Abstract
Background
Segmental duplications flanking the neurofibromatosis type 1 (NF1) gene locus on 17q11 mediate most gene deletions in NF1 patients. However, the large size of the gene and the complexity of the locus architecture pose difficulties in deletion analysis. We report the construction and application of the first NF1 locus specific microarray, covering 2.24 Mb of 17q11, using a non‐redundant approach for array design. The average resolution of analysis for the array is ∼12 kb per measurement point with an increased average resolution of 6.4 kb for the NF1 gene.
Methods
We performed a comprehensive array‐CGH analysis of 161 NF1 derived samples and identified heterozygous deletions of various sizes in 39 cases. The typical deletion was identified in 26 cases, whereas 13 samples showed atypical deletion profiles.
Results
The size of the atypical deletions, contained within the segment covered by the array, ranged from 6 kb to 1.6 Mb and their breakpoints could be accurately determined. Moreover, 10 atypical deletions were observed to share a common breakpoint either on the proximal or distal end of the deletion. The deletions identified by array‐CGH were independently confirmed using multiplex ligation‐dependent probe amplification. Bioinformatic analysis of the entire locus identified 33 segmental duplications.
Conclusions
We show that at least one of these segmental duplications, which borders the proximal breakpoint located within the NF1 intron 1 in five atypical deletions, might represent a novel hot spot for deletions. Our array constitutes a novel and reliable tool offering significantly improved diagnostics for this common disorder.
Keywords: array‐CGH, malignant peripheral nerve sheath tumours, multiplex ligation‐dependent probe amplification, neurofibromatosis type 1, segmental duplications
Neurofibromatosis type 1 (NF1) is a common autosomal dominant disorder (MIM 162200) with an incidence of 1 in 3500 individuals. The clinical features of NF1 include café au lait spots, axillary/inguinal freckling, multiple dermal neurofibromas, and Lisch nodules as well as plexiform neurofibromas and malignant peripheral nerve sheath tumours (MPNSTs).1,2 The NF1 gene from 17q11.2 has been cloned and contains 57 translated exons distributed over approximately 350 kb. About 70% of the germline mutations in the NF1 gene result in a truncated protein due to nonsense or frameshift mutations.3,4 However, 5–10% of NF1 individuals are reported to have large 17q11 deletions, encompassing the entire NF1 gene as well as several neighbouring genes.5,6,7
The large size of the NF1 gene locus that needs to be studied for deletions is a methodological obstacle in routine diagnostics of the disease. Also, the complexity of the genomic architecture of 17q11 containing the NF1 gene6,8,9,10,11 poses difficulties in the analysis of deletions. Although, no clear cut genotype‐phenotype correlation has been established for NF1 patients,12 it is known that patients with large 17q11 deletions suffer from a more severe form of the disease that includes dysmorphism, mental retardation, early onset of a large number of neurofibromas, and an increased risk for the development of MPNSTs.13,14,15 The most common type of deletion spans ∼1.5 Mb of 17q11 and hence is termed the “typical deletion”. In approximately half of the typical deletion cases, the breakpoints were mapped to large paralogous sequences (also called low copy repeats (LCRs) or segmental duplications) flanking the NF1 gene.6,8,9,10,16 There have been relatively few studies reporting atypical deletions in NF1 patients8,9,16,17,18 and this is partially due to the lack of high throughput and high resolution methodologies of deletion detection within the entire locus affected by these mutations. We here report the construction and application of an NF1 locus specific microarray, covering 2.24 Mb of 17q11, based on a previously described PCR based non‐redundant approach for array design.19 This array allows high resolution array‐CGH profiling of deletions in NF1 samples. Moreover, we present a refined bioinformatic analysis of the entire locus for identification of novel segmental duplications that might play an important role in mediating NF1 gene deletions.
