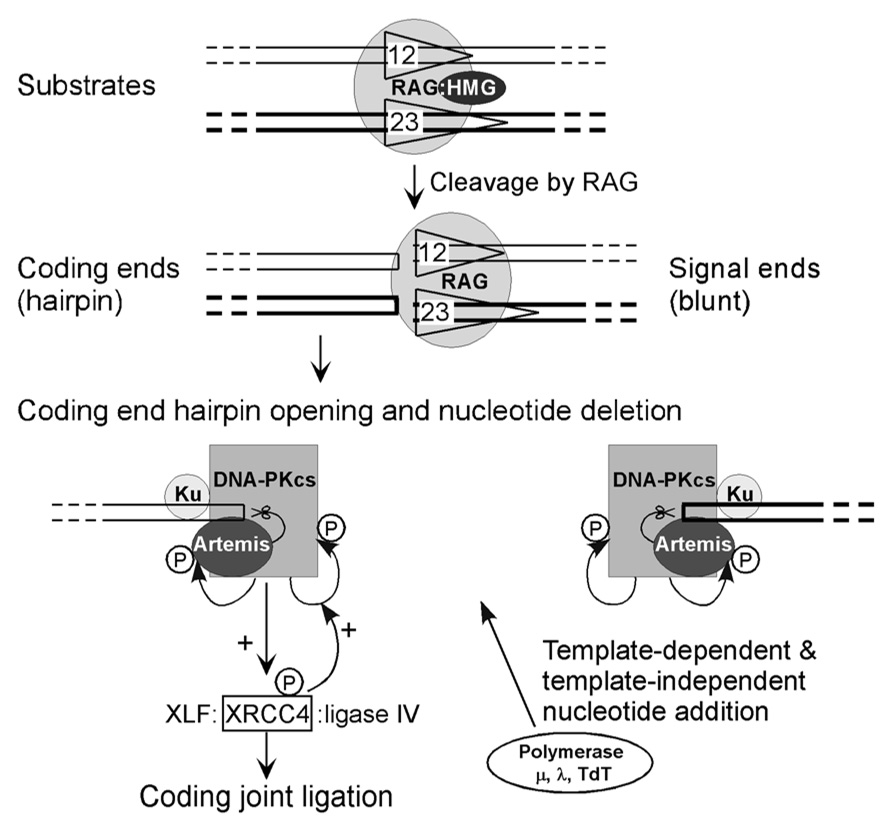
The RAG1 and 2 genes evolved from transposons that can be found among invertebrates (Chatterji et al., 2004; Kapitonov and Jurka, 2005). The lymphoid-specific RAG1, RAG2, and the ubiquitous HMGB1 protein form a complex that binds to heptamer/nonamer recombination signal sequences (RSSs) that can have either a 12-or a 23-bp spacer between the heptamer and nonamer, and are therefore referred to as a 12-RSS and a 23-RSS (Gellert, 2002; Lieber, 2007; Schatz, 2004; Swanson, 2004). Each V, D, or J segment has a 12-or 23-RSS adjacent to it. One recombination reaction involves one 12-RSS and one 23-RSS, a requirement referred to as the 12/23-rule. The RAG complex nicks at the 5' edge of the RSS, and the RAG complex uses the 3'OH of the V, D, or J coding end to carry out a nucleophilic attack on the anti-parallel strand to create a perfect hairpin at the coding end, thereby resulting in a blunt signal end that has 5'P and 3'OH (Roth, 1993; Schlissel, 1993). The coding end formerly attached to the 12-RSS can be called the 12-coding end, and the 23-coding end is named correspondingly.
The opening of the coding end hairpins requires Artemis and DNA-PKcs (Ma et al., 2002). DNA-PKcs is a serine/threonine protein kinase that must bind to DNA termini in order to become active as a kinase (Anderson and Carter, 1996). Artemis and DNA-PKcs form a complex within the cell (Ma et al., 2002). When DNA-PKcs encounters a DNA terminus, it becomes active as a kinase, autophosphorylates itself, and thereby alters the conformation of Artemis in a manner that permits Artemis to function as an endonuclease (Ma et al., 2002). The endonucleolytic properties of Artemis permit it to nick hairpins, preferably 2 nt 3' of the tip, and to nick 5' overhangs and 3' overhangs (Ma et al., 2002).
The activation of DNA-PKcs has been a point of particular uncertainty. It has been shown that the signal ends are bound so tightly by the RAG complex after coding end hairpin formation, the two signal ends cannot be ligated unless the DNA is deproteinized (Jones and Gellert, 2001). The 12-RSS is only 28 bp long, and DNA-PKcs plus Ku require 27 bp of naked DNA for DNA-PKcs to be stimulated (West et al., 1998). Hence, it is difficult to invoke the two signal ends as being sufficiently exposed to activate DNA-PKcs. Each coding end is in a hairpin conformation, and very nice work has demonstrated that hairpins do not activate DNA-PKcs for phosphorylation of p53 peptide targets (Smider et al., 1998), though the results for stimulation of autophosphorylation have been more complex (Soubeyrand et al., 2001). Hence, it has been unclear if either the signal ends or the coding ends are capable of activating DNA-PKcs. If neither can activate DNA-PKcs, then how is DNA-PKcs activated?
Once the hairpins are opened, polymerase mu or polymerase lambda can fill-in gaps or 5' overhangs in a template-dependent manner (Ma et al., 2004; NickMcElhinny and Ramsden, 2003). TdT, and to a lesser extent pol mu, can add nucleotides in a template-independent manner (Gu et al., 2007a). Artemis:DNA-PKcs can continue to resect flaps or to nick at gaps (Ma et al., 2005b). XLF:XRCC4:DNA ligase IV can carry out ligation, with XLF (also called Cernunnos) stimulating incompatible DNA end ligation (Gu et al., 2007b; Tsai et al., 2007).
The joining phase of V(D)J recombination illustrates several aspects of the flexibility of the enzymes involved. Pol mu and pol lambda can slip on the template or mis-incorporate nucleotides more frequently than high-fidelity polymerases (Ramadan et al., 2004). Pol mu can add in a template-independent manner (Gu et al., 2007a; Ramadan et al., 2004). The ligase complex can ligate the top strand independently from the bottom strand, can ligate incompatible ends, and can ligate across gaps (Gu et al., 2007a). Once activated, the Artemis:DNA-PKcs complex can not only act at 5' or 3' overhangs but can also nick at gaps; therefore, it can revise junctions that are ligated on only one strand (Ma et al., 2005b).
In summary, the lymphoid-specific components that have been identified include RAG1, RAG2 and TdT, whereas HMGB1, Ku70/86, Artemis, DNA-PKcs, pol mu, pol lambda, XLF, XRCC4, and DNA ligase IV are present in all vertebrate somatic cells. Despite identification of these proteins, reconstitution using purified components has not been described. The closest approach has been to describe some level of coding joint formation in crude extracts (Leu et al., 1997; Ramsden et al., 1997; Weis-Garcia et al., 1997). However, crude extracts can be misleading (especially for multi-step/multi-component processes) and led one group to infer a role for DNA ligase I in V(D)J recombination (Ramsden et al., 1997), rather the genetically-proven ligase IV (Grawunder et al., 1997; Schar et al., 1997; Teo and Jackson, 1997; Wilson et al., 1997). Once components are genetically-identified, reconstitution is an important aspect in understanding any complex biochemical pathway (Aboussekhra et al., 1995; Kadyrov et al., 2006; Klungland and Lindahl, 1997; Kubota et al., 1996).
Here we have succeeded in achieving coding joint formation using the purified human proteins described above. The coding joints generated by this system show the junctional features seen for coding joints formed within lymphoid cells, including N-nucleotide addition due to TdT, P-nucleotide addition due to hairpin opening by Artemis:DNA-PKcs, and nucleolytic resection due to Artemis:DNA-PKcs. The coding joint formation is abolished in the absence of RAGs, Artemis, DNA-PKcs, or XRCC4:DNA ligase IV. Using additional biochemical approaches, we have documented that the Artemis:DNA-PKcs complex can indeed be activated by DNA hairpins, that this activation is caused by the hairpin to which the complex is bound (cis activation), and that this activation results in the nicking of the hairpins. Hence, the defined system permits generation of junctional and combinatorial diversification of the vertebrate immune system using purified human proteins.