Methods
Patient material
In total, 161 samples were studied for deletions on the NF1 locus specific microarray, including validation cases 806 and 986 (tables 1 and 2). These included 148 lymphocyte derived DNA samples, 12 tumour (MPNST) derived DNA samples, and one cell line ASB4, which is a somatic cell hybrid from a patient mosaic for a deletion and containing only the deleted chromosome 17. DNA was isolated from these sources using standard methods. Patient samples were studied with the approval of the local Research Ethics Committee, Faculty of Medicine, Uppsala University.
Table 1 Summary of typical deletions detected using the NF1 locus microarray.
| Patient ID | Clinical phenotype | Previous deletion analysis | Array‐CGH results | Size | Ref | Source |
|---|---|---|---|---|---|---|
| 849 | CAL, F, NF, DF, DD | 1.5 Mb deletion, RFLP and PCR | Typical deletion (45p–155p) | 1.44–1.58 Mb | 2 | Cardiff, UK |
| 986† | CAL, F, DF, DD, LD | 1.5 Mb deletion, RFLP and PCR | Typical deletion (45p–155p) | 1.44–1.58 Mb | 32 | Cardiff, UK |
| 3M 3819 | CAL, F, LN, NF, AS | Total gene deletion, RFLP, PCR, | Typical deletion (45p–155p) | 1.44–1.58 Mb | Cardiff, UK | |
| and FISH | ||||||
| 2164 | CAL, F, LN | 1.5 Mb deletion, FISH and PCR | Typical deletion (45p–155p) | 1.44–1.58 Mb | Cardiff, UK | |
| 2M 1656 | CAL, F | 1.5 Mb deletion, FISH and PCR | Typical deletion (45p–155p) | 1.44–1.58 Mb | Cardiff, UK | |
| 637 | CAL, F, NF, DF, LD | Multi‐exonic deletion, RFLP | Typical deletion (45p–155p) | 1.44–1.58 Mb | 2 | Cardiff, UK |
| 954 | CAL, F, NF, DD, LD | 1.5 Mb deletion, RFLP and PCR | Typical deletion (45p–155p) | 1.44–1.58 Mb | 2 | Cardiff, UK |
| 1060 | CAL, F, NF, DF, DD, | 1.5 Mb deletion, RFLP and PCR | Typical deletion (45p–155p) | 1.44–1.58 Mb | 2 | Cardiff, UK |
| LD, hypotonia in | ||||||
| infancy, low IQ, | ||||||
| probable OCL | ||||||
| 3M 1369 | PNF | Total gene deletion | Typical deletion (45p–155p) | 1.44–1.58 Mb | Cardiff, UK | |
| 280988 | TMDP, CAL, F, mild | Deletion with FISH probes | Typical deletion (45p–155p) | 1.44–1.58 Mb | Leuven, Belgium | |
| MR, LAL, several CNFs | in microdeletion region* | |||||
| 222942 | TMDP, cryptorchidism, | Deletion with FISH probes | Typical deletion (45p–155p) | 1.44–1.58 Mb | Leuven, Belgium | |
| hypospadias, DD, | in microdeletion region* | |||||
| NFD, CNFs | ||||||
| 36425 | TMDP, LHF, MDW, | Deletion with FISH probes | Typical deletion (45p–155p) | 1.44–1.58 Mb | Leuven, Belgium | |
| hemifacial PNF, | in microdeletion region* | |||||
| CNFs, IQ: 71 | ||||||
| 264507 | TMDP, tall stature, | Deletion with FISH probes | Typical deletion (45p–155p) | 1.44–1.58 Mb | Leuven, Belgium | |