RESULTS
Hairpin DNA Ends Can Activate Artemis:DNA-PKcs to Nick the Hairpins
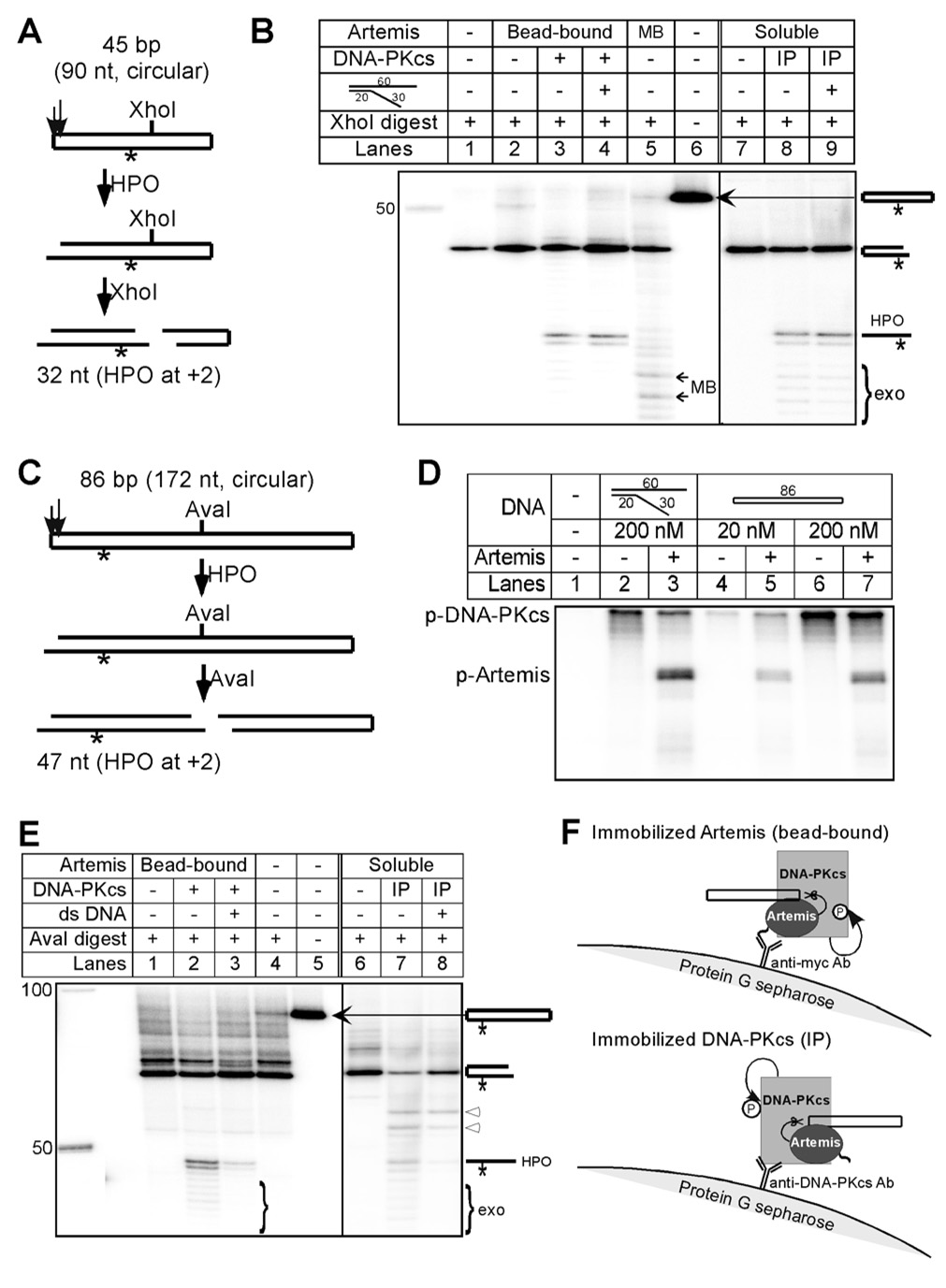
Any effort to biochemically reconstitute V(D)J recombination must first address the issue of how DNA-PKcs becomes activated. This has been a point of particular uncertainty because, after RAG cleavage, the signal ends are sequestered from DNA-PKcs by the RAG complex, and the coding ends are in a hairpin configuration which has generally been considered a poor substrate for DNA-PKcs activation (Jones and Gellert, 2001; Leu et al., 1997; Smider et al., 1998; Soubeyrand et al., 2001). To examine this issue in more detail before attempting a biochemical reconstitution of V(D)J recombination, we synthesized DNA substrates with hairpins at both ends (referred to as double hairpin substrates; Fig. 1A) and tested for hairpin opening by Artemis:DNA-PKcs. We found that the double hairpin substrate was able to activate Artemis:DNA-PKcs, as demonstrated by the hairpin opening of the substrate (Fig. 1B, lane 3). The strongest hairpin opening product was 32 nt long and resulted from hairpin opening at 2 nt 3’ of the hairpin tip (the +2 position).
Figure 1. Artemis:DNA-PKcs Can be Activated by Hairpin DNA Ends in Cis to Nick the Hairpin Structure.
(A) Hairpin opening (HPO) of the 45 bp double hairpin substrate results in a nick on one or both ends. Only the left hairpin with the radiolabeled phosphate is detected after XhoI digestion. Hairpin opening at the +2 position (2 nt 3’ to the hairpin tip) yields a product of 32 nt.
(B) The intact 45 bp double-hairpin DNA substrate migrates slightly slower than the 50 nt marker, and XhoI digestion of the substrate yielded a 56 nt (24 bp) half hairpin that is below the 50 nt marker. The DNA structures were depicted on the right corresponding to each band. The Artemis:DNA-PKcs complex (with either the Artemis or the DNA-PKcs immobilized as shown in panel F) nicked the hairpin of the 45 bp double hairpin substrate efficiently. The strongest product was from hairpin opening at the +2 position (32 nt long based on a single nucleotide ladder), and the 5’ exonuclease activity of Artemis yielded the weak shorter products. Mung bean nuclease (MB) served as a control.
(C) Hairpin opening of the 86 bp double hairpin substrate is followed by AvaI digestion. The 47 nt product indicates hairpin opening at the +2 position.
(D) DNA-PKcs with or without Artemis was incubated with the illustrated DNA (for kinase activation) in the presence of [γ-P32] ATP to allow detection of protein phosphorylation. The pseudo-Y DNA contains a 20 bp double-stranded portion, and two single stranded portions that are 30 nt and 40 nt long. DNA-PKcs autophosphorylation and Artemis phosphorylation are detected with both the pseudo-Y DNA and the double hairpin substrate.
(E) The intact 86 bp double-hairpin-end DNA substate migrates slightly faster than the 100 nt marker, and AvaI digestion yielded a 90 nt (43 bp) half hairpin. Similar to panel B, both forms of Artemis:DNA-PKcs complex efficiently nicked the hairpin of the 86 bp double hairpin substrate. The major hairpin opening product is 47 nt. In reactions 7 and 8, Artemis was not inactivated prior to digestion (it is inactivated in all other reactions). The bands indicated by white arrowheads are due to Artemis cleavage of the 5’ overhangs that resulted from the AvaI digestion.
(F) A diagram showing hairpin opening by immobilized Artemis:DNA-PKcs in cis. Upper panel: Artemis is purified using protein G sepharose with anti-myc monoclonal (MAb) antibody, and the small tail on the Artemis symbol represents the C-terminal myc-his tag. Lower panel: purified DNA-PKcs is immunoprecipated by anti-DNA-PKcs MAb on protein G sepharose.
To confirm that hairpins could indeed stimulate the Artemis:DNA-PKcs complex to perform hairpin opening, a longer double hairpin substrate was made (Fig. 1C). Upon incubation, we found that the hairpin was cleaved by Artemis:DNA-PKcs in the absence of any stimulatory linear or pseudo-Y DNA (Fig. 1E, lane 2). In fact, addition of linear duplex DNA reduced the efficiency of hairpin opening (Fig. 1E, lane 3). Consistent with the 45 bp substrate study above, this 86 bp hairpin was also nicked at the +2 position. In the kinase activity assay, the double hairpin substrate stimulated DNA-PKcs in a manner proportional to its concentration (Fig. 1D, lanes 4–7). Indeed, the double hairpin substrate stimulated the autophosphorylation of DNA-PKcs to a greater extent than the pseudo-Y structure DNA (Fig. 1D, lanes 2 & 3 versus lanes 6 & 7). Artemis phosphorylation by DNA-PKcs was also observed, although with the double hairpin substrate, this was slightly weaker than with the pseudo-Y structure DNA. Therefore, DNA hairpin ends can efficiently stimulate DNA-PKcs autophosphorylation, and this activates Artemis to nick the hairpins at the physiologic position.
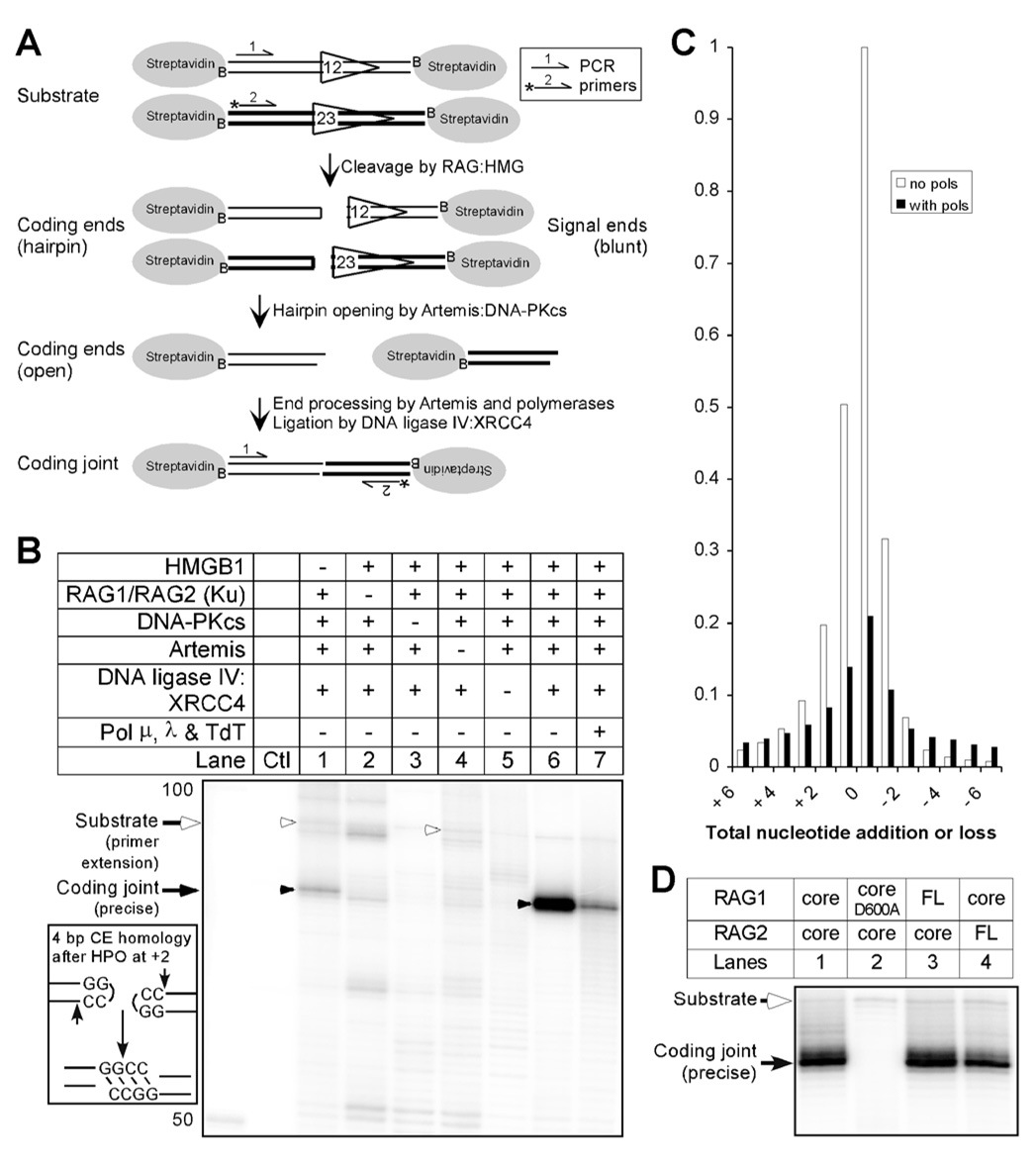
Figure 2. Coding Joint Formation Dependence on V(D)J Recombination Enzymes.
(A) In this coding joint formation assay, two PCR primers (the * indicates the P32 phosphate at the 5’ end) specifically amplify the coding joints. Uncleaved substrates also become amplified by primer extension.
(B) Without polymerases, the V(D)J recombination system yielded a strong precise coding joint (lane 6, black arrowhead). A very low amount of Ku copurified with the RAG complex (estimated molar ratio less than 1 Ku :1000 RAG complexes). Leaving out HMGB1, greatly reduced coding joint efficiency, whereas omitting the RAG complex, Artemis, DNA-PKcs or XRCC4:DNA ligase IV abolished coding joint formation. Addition of polymerases (pol mu, pol lambda, and TdT, lane 7) resulted in a broader range of coding joint products, seen as the slightly darker range of bands above and below the primary joining product.
(C) From the gel in panel B, the bands in lanes 6 and 7 corresponding to the positions of precise coding joint −6 ~ +6 nt are individually quantified and the density was plotted as the fraction of the precise coding joint in lane 6. Addition of polymerases reduced the precise coding joints but increased the diversity of coding joint length.
(D) MBP-tagged core RAG1/core RAG2, full-length RAG1/core RAG2 and core RAG1/full length RAG2 are adjusted to have the same amount of RAG1, and compared for coding joint formation. All other V(D)J recombination proteins (as in panel B lane 7) are included in each reaction. An inactive mutant RAG1 D600A abolished coding joint formation.
Artemis:DNA-PKcs Can be Activated by the Hairpin DNA End to Which it is Bound
In order for coding joints to form, both coding end hairpins generated by a RAG complex would need to be nicked by Artemis:DNA-PKcs. Both cis and trans autophosphorylation of DNA-PKcs have been observed (Meek et al., 2007). Does the Artemis:DNA-PKcs complex bind to the 23-coding end but phosphorylate and activate the Artemis:DNA-PKcs bound to the 12-coding end (in trans)? Or is it activated by the same 23-coding end to which it is bound (in cis)? Again, to gain insight for the optimal design of a system for coding joint formation, we examined these issues in more detail.
We immobilized DNA-PKcs on protein G Sepharose beads using anti-DNA-PKcs antibodies (Fig. 1F). The amount of protein bound to the Sepharose beads was less than 1% of the binding capacity of the immunobeads, ensuring that molecules of DNA-PKcs are physically well separated. Soluble Artemis was mixed with the protein G-bound DNA-PKcs with constant rotation. With both the 45 and 86 bp DNA substrates, the hairpins were opened efficiently (Fig. 1B, lane 8 and Fig. 1E, lane 7). Addition of pseudo-Y structure DNA or linear duplex DNA was not needed to activate the Artemis:DNA-PKcs for the hairpin opening (Fig. 1B, lane 9 and Fig. 1E, lane 8). In additional studies using soluble Artemis and soluble DNA-PKcs, double hairpin substrate nicking was also observed as expected (data not shown). With immobilization of DNA-PKcs, the collision frequency of individual DNA-PKcs molecules with one another is markedly reduced. But we find that the amount of hairpin opening is very similar to that of free DNA-PKcs (and to bead-bound Artemis) (Fig. 1B, lane 3 versus 8). The hairpin opening by immobilized Artemis:DNA-PKcs complexes is most easily understood if Artemis:DNA-PKcs complexes are activated and nick the hairpin ends to which they are bound. The relevance of this for V(D)J recombination is that it indicates that the Artemis:DNA-PKcs complex bound to a given hairpin DNA end can be activated by that hairpin and can open it.
The Known V(D)J recombination Factors in a Biochemically Defined System for Coding Joint Formation
Having clarified the above points, we sought to reconstitute the coding joint formation of V(D)J recombination. The thirteen known polypeptides for V(D)J recombination are RAG1, RAG2, HMGB1, Ku70, Ku86, DNA-PKcs, Artemis, polymerase mu, polymerase lambda, TdT, XRCC4, DNA ligase IV and XLF. The mammalian proteins were purified and tested for activity individually (Suppl. Table 1. & Suppl. Fig. 1). In the course of optimization of the reaction conditions, we found that activated DNA-PKcs consistently improved the activity of the RAG complex and the ligase complex, and the latter increased autophosphorylation of DNA-PKcs (Suppl. Figs. 2 & 3). The DNA substrates for in vitro V(D)J recombination are two 88 to 95 bp double-stranded linear molecules with a 3’ biotin group on each strand (one at each end). Each duplex substrate contains a 12-or 23-RSS and 41 to 45 bp of DNA corresponding to the coding segments (Fig. 2A). Streptavidin was added to the DNA substrates to block their DNA ends.
In this biochemical system, the 12- and 23-RSS substrates are incubated with the V(D)J recombination factors (Fig. 2A). Cleavage at the signals by the RAG complex results in four DNA ends: two blunt signal ends that are tightly bound to the RAG complex, and two hairpin coding ends (Suppl. Fig. 2 & data not shown). The hairpins are opened by Artemis:DNA-PKcs and may undergo further deletion due to Artemis exo-and endonuclease activity (Ma et al., 2002). Polymerases and ligases complete the joining. For detection of coding joints, 30 cycles of PCR are done using primers specific to the coding ends. The PCR products are then resolved by denaturing PAGE.
We found that a subset of the enzymes (the RAG/HMGB1 complex, Artemis, DNA-PKcs, and XRCC4:DNA ligase IV) were able to carry out V(D)J recombination and generate coding joints (Fig. 2B, lane 6). Low amounts of endogenous Ku70/Ku86 commonly copurify with the RAG complex (data not shown and (Raval et al., 2008)), but joining in reactions with stringently purified RAGs showed no dependence on added purified Ku (see Suppl. Figs. 4 & 5 and Discussion). HMGB1 facilitates DNA binding and cleavage by the RAG complex, and consistently, omission of HMGB1 diminished the coding joint products about 10-fold (Fig. 2B, lane 1). Importantly, omitting the RAG complex, DNA-PKcs, Artemis or XRCC4:DNA ligase IV reduced coding joint formation to background levels (Fig. 2B, lane 2–5). The low level of molecules responsible for the background signal in reactions without the RAG complex, Artemis, or XRCC4:DNA ligase IV were sequenced and were PCR products from nonspecific annealing of the substrates (data not shown). Full-length (FL) RAG1/core RAG2 and core RAG1/FL RAG2 showed comparable coding joint efficiency to core RAG1/core RAG2, and the pattern of coding joint products was also indistinguishable (Fig. 2D, lane 1 versus lanes 3 & 4). A catalytically inactive mutant RAG1 D600A abolished coding joint formation (Fig. 2D, lane 2). With the relatively high concentration of XRCC4:DNA ligase IV necessary for the reactions, there was little dependence on XLF (Suppl. Fig. 4). Polymerases mu, lambda and TdT are preferred for the purpose of V(D)J recombination (addition of the Klenow fragment of E. coli polymerase I to the reaction resulted in coding joints that are shorter than those found in vivo (Suppl. Fig. 6)), and the system showed specificity for XRCC4:DNA ligase IV (Fig. 3C) (Tomkinson et al., 2006). And very importantly, the coding joint formation conforms to the 12/23 rule (Suppl. Fig. 7B, panel D).