| LHF | in microdeletion region* | |||||
| F87 | CAL, F, LAL, DD, HJ, | Total gene deletion | Typical deletion (45p–155p) | 1.44–1.58 Mb | Birmingham, | |
| soft loose skin | AL, USA | |||||
| F110 | CAL, F, NFs, LN, | Total gene deletion | Typical deletion (45p–155p) | 1.44–1.58 Mb | Birmingham, | |
| puberty before age | AL, USA | |||||
| 10 years | ||||||
| F192 | CAL, F, | Total gene deletion | Typical deletion (45p–155p) | 1.44–1.58 Mb | Birmingham, | |
| no further data | AL, USA | |||||
| F249 | CAL, F, DD | Total gene deletion | Typical deletion (45p–155p) | 1.44–1.58 Mb | Birmingham, | |
| AL, USA | ||||||
| F256 | CAL, F, subCNFs, | Total gene deletion | Typical deletion (45p–155p) | 1.44–1.58 Mb | Birmingham, | |
| mild MR | AL, USA | |||||
| F320 | CAL, F, no | Total gene deletion | Typical deletion (45p–155p) | 1.44–1.58 Mb | Birmingham, | |
| other data | AL, USA | |||||
| F321 | CAL, F, no NF, | Total gene deletion | Typical deletion (45p–155p) | 1.44–1.58 Mb | Birmingham, | |
| SOG, DD, DF | AL, USA | |||||
| 2292 | CAL, dermal NFs, LN | NS | Typical deletion (45p–155p) | 1.44–1.58 Mb | Cardiff, UK | |
| 2337 | CAL, dermal NFs, LN, | NS | Typical deletion (45p–155p) | 1.44–1.58 Mb | Cardiff, UK | |
| PNF, spinal NF | ||||||
| T127 | CAL, F, NFs, MPNST | LOH identified by RFLP | Typical deletion (45p–155p) | 1.44–1.58 Mb | Cardiff, UK | |
| 1429 | CALs | NS | Typical deletion (45p–155p) | 1.44–1.58 Mb | Uppsala, Sweden | |
| 1962 | CALs, PNF on left leg, | NS | Typical deletion (45p–155p) | 1.44–1.58 Mb | Uppsala, Sweden | |
| mild PD | ||||||
| 3134 | Uncountable CALs, | NS | Typical deletion (45p–155p) | 1.44–1.58 Mb | Uppsala, Sweden | |
| mild PD |
*No PCR junction fragment for deletion detected following the methodology described in Lopez‐Correa et al6; †sample not blinded.
AS, aqueduct stenosis; CAL, café au lait spots; CNF, cutaneous neurofibroma; DD, developmental delay; DF, dysmorphic features; F, freckling; FISH, fluorescence in situ hybridisation; HJ, hypermobile joints; LAL, legs of asymmetric length; LD, learning disability; LHF, large hands and feet; LN, Lisch nodules; LOH, loss of heterozygosity; MDW, Madelung deformity wrist; MPNST, malignant peripheral nerve sheath tumours; MR, mental retardation; NF, neurofibromas; NFD, Noonan facial dysmorphism; NS, not studied previously; OCL, optic chiasm lesion; PD, pyschomotor delay; PNF, plexiform neurofibroma; RFLP, restriction fragment length polymorphism; SOG, symptomatic optic glioma; TMDP, typical microdeletion phenotype.
Table 2 Summary of atypical deletions detected using the NF1 locus microarray.