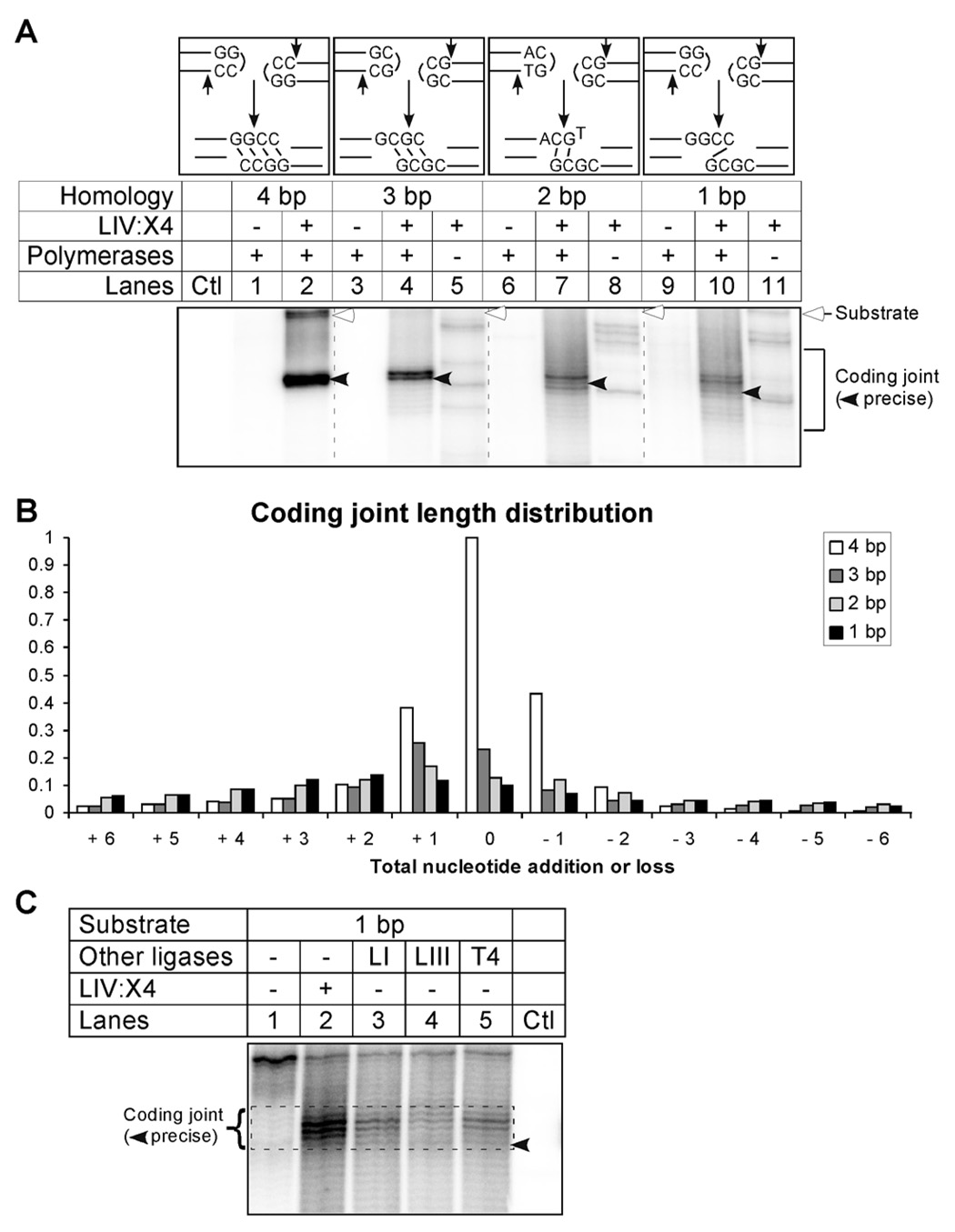
Figure 3. Coding Ends with Different Extents of Microhomology Show Different Extents of Coding Joint Diversity.
(A) The last two base pairs of the coding end sequences from the four different sets of substrates are displayed. The strongest hairpin opening product at the +2 position is depicted on the top. The microhomologies at the coding ends are 4 bp, 3 bp, 2 bp and 1 bp, respectively. All V(D)J recombination reactions contain the RAG complex (with Ku), HMGB1, and Artemis:DNA-PKcs. XRCC4:DNA ligase IV and the polymerases (pol mu, pol lambda and TdT) are added where indicated. The shift towards longer coding joints and higher diversity indicates stronger polymerase action on the low homology substrates.
(B) The coding joints in lanes 2, 4, 7 and 10 of panel A (reactions with ligase and polymerases) between the length of the precise coding joint ± 6 nt are individually quantified. The density is plotted as a fraction of the precise coding joint from the 4 bp substrate in lane 2. Coding joints from the 4 bp homology substrate are shown in open bars, whereas the dark gray bars indicate coding joints from the 3 bp substrate, light gray bars from the 2 bp substrate, and black bars from the 1 bp substrate. As the homology is reduced, the diversity of the coding joints increases.
(C) T4 DNA ligase, DNA ligase I and III are adjusted to have the same nick ligation activity, and then used in place of XRCC4:DNA ligase IV for the V(D)J recombination reactions. Coding joint ligation by XRCC4:DNA ligase IV in lane 2 showed the most efficient coding joint formation for the 1 bp homology substrates.
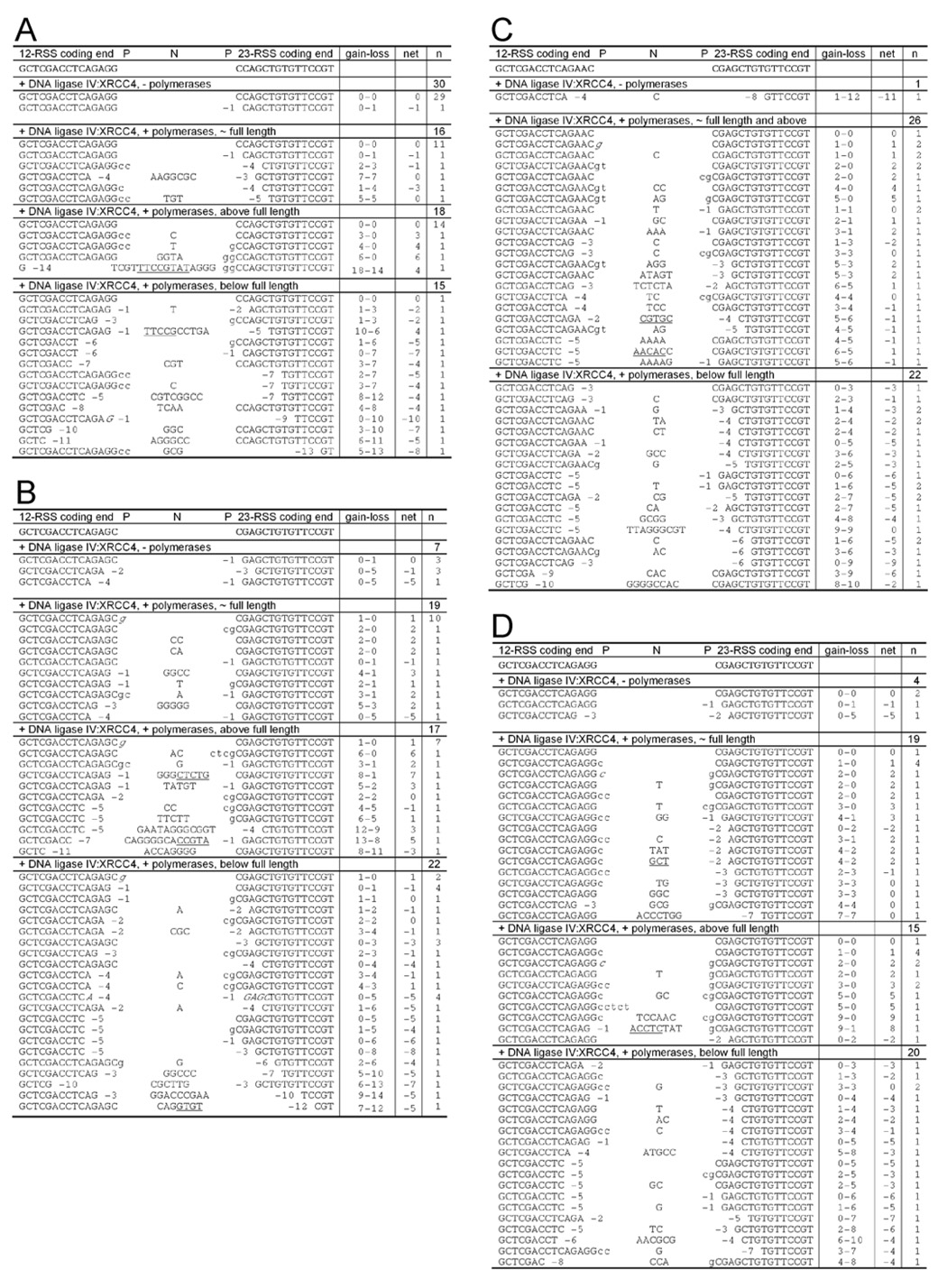
The coding end microhomology (depicted to the left of the gel in Fig. 2B) influenced the nature of the coding joint that was formed. The primary hairpin opening position by Artemis:DNA-PKcs is 2 nt 3’ to the hairpin tip, resulting in two 4 nt overhangs that are fully compatible (Lu et al., 2007b). Ligation of this pair of compatible ends by XRCC4:DNA ligase IV is extremely efficient (Gu et al., 2007a; Ma et al., 2004). The major product has no deletion or addition (Fig. 2B, lane 6), and this 'precise' coding joint, was confirmed by sequencing of the product (Fig. 4A). This constraint of diversity by terminal microhomology corresponds to in vivo V(D)J recombination (Bertocci et al., 2006; Feeney, 1992; Gerstein and Lieber, 1993b). Addition of polymerases reduced the amount of precise coding joint product, but increased the diversity of the coding joints that have shorter or longer lengths, as manifested by the range of darker bands above and below the primary joining product (Fig. 2B, lane 7 and Fig. 2C). This increase in diversification upon addition of polymerases is similar to what is seen in vivo (Bertocci et al., 2006). These diverse coding joint sequences included short deletions of up to 14 nt (sum of nts resected from the two ends) and nucleotide additions of up to 16 nt. P nucleotides were present at 25% of the junctions (see the coding joint sequences in Fig. 4A).
Figure 4. Coding Joint Sequences.
(A) Coding joint sequences from the 4 bp homology substrate. The coding end sequences flanking the 12- and 23- signals are shown on the top in bold and capitalized. The first group of sequences is from Figure 2B lane 6 with ligase but no polymerases, and the rest are from Figure 2B lane 7. The coding joints are divided into three regions according to the size of the bands: full length ±1 nt, full length +2 ~ +6 nt and full length −2 ~ −6 nt. The number of molecules sequenced is shown in the last column on the right. Deletions are shown in numbers next to the coding ends. The P nucleotides are shown adjacent to the coding ends in lower case. N nucleotides are shown in the middle, with the direct and indirect repeats underlined. The sequences in italic indicate microhomology usage. The total nucleotide gain and loss and the net change compared to precise coding joint are shown on the right of the sequences.