| Patient ID | Clinical phenotype | Previous deletion analysis | Array‐CGH results | Size | Ref | Source |
|---|---|---|---|---|---|---|
| 806† | CAL, F, DF, DD | 7 cM deletion | All data points | >2.2 Mb | 22 | Cardiff, UK |
| on the array | ||||||
| T165 | CAL, F, NFs, MPNST‐pelvic | LOH identified by RFLP | All data points | >2.2 Mb | Cardiff, UK | |
| metastases | on the array | |||||
| 282775 | No NFs, uncountable CALs, NFD, | FISH: deletion of NF1 gene | 80p–end of array | >1.33 Mb | Leuven, | |
| PD, language disorder | probes‡ | Belgium | ||||
| T145 | CAL, F, NFs, MPNST, PNF, | LOH identified by RFLP | 21p–155p | 1.61–1.75 Mb | Cardiff, UK | |
| epilepsy | ||||||
| ASB4* | Mild phenotype, 1 CAL, NF, LN, | One breakpoint in intron 8 of | 45p–144p | 1.07 Mb | Leuven, | |
| mosaicism for deletion in blood | JJAZ1 pseudogene, other | Belgium | ||||
| breakpoint probably around | ||||||
| position 148000 of | ||||||
| AC007923 | ||||||
| NF1_619 | NF, facial DF, severe MR, | Deleted (size unknown); | Two deletions | 875 kb and | 16 | Hamburg, |
| cerebral astrocytoma | microsatellite markers | (45p–121p | >920 kb | Germany | ||
| and FISH | and 138–end) | |||||
| F538 | CAL, F, subCNFs | Intron 1 to intron 56; | 80p–113p (intron | 223 kb | Birmingham, | |
| long range RT‐PCR and MLPA | 1 to intron 56) | AL, USA | ||||
| 119688 | Classical NF1 but not TMDP, | Intron 1 to intron 56; | 80p–113p (intron | 223 kb | 6 | Leuven, |
| UDE, no CNFs at age 18, IQ: 77 | breakpoints are known | 1 to intron 56) | Belgium | |||
| 193 | CAL, F, LN, NF, scoliosis, | 90 kb deletion | 88p–96p (intron | 93 kb | 31 | Cardiff, UK |
| above average intelligence, | 8 to intron 35) | |||||
| NFs in the gum | ||||||
| 2338 | Clinical data not available | NS | 80p–89p (intron | 66.4 kb | Cardiff, UK | |
| 1 to intron 11) | ||||||
| 162 | CAL, F, LN, NF | 50 kb deletion | 80p–83p (intron | 25.8 kb | Cardiff, UK | |
| 1 to intron 4) | ||||||
| F548 | 4 year old with CAL, F, mild | Intron 5 to intron 8; | 88p (intron 7 | 7.1 kb | Birmingham, | |
| scoliosis, coarse facies, SOG | long range RT‐PCR and MLPA | to intron 8) | AL, USA | |||
| F486 | 6 year old with CAL, F, | Negative by long range RT‐PCR, | 74p (5′ UTR | 6 kb | Birmingham, | |
| subCNFs, PNF | exon 1 deletion by MLPA | of exon 1) | AL, USA |
*ASB4 is a somatic cell hybrid containing monosomy 17; †sample not blinded; ‡deletion with PACs 962N3 and 984G23, but not with 409L16. Centromeric breakpoint is in 100 kb fragment proximal to the NF1 gene and telomeric breakpoint is further than 984G23. 984G23 is located distal to the telomeric NF1REP and is 800 kb telomeric from the NF1 gene.
CAL, café au lait spots; CNF, cutaneous neurofibroma; DD, developmental delay; DF, dysmorphic features; F, freckling; FISH, fluorescence in situ hybridisation; LOH, loss of heterozygosity; LN, Lisch nodules; MLPA, multiplex ligation‐dependent probe amplification; MPNST, malignant peripheral nerve sheath tumours; MR, mental retardation; NF, neurofibromas; NFD, Noonan facial dysmorphism; NS, not studied previously; PD, pyschomotor delay; PNF, plexiform neurofibroma; RFLP, restriction fragment length polymorphism; RT‐PCR, real time PCR; SOG, symptomatic optic glioma; TMDP, typical microdeletion phenotype; UDE, unilateral deafness and epilepsy; UTR, untranslated region.
Bioinformatic analysis
The 2.24 Mb genomic sequence, encompassing the NF1 gene, was derived from the chromosome 17 contig NT_010799.14 (human genome build 35.1) from 25587134 to 27827134 bp. As we used the sequence available from GenBank and Ensembl, we also used the same annotations for all exons of the NF1 gene numbering them consecutively 1 to 57. The GenBank accession number for sequence retrieval of NF1 gene exons was NM_000267. A comparison of the NF1 exons nomenclature according to NCBI and the Neurofibromatosis (NF) Consortium is given in supplementary table 1 available at http://www.jmedgenet.com/supplemental. The Sequence Allocator program (abbreviated as Allocator) combines and automates three bioinformatic protocols: (i) RepeatMasker (http://www.repeatmasker.org) for identifying common repeats; (ii) BlastN at the NCBI server (http://www.ncbi.nlm. nih.gov/blast) to detect other similar sequences, with a threshold of greater than 80% identity over 50 bp, in the human genome; and (iii) Primer3 (http://frodo.wi.mit.edu/cgi‐bin/primer3/primer3_www.cgi) software for designing primer pairs in the unique genomic sequences.
Design of NF1 diagnostic array
The array was constructed across a 2.24 Mb interval of 17q11.2 using the previously reported PCR based non‐redundant approach for construction of microarrays19 which excludes all common repeats and other redundant sequences from the array. Genomic sequence of the NF1 locus with flanking regions on chromosome 17, from 25590 to 27890 kb (according to NCBI build 35.1), was processed by Allocator. In total, oligonucleotide primers for amplification of 548 PCR products ranging in size from 200 to 998 bp (average 637 bp) were selected. Additionally, 16 control genomic loci from chromosome X and 23 autosomal control loci derived from non‐chromosome 17 autosomes were treated by Allocator analogously and incorporated into the array set up.
Preparation and pooling of PCR products
Initially, PCR amplification of all fragments was done by conventional PCR using the fragment specific upper and lower primer pairs. All upper primers included a 5′ universal priming site (5′‐TGACCATG‐3′). This site allowed a second round PCR amplification using a universal primer with an amine anchor attached to the 5′ end. The amine anchor is necessary for binding of the PCR fragments to the array slide surface. Each PCR amplification reaction was done in a 100 µl volume. The size of amplified fragments was checked by agarose gel electrophoresis. Post reaction mixtures were purified using PCR Cleanup Filter Plates, Montage PCR96 (Millipore, Billerica, MA, USA). Products were eluted in 50 µl deionised water. Individual PCR products were combined to form pools of 1.5 kb size on average (see supplementary table 2 at http://www.jmedgenet.com/supplemental) and suspended in spotting buffer (0.25 M phosphate buffer, pH 8.5; 0.00025% sarkosyl).
Array printing, hybridisation, scanning, and image analysis
The methods used for blocking CodeLink slides, DNA labelling, hybridisation, and post‐hybridisation processing are detailed in the supplementary information (available at http://www.jmedgenet.com/supplemental). Briefly, each PCR pool was spotted on the CodeLink aldehyde slides (GE HealthCare, Slough, UK) in triplicate as a single measurement point. The slides were blocked by sodium borohydride treatment and prehybridised (5× SSC, 0.1% SDS, 0.4% BSA; 1 h, 45°C). Sonicated test and reference DNA was labelled by random priming with Cy‐3 dCTP (cat. no. PA53021, GE HealthCare) and Cy‐5 dCTP (cat. no. PA55021, GE HealthCare), respectively, using the BioPrime Array CGH Genomic Labeling System with purification module (cat. no. 18095‐011; Invitrogen, Carlsbad, CA). Labelled DNA was suspended in the hybridisation buffer (4× SSC, 0.1% SDS, 30% formamide) and hybridised to the array (36 h, 45°C). Post‐hybridisation processing consisted of four washing steps: (i) 2× SSC, 0.1% SDS, 25% formamide; 20 min, 45°C; (ii) 1× PBS; 10 min, RT; (iii) 0.2× SSC; 15 s, RT; (iv) deionised water; 15 s, RT. The slides were dried with compressed air. Image acquisition was performed using the GenePix 4000B scanner (Axon Instruments, Union City, CA). Analysis of hybridisation intensity was carried out using the GenePixPro image analysis software (Axon Instruments). The ratio of the intensities between test DNA and reference DNA was calculated using “the ratio of means” formula. The GenePixPro software subtracts the local background from the signal intensities for each spot. The average and standard deviation of the three replicas for each PCR pool were also calculated. Clones displaying a standard deviation greater than 5% of the average between a minimum of two replica spots were discarded from further analysis (unsuccessfully scored loci). The average ratio from the autosomal controls was used in the normalisation of all average ratio values from all clones on the array in each hybridisation experiment. The average normalised inter‐locus fluorescence ratio (ANILFR) values were calculated in order to assess the inter‐locus variation, representing a region(s) on the array. The normalised ratio for successfully scored loci from a certain, continuous region on the array was used to calculate the ANILFR value and standard deviation.