(B) Coding joint sequences from the 3 bp homology substrate. The first group of sequences is from Figure 3A, lane 5 with ligase but no polymerases, and the rest are from Figure 3A lane 4. The coding joints are divided into three regions according to the size of the bands: full length −1 ~ +2 nt, full length +3 ~ +7 nt and full length −2 ~ −6 nt.
(C) Coding joint sequences from the 2 bp homology substrate. The first group of sequences is from Figure 3A, lane 8 with ligase but no polymerases, and the rest are from Figure 3A lane 7. The coding joints are divided into two regions according to the size of the bands: full length −1 ~ +4 nt and full length −2 ~ −6 nt.
(D) Coding joint sequences from the 1 bp homology substrate. The first group of sequences is from Figure 3A, lane 11 with ligase but no polymerases, and the rest are from Figure 3A lane 10. The coding joints are divided into three regions according to the size of the bands: full length −1 ~ +2 nt, full length +3 ~ +7 nt and full length −2 ~ −6 nt.
For in vivo V(D)J recombination junctions that have junctional additions, some portions of some of the additions are direct or inverted repeats of the sequence of one coding end (these have sometimes been called T nucleotides) (Gauss and Lieber, 1996; Jaeger et al., 2000; Lieber et al., 1988; Ma et al., 2005a). In the biochemical system with different substrates, we find several examples of junctional additions that include direct or inverted repeats of 4 bp or longer, from either of the two coding ends (Fig. 4, underlined sequences). For the 4 bp microhomology substrate, one of these is 8 nt long. With respect to this feature, the junctions are indistinguishable from those that have been described at coding joints formed in vivo (Gauss and Lieber, 1996; Jaeger et al., 2000; Lieber et al., 1988; Ma et al., 2005a).
Coding Joint Diversity is Influenced by Coding End Microhomology
For in vivo V(D)J recombination, small variations in coding end sequence can cause marked changes in coding joint diversity, and it is difficult to dissect the mechanistic basis for these changes using in vivo approaches (Gerstein and Lieber, 1993a). To assess coding joint diversity in vitro, substrates with less coding end homology than the ones described above were tested. The last two base pairs of the substrates were varied such that after hairpin opening at the 2 nt position 3’ to the hairpin tip, the possible microhomology at the end would be 3, 2 or 1 bp (Fig. 3A). The RAG complex (with Ku), HMGB1, Artemis:DNA-PKcs were included in all reactions. Specified reactions additionally contained XRCC4:DNA ligase IV and the polymerases mu, lambda and TdT. As the homology decreased from 4 bp to 1 bp, the precise coding joint product became much weaker, and the major products shifted to higher positions, indicating gap filling by the polymerases (Fig. 3A, lanes 2, 4, 7, and 10). Similar to in vivo V(D)J recombination, the diversity of the products also increased, as shown by the difference in the length distribution of coding joints (Fig. 3B). When polymerases were omitted, the amount and diversity of coding joints was greatly reduced (Fig. 3A, lanes 5, 8 and 11), illustrating the requirement for fill-in synthesis by either pol mu or pol lambda.
We previously showed that DNA ligase I and III have weaker ligation activity on incompatible double-stranded DNA ends than XRCC4:DNA ligase IV. These ligases were tested in the biochemically defined V(D)J recombination system here using the 1 bp microhomology substrates described above, because they are more difficult to ligate. We find that DNA ligase I and III yielded substantially less coding joint product than XRCC4:DNA ligase IV (Fig. 3C, lane 2 vs. lanes 3–5). The 2 bp microhomology substrate was also tested and showed similar results (data not shown). Therefore, DNA ligase IV supports the strongest ligation in the biochemical system for coding joint formation here.
This biochemically defined system for coding joint formation recapitulates many features of the cellular V(D)J recombination process: P nucleotides, N nucleotide addition, microhomology usage, nucleolytic resection of the ends, and occasional direct or inverted repeats within the junctional additions (see the 252 coding joint sequences in Fig. 4). Moreover, the length and frequency of these features is within the range of that seen in vivo (Bertocci et al., 2006; Gauss and Lieber, 1996). Coding joint formation was dependent on the RAG/HMGB1 complex, on Artemis, on DNA-PKcs, and on XRCC4:DNA ligase IV. Similar to the in vivo situation, the joining did not require the Pol X polymerases mu, lambda or TdT, but the junctional diversity was substantially increased by these polymerases (Bertocci et al., 2006; Gilfillan, 1993; Komori, 1993). The similarity of the junctional sequences to those in vivo suggests that many aspects of the coordination between the various protein factors is recapitulated.
DISCUSSION
Which DNA Ends in V(D)J Recombination Activate Artemis:DNA-PKcs for Hairpin Opening?
In V(D)J recombination, the coding end hairpins generated by RAG cleavage must be nicked open by Artemis:DNA-PKcs before any joining can occur. The source of activation of DNA-PKcs has been a debated issue (Smider et al., 1998; Soubeyrand et al., 2001). The signal ends are thought to be blocked by the tight binding of the RAG complex. The RAG binding to the signals is sufficiently tight that it prevents any nucleolytic resection. Therefore, the exposed length of naked DNA at the signal ends is unlikely to be adequate to stimulate DNA-PKcs. The coding ends are in a hairpin configuration, and the hairpin configuration has been thought to be inadequate to activate DNA-PKcs (Smider et al., 1998). This latter inference was based on differences in stimulation between hairpin and single-stranded DNA ends for p53 peptide phosphorylation by DNA-PKcs. Whether DNA-PKcs autophosphorylation is stimulated by hairpins has been less clear. Hence, the identity of the DNA ends that activate hairpin opening has been one of the key remaining problems in the mechanism of V(D)J recombination.
There are several points that our studies clarify. First, we have demonstrated autophosphorylation of DNA-PKcs after stimulation by double hairpin DNA. We find that Artemis:DNA-PKcs is activated by hairpin ends to open those hairpins. Second, when interaction between different molecules of DNA-PKcs is prevented by immobilization at dilute concentration on the surface of rotated beads, the bound Artemis:DNA-PKcs complex efficiently opens the hairpins. Because the DNA-PKcs molecules are immobilized, this is compelling evidence that the hairpins are opened by the individual Artemis:DNA-PKcs complex to which they bind. Third, each activated DNA-PKcs autophosphorylates itself in cis under these circumstances because the DNA-PKcs molecules are unable to contact each other. These points lead to a V(D)J recombination model in which an Artemis:DNA-PKcs complex binds to either or both coding end hairpins, are activated by those ends in cis, and then nick those hairpin ends. After hairpin opening, it is quite conceivable that additional steps during coding end processing would involve additional cis and trans phosphorylation events.
DNA-PKcs not only is essential for coding end hairpin opening by Artemis, but also stimulates coding end ligation by XRCC4:DNA ligase IV. DNA-PKcs may regulate and coordinate the cleavage, coding end processing, and ligation (Fig. 5). The increase in DNA-PKcs autophosphorylation by XRCC4:DNA ligase IV could either be positive feedback for stimulation of DNA-PKcs kinase activity, or it could be due to phosphorylation of additional sites which may lead to DNA-PKcs dissociation from DNA ends.
Figure 5. The Role of Cis Artemis:DNA-PKcs Activation by Hairpin DNA Ends in the V(D)J Recombination System.
Each hairpin coding end can activate the Artemis:DNA-PKcs complex to which it is bound and can be nicked by the same complex in cis. Activated DNA-PKcs enhances the activity of DNA ligase IV:XRCC4. XRCC4:DNA ligase IV also stimulates the autophosphorylation of DNA-PKcs, thereby regulating the activity of DNA-PKcs. The dashed line on each side of the DNA represents DNA flanking the coding ends and signal ends. Arrows with “+” represent stimulation. Arrows with a letter “P” in a circle indicate phosphorylation.
The Known Enzymes of V(D)J Recombination are Sufficient for Coding Joint Formation in a Biochemically Defined System
Despite some limitations of the system (see Supplementary text), this biochemically defined system for coding joint formation recapitulates many features of the cellular V(D)J recombination process: P nucleotides, N nucleotide addition, microhomology usage, nucleolytic resection of the ends, and occasional direct or inverted repeats within the junctional additions. Moreover, the length and frequency of these coding end or junctional changes is within the range of that seen in vivo. Coding joint formation was dependent on the RAG/HMGB1 complex, on Artemis, on DNA-PKcs, and on XRCC4:DNA ligase IV. Very similar to the in vivo situation, the joining did not require the Pol X polymerases mu, lambda or TdT, but the junctional diversity was substantially modified by them. The similarity of the junctional sequences to those in vivo suggests that the coordination between the protein factors is recapitulated.
In the biochemically defined system, the efficiency of conversion from starting substrate to final coding joints is estimated to be between 0.1% and 0.5% (Suppl. Fig. 9). This may seem low, but the final yield of products from extrachromosomal substrates in the nucleus of pre-B cell lines are often no higher than this (Lieber et al., 1987). It is likely that there are additional protein factors that function in V(D)J recombination. These factors could remodel the chromatin structure, modify the enzymatic proteins, and regulate the expression as well as the cellular localization of the components. The system here, with the known components, provides a basis for incorporating these elements.
In vivo and in vitro, Ku improves the binding of the nuclease, polymerase and ligase activities for NHEJ. However, the nuclease, polymerases, and ligase function even in the absence of Ku on DNA ends, and the concentration of Ku relative to the concentration of DNA ends in an in vitro system determines whether Ku stimulation is observable, as we have seen previously (Gu et al., 2007a; Ma et al., 2004). Even for in vivo coding end combinations on intrachromosomal substrates, the Ku-dependence for joining can be as small as 1.5- to 8.5-fold (Schulte-Uentrop et al., 2008; Weinstock et al., 2007), illustrating that the nuclease, polymerases, and ligase components can function in coding joint formation without Ku. In vitro, Ku stimulation of ligation is only observed when their concentration is comparable to that of the DNA ends (Gu et al., 2007a). We could lower the Ku concentration to a level equal to that of the coding ends, but then we would need to lower the concentration of XRCC4:DNA ligase IV in order to have any stimulatory effect, and this places the joining outside of the measurable range. The much stronger Ku-dependence for primary pre-B and pre-T cell differentiation is well-known (Gu et al., 1997; Zhu et al., 1996) and presumably reflects a greater reliance on Ku for loading proteins in vivo and the multiple sequential steps of V(D)J recombination on the two chains of the antigen receptors.