Multiplex ligation‐dependent probe amplification analysis
Confirmation of array data was performed using the newly developed multiplex ligation‐dependent probe amplification (MLPA) technique20 with the SALSA P081/082 NF1 MLPA kit (MRC Holland, Amsterdam, The Netherlands). Approximately 50 ng of genomic DNA extracted from peripheral blood lymphocytes was used for each probe mix. Hybridisation, ligation, and amplification were performed in thermocyclers with a heated lid (PCR Sprint, Thermo Electron, Milford, MA and GeneAmp PCR system 2700, Applied Biosystems, Foster City, CA) as previously described.20 A 1 μl sample of the amplification product was analysed using an ABI 3100 Genetic Analyzer (Applied Biosystems) using ROX‐500 (ABI, Warrington, UK) as an internal size standard. Analysis was performed with GeneMapper software 3.7.
Results
The aim was to construct a high resolution genomic microarray spanning the NF1 gene, including large genomic margins on both sides of this gene, and to apply this array in analysis of a representative cohort of NF1 derived samples. The underlying strategy for array design was the PCR based and entirely non‐redundant approach previously reported for the NF2 gene,19 including the application of improved surface chemistry (CodeLink, GE HealthCare) for spotting of the arrays.21
Construction of the NF1 locus microarray
The construction and design of the NF1 locus microarray demanded extensive bioinformatic analysis of the 17q11 locus, especially in view of previous reports indicating a complex genomic architecture as regards abundant segmental duplications (abbreviated as “segmentons”).6,8,9 The aims of bioinformatic analysis were twofold. First, we hoped to identify the unique sequences in the 2.24 Mb sequence, thus allowing construction of a non‐redundant genomic array. Second, we intended to perform a refined analysis of the locus in order to discover potential new segmentons. Both tasks were facilitated by use of our recently developed program, called Allocator, which allows automation of these bioinformatic procedures (de Bustos et al, manuscript in preparation; a program demo is available at http://puffer.genpat.uu.se/). Allocator identifies unique regions in the input sequence by separating non‐redundant and redundant sequences, the latter being common repeats, segmentons, and other similar sequences (for example, close paralogs and pseudogenes). The program automatically designs primer pairs for PCR amplification in the identified unique regions. The redundant regions filtered out by Allocator were further analysed for the presence of segmentons, as described below.
Following analysis of the 2.24 Mb sequence, 941 unique sequence segments were identified by Allocator, including 232 non‐redundant stretches spanning the NF1 gene. The graphical output of Allocator, indicating the sizes of unique sequences, is shown in fig 1. Of the 941 unique segments, we selected 548 fragments for PCR amplification. The overall frequency of successful amplification was ∼90% using total human DNA as template. After quality control on agarose gels, 444 genomic fragments, ranging in size from 200 to 998 bp (average 637 bp), were chosen for further construction of the array. The details of the primer pairs are provided in supplementary table 2 at http://www.jmedgenet.com/supplemental.
Figure 1 Graphical output from bioinformatic analysis of the 2.24 Mb sequence from chromosome 17, including the NF1 gene, using Allocator. The approximate positions of the NF1 gene and previously reported segmentons (also called LCRs), such as LCR‐proximal (LCR‐P), JJAZ1 pseudogene (JJAZ1‐P), and LCR‐distal (LCR‐D), are shown as black horizontal bars.16 The shaded regions in the graph represent the large genomic segments within the studied locus, for which Allocator did not identify any unique sequences longer than 200 bp using the chosen criteria (see Methods). The x axis denotes the analysed sequence while the y axis describes the length of non‐redundant sequence (200–1000 bp) identified by the program. Each point in the graph represents the length of unique sequence that could be used as a template for PCR amplification during construction of the NF1 locus specific microarray.
We aimed at maximum resolution and robust performance of array‐CGH analysis in covering this locus, especially with regard to the strength of fluorescent signals from each feature on the array. Hence, for the majority of the loci we pooled two to six neighbouring PCR products and spotted them as single measurement points on the array. However, nine PCR products were not pooled and were printed individually. In total, we spotted 222 PCR pools (see supplementary table 2 available at http://www.jmedgenet.com/supplemental) on the NF1 locus microarray, including 183 measurement points from 17q11 (ID 1p to 183p), 16 pools from chromosome X (ID 184p to 199p), and 23 PCR pools from non‐chromosome 17 autosomal control loci (ID 200p to 222p) (fig 2). The amount of unique sequence in each pool varied from 0.49 to 2.41 kb (average 1.5 kb). The genomic coverage of these pools varied from 0.49 to 245.87 kb. Hence, the average resolution of analysis for the NF1 locus microarray is ∼12 kb per measurement point. The NF1 gene was covered using 44 pools (ID 75p to 118p; fig 2B) conferring an average resolution of 6.4 kb per measurement point.
Figure 2 Map of the studied 2.24 Mb genomic segment from 17q11 and summary of all identified deletions. All elements of the figure are drawn to scale. (A) The figure displays, from top to bottom: scale of locus, known genes, PCR pools used in the construction of the array, location of segmentons, and the extent of typical as well as atypical deletions observed in NF1 derived samples. In total, 183 array measurement points were included and are shown as black rectangles in two alternating tiers. Two PCR pools (45p and 155p), which are located in the vicinity of breakpoints for the typical deletion in NF1 patients, are labelled. The light grey bars indicate the size and position of segmentons identified through bioinformatic analysis using the Allocator program using 3 kb threshold analysis for the size of segments. The dark grey bars indicate the extent of 26 typical and 13 atypical deletions that were observed by array‐CGH profiling of DNA derived from 161 NF1 derived samples (tables 1 and 2). The 1.44–1.58 Mb typical deletion (25890 to 27346 kb) spans the PCR pools 45p to 155p on the array. Owing to the redundancy in the 124 kb sequence between pools 155p and 156p, we could not cover this region with any array measurement points. Hence, the distal breakpoints, in the typical deletion and the atypical deletion observed in case T145, cannot be exactly defined. This is displayed as a discontinuous bar at the end of the respective deletions. (B) Detailed view of the 350 kb locus (positions 26390 to 26740 kb) encompassing the NF1 gene. Three genes (OMG, EVI2B, and EVI2A) located within the NF1 gene intron 35 and transcribed on the reverse strand are shown. Fifty seven exons of the NF1 gene, spanning 280.5 kb, are indicated in light grey. The black rectangles indicate the PCR pools covering the NF1 gene. In total, 48 pools (from 72p to 118p) are shown in the figure, of which 44 pools (75p to 118p) cover the NF1 gene, including the promoter and the 5′ as well as 3′ untranslated regions (UTRs). The segmentons within the locus, which were identified using a 3 kb threshold setting, are shown as medium grey bars. The dark grey bars indicate the atypical deletion profiles, in which one of the breakpoints was located in the NF1 gene. In total, seven deletion profiles are displayed, including the 7.1 and 6 kb microdeletions in patients F548 and F486.