XLF is stimulatory in vivo (Buck et al., 2006) and in vitro (Gu et al., 2007b; Tsai et al., 2007).; however, substantial levels of V(D)J recombination are observed in humans that are mutant for XLF (Buck et al., 2006). Mice which are knocked out for XLF show near-normal levels of V(D)J recombination (F. Alt, personal communication). Therefore, biochemical findings of V(D)J recombination coding joint formation in our defined system are consistent with these genetic studies. Among its functions, XLF, and the S. cerevisiae homologue, NEJ1, may participate in nuclear localization of the XRCC4:DNA ligase IV ligase complex, which would not be as relevant to all aspects of the end joining monitored in our assay system here (Lu et al., 2007a; Valencia et al., 2001).
Post-Cleavage Retention of the Coding Ends is Not Required for the Basic Enzymatic Processes of Coding Joint Formation and May Involve Additional Factors or Chromatin
Post-cleavage retention of the coding ends by the RAG complex has been suggested to have an important role in V(D)J recombination. The post-cleavage retention of the coding ends could conceivably occur primarily at the chromatin level or could be due to coding end DNA binding by the RAG complex (Leu et al., 1997). Binding of the RAG complex to the signal ends is very strong (Jones and Gellert, 2001), but the binding to the coding ends is thought to be exceedingly weak (Jones and Gellert, 2001; Tsai et al., 2002), and we have confirmed this (NS and ML, unpublished). We tested this issue functionally in our V(D)J recombination assay. The core RAG complex was pulled out from the reaction after cleavage occurred, and before addition of all of the other V(D)J recombination factors; however, we found that coding joint production was not affected (data not shown). We tested for coding end holding by the core RAG complex more formally in two additional experimental designs. In one, we did the same type of experiment as in Figure 2, but we assayed for the joining of two 12-coding ends rather than of the 12-coding end with the 23-coding end. We used a mixture of two 12-substrates (with different coding ends) and a 23-substrate. Without the addition of a 23-substrate, we observe no joining between the two 12-coding ends, in accord with the 12/23 rule (Suppl. Fig. 7B, panel D). With the 23-RSS present, 12-12 coding end joining was within two-fold of 12–23 coding end joining (Suppl. Fig. 7B, panel A vs. B). Therefore, there was no indication that the coding ends were being held by the RAG complex after cleavage. In the second type of experiment, the substrates had a 12-RSS and a 23-RSS on the same 235 bp duplex (Suppl. Fig. 8). A second substrate (237 bp) also had a 12-RSS and a 23-RSS on the same duplex, but the coding ends of the 237 bp substrate were different from those of the 235 bp substrate. The 235 bp and the 237 bp substrates were incubated separately, with the RAG complex, HMGB1, Artemis:DNA-PKcs for 90 min. Then two reactions were pooled and DNA ligase IV:XRCC4 were added to permit ligation for only 20 min. We then assayed for coding joint formation by coding ends from the same duplex versus from the two different duplexes. We found that the coding ends could freely join between the two different substrates in the reaction mixture (Suppl. Fig. 8). Therefore, we see no evidence that the core RAG complex can hold onto naked DNA coding ends. We also see no evidence of full-length RAG holding of coding ends (Suppl. Fig. 7C), and this finding is consistent with the fact that we have not observed joining efficiency differences for these different forms of RAGs (Fig. 2D). Lack of tight holding of naked coding end DNA by the RAG complex may make evolutionary sense. Tight holding of the naked coding ends by the RAG complex might preclude rounds of enzymatic modification by the Artemis:DNA-PKcs complex, pol mu, pol lambda, TdT, and the DNA ligase IV complex.
Since the RAG complex does not appear to bind to naked coding ends strongly, the post-cleavage complex may largely be stabilized at the chromatin level. Recent studies have suggested binding between full-length RAG2 and trimethylated K4 of histone H3 (Liu et al., 2007; Matthews et al., 2007). This interaction could mediate the coding end holding within the post-cleavage complex. Coding end holding by RAGs via the nearest nucleosome at each coding end would permit access by the various hairpin opening and NHEJ enzymatic activities without releasing the coding ends from the RAG complex. To demonstrate the effect of such post-cleavage complex, the naked DNA substrates for the V(D)J recombination assay need to be replaced with reconstituted nucleosomes, and the system described here will facilitate such efforts in the field over the next several years.
Signal Joint Formation Requires Removal of the RAG Complex
In this defined V(D)J recombination system, we were unable to detect any signal joint product using the known protein factors. Similar attempts with either crude extracts or partial NHEJ systems also failed to detect signal joints (Jones and Gellert, 2001; Leu et al., 1997; Weis-Garcia et al., 1997). In fact, signal end ligation could only be detected when the RAG cleavage products were deproteinated before ligation. This is generally agreed to be due to the tight binding of the RAG complex to the signal ends. The N-terminal noncore region of full-length RAG1 contains an E3 ubiquitin ligase domain and ubiquitination of the RAG complex has been observed in vivo. We suspect that the RAG complex must be degraded to release the signal ends for ligation. Full-length RAG1/core RAG2 complex was tested for signal joint formation in our biochemically defined system, with the addition of ubiquitination components with or without a proteasome fraction. However, no signal joint formation was detected, even though coding joint formation remained unaltered by adding these additional proteins late in the reaction (data not shown). For the signal ends to be released, ubiquitination of RAG may not be the essential step, because core RAG1/core RAG2 is capable of mediating signal joint formation in cellular V(D)J recombination assays. There could be a factor that mediates the dissociation of the RAG complex from the signal ends. Further studies are required to reveal the mechanism of signal end release by the RAG complex.
Concluding Comments
Our findings lead to a V(D)J recombination model in which an Artemis:DNA-PKcs complex binds to either or both coding end hairpins, are activated by those ends in cis, and then nick those hairpin ends in cis (Fig. 5). We have shown that the proteins in a biochemically defined system are capable of recapitulating the key coding joint formation features of V(D)J recombination, including P nucleotide formation, nucleotide end resection, and junctional addition. This system will permit incorporation of additional components when and if any are identified. In our system, core or full-length RAG protein holding of naked coding ends was inefficient, suggesting that RAG contacts with nucleosomes might be critical for coding end holding in a post-synaptic complex H3 (Liu et al., 2007; Matthews et al., 2007). Functional testing for this can now be done by building upon this defined system. Furthermore, the system will be useful for testing inhibitors that would be of therapeutic value in RAG-positive acute lymphoblastic lymphomas (Bories et al., 1991), where interruption of V(D)J recombination would create double-strand breaks.
EXPERIMENTAL PROCEDURES
Double Hairpin Substrates and Substrates for V(D)J Recombination
The 45 bp double hairpin substrate was ligated after allowing a 90 nt long oligonucleotide HL-157 to self-anneal. 50 pmol HL-157 was kinased with 1 uM [gamma-P32] ATP and 25 units of T4 polynucleotide kinase (PNK) (New England Biolabs) in 50 ul NEB buffer 4 with 5% PEG at 37°C for 40 min, and then 1 nmol ATP was added and the reaction was incubated at 37°C for another 20 min. T4 PNK was heat denatured at 72°C for 20 min, and the reaction mixture was desalted using a microspin G25 column (GE). For 40 pmol of kinased HL-157, 10× T4 DNA ligase buffer was added to a final concentration of 1×, heated at 100°C for 5 min, chilled on ice, and 800 units of T4 DNA ligase (New England Biolab) was added. The ligation reaction was incubated at 16°C for 20 hr. Then an 15 additional 800 units of T4 DNA ligase were added and ligated at 20°C for 2 hr. The ligation products were resolved on 40% formamide-6 M urea-6% PAGE. The gel piece with ligated monomer product was cut out and crushed, extracted at 37°C overnight in TE with 500 mM NaCl, followed by freeze and thaw at −80°C. In the end, DNA was precipitated from the supernatant. The purity of the ligated DNA was >99%, as judged by denaturing PAGE and restriction digestion (data not shown).
The 86 bp double hairpin substrate was ligated from three oligonucleotides HL-150, HL-151 and HL-152. For the radioactive substrate, HL-150 was kinased with 1 uM [gamma- P32] ATP the same way as HL-157. 1 nmol of HL-151, HL-152 and HL-150 were separately kinased with 0.1 mM ATP and 50 units T4 PNK in 200 ul of NEB buffer 4 at 37°C for 1 hr and phenol/chloroform extracted and precipitated by ethanol. 40 pmol of radioactively labeled HL-150, with 5’ phosphorylated HL-151 and HL-152 were ligated in the same way as for HL-157. To make unlabeled double hairpin substrate, 0.96 nmol of 5’ phosphorylated HL-150, HL-151 and HL-152 were resuspended in 1× T4 DNA ligase buffer, heated and cooled to anneal, and then 2,000 units of T4 DNA ligase were added. After ligation, the radioactively labeled and unlabeled ligation products were purified the same way as for the 45 bp substrate. The purity of the ligated 86 bp double hairpin substrate was above 95%, as assessed by denaturing PAGE and exonuclease treatment (data not shown).
The 4 bp homology substrates for V(D)J recombination (the 4 bp substrate) consisted of a 12-substrate that was annealed from HL-133 and HL-134, and a 23-substrate that was annealed from HL-135 and HL-136. The 3 bp, 2 bp and 1 bp substrates have the same sequences as the 4 bp substrate except for the last two base pairs of the coding ends, as illustrated in Figure 3A. All DNA oligonucleotides have covalently linked 3’ biotin TEG. The primers for coding joint PCR were HL-66 and HL-68. The HL-66 primer was radioactively labeled at the 5’ end with [gamma-P32] ATP and T4 PNK.
The pseudo-Y DNA for the hairpin opening assay has been described (Lu et al., 2007b), as has the ligation substrate (Lu et al., 2007a). The sequences of the oligonucleotides are shown in the Supplementary Methods.
Protein Purification
Expression and purification of proteins are summarized in Supplementary Table 1. Core RAG1 consists of aa 384–1040, and core RAG2 consists of aa 1-383. GST-core RAG1/2, HMGB1, Ku70/Ku86, Artemis, DNA-PKcs, XRCC4:DNA ligase IV, XLF, polymerase mu and lambda are purified as described in the references listed in Suppl. Table 1 (Bergeron et al., 2006; Chan et al., 1996; Dominguez et al., 2000; Lu et al., 2007a; Ma et al., 2002; NickMcElhinny et al., 2000; Shimazaki et al., 2002; Yaneva et al., 1997; Yu et al., 2002). Human TdT was a gift from Dr. Fred Bollum and Dr. Lucy Chang (Chang et al., 1988). Artemis-His baculovirus was generously provided by Drs. John Harrington and Steve Murphy at Arthersys Inc. (Cleveland, OH). Sf21 insect cells infected with the Artemis-His baculovirus was lysed in lysis buffer (50 mM NaH2PO4(pH 7.8), 0.5 M NaCl, 2 mM 2-mercaptoethanol, 10% glycerol, 0.1% Triton X-100, 20 mM imidazole, 0.1 mM phenylmethylsulfonyl fluoride, 1 µg/ml aprotinin, pepstatin A, and leupeptin). And the cell extracted was purified using Ni-NTA agarose (Qiagen). Fractions containing Artemis were dialyzed against Mono Q buffer (50 mM Tris-HCl (pH 7.5), 10% glycerol, 2 mM EDTA, 1 mM DTT, and 0.02% NP-40, containing 0.1 M NaCl), loaded onto a Mono Q column (Amersham Biosciences), and eluted with a linear gradient of 0.1–0.5 M NaCl. Fractions containing Artemis were dialyzed against storage buffer (50 mM Tris-HCl (pH 7.5), 20% glycerol, 1 mM DTT, and 0.1 M KCl) and stored at −80 °C.
Kinase Assay and Hairpin Opening Assay
The kinase assay for DNA-PKcs has been described (Ma and Lieber, 2002). The hairpin opening assay by Artemis:DNA-PKcs was modified from the one described by increasing the KCl concentration to 60 mM. After hairpin opening, the reactions were heated at 65°C for 20 min to inactivate Artemis:DNA-PKcs. Then an equal volume (10 ul) of NEB buffer for restriction digestion was added, followed by the addition of restriction enzymes. Hairpin opening products by mung bean nuclease were extracted with phenol/chloroform, precipitated and dissolved in the restriction buffer with enzyme. The digestions were incubated at 37 °C for 2 hr for AvaI and 1 hr for XhoI, and the products were resolved using 7 M urea-10% PAGE (for Fig. 1E, lanes 6–8, samples were resolved on a 7 M urea-12% PAGE gel). For immobilization of DNA-PKcs to anti-DNA-PKcs immunobeads, 2.4 pmol of DNA-PKcs protein was mixed with a total of 5 pmol of monoclonal anti-DNA-PKcs antibodies (clone 42–27, 25–4, and 18–2) in a total volume of 30 ul with a buffer composition of 25 mM HEPES-KOH (pH 7.5), 30 mM KCl, 10 mM MgCl2, 5% glycerol, 1 mM DTT and 0.1 mg/ml of BSA and incubated for 2 hours. Then, 10 ul of protein G beads, which have 100-fold more binding capacity of antibodies to the amount of antibodies used here, were mixed in and rotated for 1 hour. The beads were washed three times with same buffer without BSA and then washed two times with 1xnuclease assay buffer. More than 65 % of DNA-PKcs was estimated to be captured on anti-DNA-PKcs immunobeads by SDS-PAGE and coomassie staining and a quarter of the reaction containing at least 0.4 pmol of DNA-PKcs was used in each nuclease assay.
V(D)J Recombination Assay
Various molar ratios of streptavidin to biotin-DNA ends were tested and a 2.4:1 ratio was needed for maximum binding to the biotin-DNA ends. Streptavidin is a tetramer, and multiple DNA end binding to the same molecule of streptavidin could hinder proper alignment of the 12 and 23-RSS for synapsis. Therefore, an even higher ratio of 12:1 streptavidin to biotin-DNA ends was used. Each double-stranded substrate (9 pmol) was first incubated with 9.6 ug streptavidin (Sigma) in 10 mM Tris, pH 8.0, 70 mM NaCl at room temperature for 40 min. Then 10 ul streptavidin MagneSphere paramagnetic particles (Promega) were added for 10 min to bind any biotin-DNA that was not bound by streptavidin, and the supernatant was collected. 0.3 pmol each of the 12-and 23-substrate was incubated with 2 pmol GST-core RAG1/2 (with low amount of Ku), 5 pmol of HMGB1, 1 pmol of Artemis and 0.3 pmol of DNA-PKcs in a total volume of 10 ul of reaction buffer (25 mM HEPES, pH 7.5, 60 mM KCl, 5 mM MgCl2, 250 uM ATP, 1 mM DTT, 5% PEG (Mr = 8000)). 3 pmol each of polymerase mu, lambda and TdT are added where indicated. The reaction was incubated at 37°C for 20 min before addition of XRCC4:DNA ligase IV. 2 pmol total of XRCC4:DNA ligase IV was added every 30 min for a total of three additions. The total incubation time at 37°C is 2 hr. Then 0.5 ul of each reaction product is used as a template for coding joint PCR (94°C, 45 sec, 59°C, 25 sec, and 72°C, 30 sec; 30 cycles). The PCR products were resolved by 7 M urea-10%PAGE. The gel was dried, exposed in a PhosphorImager cassette, and the screen was scanned using a PhosphorImager SI445 (Amersham Biosciences, Piscataway, NJ).
Sequencing of Coding Joint PCR Products
After the gel image was obtained, bands of desired size range were excised from the dried gel and eluted by TE (10 mM Tris, 1 mM EDTA, and [pH 8.0]). The coding joint sequences were derived from products ranging from 71–85 nts of Figure 2B and Figure 3A. The eluted DNA was amplified in a second round of PCR (10 cycles) with unlabeled primers, and then cloned into the Topo TA cloning vector pCR2.1 (Invitrogen) according to the manufacturer’s recommendation. Individual clones were then sequenced on a Li-Cor sequencer (Li-Cor) following the manufacturer’s instructions.
Oligonucleotides
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aboussekhra A, Biggerstaff M, Shivji M, Vilpo J, Moncollin V, Podust V, Protic M, Hubscher U, Egly JM, Wood RD. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- Anderson CW, Carter TH. The DNA-Activated Protein Kinase-DNA-PK. In: Jessberger R, Lieber MR, editors. Molecular Analysis of DNA Rearrangements in the Immune System. Heidelberg: Springer-Verlag; 1996. pp. 91–112. [Google Scholar]
- Bergeron S, Anderson DK, Swanson PC. RAG and HMGB1 proteins: purification and biochemical analysis of recombination signal complexes. Methods Enzymol. 2006;408:511–528. doi: 10.1016/S0076-6879(06)08032-3. [DOI] [PubMed] [Google Scholar]
- Bertocci B, DeSmet A, Weill J-C, Reynaud CA. Non-overlapping functions of polX family DNA polymerases, pol m, pol l, and TdT, during immunoglobulin V(D)J recombination in vivo. Immunity. 2006;25:31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Bories JC, Cayuela JM, Loiseau P, Sigaux F. Expression of human recombination activating genes (RAG1 and RAG2) in neoplastic lymphoid cells: Correlation with cell differentiaion and antigen receptor expression. Blood. 1991;78:2053–2061. [PubMed] [Google Scholar]
- Buck D, Malivert L, deChasseval R, Barraud A, Fondaneche M-C, Xanal O, Plebani A, Stephan J-L, Hufnagel M, LeDiest F, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- Chan DW, Mody CH, Ting NS, Lees-Miller SP. Purification and characterization of the double-stranded DNA-activated protein kinase, DNA-PK, from human placenta. Biochem Cell Biol. 1996;74:67–73. doi: 10.1139/o96-007. [DOI] [PubMed] [Google Scholar]
- Chang LM, Rafter E, Rusquet-Valerius R, Roy NK, Cheung LC, Bollum FJ. Expression and processing of recombinant human terminal transferase in the baculovirus system. J Biol Chem. 1988;263:12509–12513. [PubMed] [Google Scholar]
- Chatterji M, Tsai C-L, Schatz DG. New concepts in the regulation of an ancient reaction: transposition by RAG1/RAG2. Immunological Rev. 2004:261–271. doi: 10.1111/j.0105-2896.2004.00167.x. [DOI] [PubMed] [Google Scholar]
- Dominguez O, Ruiz JF, Lain de Lera T, Garcia-Diaz M, Gonzalez MA, Kirchhoff T, Martinez AC, Bernad A, Blanco L. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. Embo J. 2000;19:1731–1742. doi: 10.1093/emboj/19.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney AJ. Predominance of VH-D-JH junctions occurring at sites of short sequence homology results in limited junctional diversity in neonatal antibodies. J Immunol. 1992;149:222–229. [PubMed] [Google Scholar]
- Gauss GH, Lieber MR. Mechanistic constraints on diversity in human V(D)J recombination. Mol Cell Biol. 1996;16:258–269. doi: 10.1128/mcb.16.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M. Molecular analysis of V(D)J recombination. Annual Review of Genetics. 1992;26:425–446. doi: 10.1146/annurev.ge.26.120192.002233. [DOI] [PubMed] [Google Scholar]
- Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- Gerstein RM, Lieber MR. Coding end sequence can markedly affect the initiation of V(D)J recombination. Genes and Development. 1993a;7:1459–1469. doi: 10.1101/gad.7.7b.1459. [DOI] [PubMed] [Google Scholar]
- Gerstein RM, Lieber MR. Extent to which homology can constrain coding exon junctional diversity in V(D)J recombination. Nature. 1993b;363:625–627. doi: 10.1038/363625a0. [DOI] [PubMed] [Google Scholar]
- Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science. 1993;261:1755–1759. doi: 10.1126/science.8356452. [DOI] [PubMed] [Google Scholar]
- Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- Gu J, Lu H, Tippin B, Shimazaki N, Goodman MF, Lieber MR. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. Embo J. 2007a;26:1010–1023. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Lu H, Tsai AG, Schwarz K, Lieber MR. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: influence of terminal DNA sequence. Nucleic Acids Res. 2007b;35:5755–5762. doi: 10.1093/nar/gkm579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Seidl K, Rathbun GA, Zhu C, Manis JP, vanderStoep N, Davidson L, Cheng HL, Sekiguchi J, Frank K, et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7:653–665. doi: 10.1016/s1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- Jaeger U, Bocskor S, Le T, Mitterbauer G, Bolz I, Chott A, Kneba A, Mannhalter C, Nadel B. Follicular lymphomas BCL-2/IgH junctions contain templated nucleotide insertions: novel insights into the mechanism of t(14;18) translocation. Blood. 2000;95:3520–3529. [PubMed] [Google Scholar]
- Jones JM, Gellert M. Intermediates in V(D)J recombination: a stable RAG1/2 complex sequesters cleaved RSS ends. Proc Natl Acad Sci. 2001;98:12926–12931. doi: 10.1073/pnas.221471198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klungland A, Lindahl T. Second pathway for completion of human DNA base excision repair: reconstitution with purified proteins and requirement for DNase IV (FEN-1) EMBO J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Okada A, Stewart V, Alt F. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–1175. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. Embo J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- Leu TM, Eastman QM, Schatz DG. Coding joint formation in a cell-free V(D)J recombination system. Immunity. 1997;7:303–314. doi: 10.1016/s1074-7613(00)80532-4. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of human nonhomologous DNA end joining. J Biol Chem. 2007 doi: 10.1074/jbc.R700039200. [DOI] [PubMed] [Google Scholar]
- Lieber MR, Hesse JE, Mizuuchi K, Gellert M. Developmental stage specificity of the lymphoid V(D)J recombination activity. Genes & Dev. 1987;1:751–751. doi: 10.1101/gad.1.8.751. [DOI] [PubMed] [Google Scholar]
- Lieber MR, Hesse JE, Mizuuchi K, Gellert M. Lymphoid V(D)J recombination: nucleotide insertion at signal joints as well as coding joints. Proc Natl Acad Sci. 1988;85:8588–8592. doi: 10.1073/pnas.85.22.8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Pannicke U, Schwarz K, Lieber MR. Length-dependent binding of human XLF to DNA and stimulation of XRCC4.DNA ligase IV activity. J Biol Chem. 2007a;282:11155–11162. doi: 10.1074/jbc.M609904200. [DOI] [PubMed] [Google Scholar]
- Lu H, Schwarz K, Lieber MR. Extent to which hairpin opening by the Artemis:DNA-PKcs complex can contribute to junctional diversity in V(D)J recombination. Nucleic Acids Res. 2007b doi: 10.1093/nar/gkm823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Lu H, Schwarz K, Lieber MR. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle. 2005a;4:1193–2000. doi: 10.4161/cc.4.9.1977. [DOI] [PubMed] [Google Scholar]
- Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh C-L, Schwarz K, Lieber MR. A Biochemically Defined System for Mammalian Nonhomologous DNA End Joining. Molecular Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis:DNA-PKcs complex in V(D)J recombination and in nonhomologous end joining. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- Ma Y, Schwarz K, Lieber MR. The Artemis:DNA-PKcs Endonuclease Can Cleave Gaps, Flaps, and Loops. DNA Repair. 2005b;4:845–851. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek K, Douglas P, Cui X, Ding Q, Lees-Miller SP. trans Autophosphorylation at DNA-dependent protein kinase's two major autophosphorylation site clusters facilitates end processing but not end joining. Mol Cell Biol. 2007;27:3881–3890. doi: 10.1128/MCB.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NickMcElhinny SA, Ramsden DA. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol Cell Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NickMcElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku Recruits the XRCC4-Ligase IV Complex to DNA Ends. Mol Cell Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan K, Shevelev IV, Maga G, Hubscher U. De novo DNA synthesis by human DNA polymerase lambda, DNA polymerase mu, and terminal deoxynucleotidyl transferase. J Mol Biol. 2004;339:395–404. doi: 10.1016/j.jmb.2004.03.056. [DOI] [PubMed] [Google Scholar]
- Ramsden D, Paull T, Gellert M. Cell-free V(D)J recombination. Nature. 1997;388:488–491. doi: 10.1038/41351. [DOI] [PubMed] [Google Scholar]
- Raval P, Kriatchko AN, Kumar S, Swanson PC. Evidence for Ku70/80 association with full-length RAG-1. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn049. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revy P, Buck D, le Deist F, de Villartay JP. The repair of DNA damages/modifications during the maturation of the immune system: lessons from human primary immunodeficiency disorders and animal models. Adv Immunol. 2005;87:237–295. doi: 10.1016/S0065-2776(05)87007-5. [DOI] [PubMed] [Google Scholar]
- Roth DB, Zhu C, Gellert M. Characterization of broken DNA molecules associated with V(D)J recombination. Proc Natl Acad Sci. 1993;90:10788–10792. doi: 10.1073/pnas.90.22.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schar P, Herrmann G, Daly G, Lindahl T. A newly identified DNA ligase of S. cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes & Dev. 1997;11:1912–1924. doi: 10.1101/gad.11.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz DG. V(D)J recombination. Immunol Rev. 2004;200:5–11. doi: 10.1111/j.0105-2896.2004.00173.x. [DOI] [PubMed] [Google Scholar]
- Schlissel M, Constantinescu A, Morrow T, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5'-phosphorylated, RAG-dependent, and cell cycle regulated. Genes & Dev. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- Schulte-Uentrop L, El-Awady RA, Schliecker L, Willers H, Dahm-Daphi J. Distinct roles of XRCC4 and Ku80 in non-homologous end-joining of endonuclease- and ionizing radiation-induced DNA double-strand breaks. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz K, Gauss GH, Ludwig L, Pannicke U, Li Z, Lindner D, Friedrich W, Seger RA, Hansen HT, Desiderio S, et al. RAG mutations in human B cell-negative SCID. Science. 1996;274:97–99. doi: 10.1126/science.274.5284.97. [DOI] [PubMed] [Google Scholar]
- Shimazaki N, Yoshida K, Kobayashi T, Toji S, Tamai K, Koiwai O. Over-expression of human DNA polymerase lambda in E. coli and characterization of the recombinant enzyme. Genes Cells. 2002;7:639–651. doi: 10.1046/j.1365-2443.2002.00547.x. [DOI] [PubMed] [Google Scholar]
- Smider V, Rathmell WK, Brown G, Lewis S, Chu G. Failure of Hairpin-Ended and Nicked DNA to Activate DNA-Dependent Protein Kinase: Implications for V(D)J Recombination. Mol Cell Biol. 1998;18:6853–6858. doi: 10.1128/mcb.18.11.6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubeyrand S, Torrance H, Giffin W, Gong W, Schild-Poulter C, Hache RJ. Activation and autoregulation of DNA-PK from structured single-stranded DNA and coding end hairpins. Proc Natl Acad Sci. 2001;98:9605–9610. doi: 10.1073/pnas.171211398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson PC. The bounty of RAGs: recombination signal complexes and reaction outcomes. Immunolog Rev. 2004;200:90–114. doi: 10.1111/j.0105-2896.2004.00159.x. [DOI] [PubMed] [Google Scholar]
- Teo SH, Jackson SP. Identification of S. cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J. 1997;16:4788–4795. doi: 10.1093/emboj/16.15.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkinson AE, Vijayakumar S, Pascal JM, Ellenberger T. DNA ligases: structure, reaction mechanism, and function. Chem Rev. 2006;106:687–699. doi: 10.1021/cr040498d. [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Kim SA, Chu G. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc Natl Acad Sci U S A. 2007;104:7851–7856. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CL, Drejer AH, Schatz DG. Evidence of critical architectural function for the RAG proteins in end processing, protection, and joining in V(D)J recombination. Genes Dev. 2002;16:1934–1949. doi: 10.1101/gad.984502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia M, Bentele M, Vaze MB, Herrmann G, Kraus E, Lee SE, Schar P, Haber JE. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature. 2001;414:666–669. doi: 10.1038/414666a. [DOI] [PubMed] [Google Scholar]
- Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol. 2007;9:978–981. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis-Garcia F, Besmer E, Sawchuk DJ, Yu W, Hu Y, Cassard S, Nussenzweig MC, Cortes P. V(D)J recombination: in vitro coding joint formation. Mol Cell Biol. 1997;17:6379–6385. doi: 10.1128/mcb.17.11.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RB, Yaneva M, Lieber MR. Productive and Nonproductive Complexes of Ku and DNA-PK at DNA Termini. Mol Cell Biol. 1998;18:5908–5920. doi: 10.1128/mcb.18.10.5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TE, Grawunder U, Lieber MR. Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- Yaneva M, Kowalewski T, Lieber MR. Interaction of DNA-dependent protein kinase with DNA and with Ku: biochemical and atomic-force microscopy. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Taghva A, Lieber MR. The cleavage efficiency of the human immunoglobulin heavy chain VH elements by the RAG complex. J Biol Chem. 2002;277:5040–5046. doi: 10.1074/jbc.M109772200. [DOI] [PubMed] [Google Scholar]
- Zhu C, Bogue MA, Lim D-S, Hasty P, Roth DB. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.