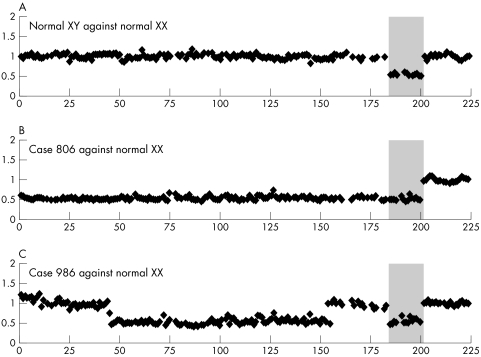
Validation of the array
To validate the performance of the NF1 locus microarray, we carried out three array‐CGH experiments (fig 3). The hybridisation profile of DNA from a normal male (XY) versus reference DNA from normal female (XX) is shown in fig 3A. As expected, all the pools covering autosomal loci, including 17q11, showed fluorescence ratios for two DNA copies. The PCR pools covering chromosome X loci displayed normalised fluorescence ratios for one DNA copy. The following two experiments were performed using DNA derived from male NF1 patients in which the 17q11 deletions were previously characterised. Sample 806 is known to contain a 7 cM deletion on 17q11 encompassing the NF1 gene.22 Upon hybridisation of this sample to our array, we observed that all the data points spanning chromosome 17 displayed normalised fluorescence ratios consistent with a heterozygous deletion, that is, at the level of chromosome X controls (fig 3B). DNA from the male NF1 patient 986 was previously characterised to contain the typical ∼1.5 Mb deletion.32 As seen in fig 3C, the array‐CGH profile of this sample displays a heterozygous deletion between ID 45p and 155p (1.44–1.58 Mb). The breakpoints of the deletion were observed to be delimited by previously known segmentons and these results correspond well with previous studies.6,8,9,10,16 From the above three experiments, we considered the sensitivity and specificity of the NF1 locus microarray for detection of single copy number deletions in the 17q11 locus to be positively established.
Figure 3 Validation of the NF1 locus microarray in three experiments. The 222 array measurement points are shown on the x axis and the y axis represents the normalised, average ratio between fluorescent signals for each locus, which are derived from three independent replicate spots on the array. DNA from 10 normal female controls was pooled and used as reference DNA in all array‐CGH experiments. (A) Array‐CGH profile from hybridisation of DNA from a normal male (XY) versus normal female (XX). The autosomal controls displayed an ANILFR±standard deviation value of 0.99±0.05 and the chromosome X pools provided the sensitivity to detect levels of one gene copy (0.54±0.03). (B) Graph displaying the hybridisation of DNA derived from a previously characterised male patient (sample 806) and normal XX DNA to NF1 locus microarray. As seen in the figure, the ratios of fluorescence for all 17q derived measurement points (0.53±0.04) are at the level of chromosome X pools (0.52±0.04) confirming the presence of a heterozygous deletion in this patient across the entire array. The non‐chromosome 17 autosomal control pools display ANILFR for two gene copies (1.00±0.05). (C) Array‐CGH profile of hybridisation of DNA from previously characterised case 986 and normal female DNA. We identified the 1.44–1.58 Mb deletion from 45p to 155p (0.53±0.07). The ANILFR values for chromosome X and autosomal controls were as expected (0.55±0.06 and 1.00±0.04, respectively).
Application of the array to profile NF1 related samples
In order to perform a comprehensive analysis of the deletion spectrum in NF1 patients, we totally profiled 159 additional patient derived DNA samples. There was a bias in the selection of these samples. A large proportion of this cohort (96 samples, 60%) has previously been studied for point mutations in the NF1 gene, and in 44 of these samples no point mutations have been detected (not shown). The majority of the samples (100/161) have not previously been studied for their deletion status in 17q11, while the remaining 61 samples have been analysed for DNA copy number changes by various methods and deletions larger than 5 kb were found in 30 cases (not shown). However, as we aimed for an unbiased approach towards scoring deletions by array‐CGH, the details of previously characterised deletions were withheld from the group performing array‐CGH. Furthermore, in several instances the DNA series from our collaborators included patient samples which were already known not to contain any deletions, but these details were also not disclosed to the array‐CGH investigators. In this cohort of samples, a proportion of which was previously characterised for deletions using alternative and low resolution approaches, we found heterozygous deletions of various sizes in 39 out of 161 studied cases (table 3). The observed deletions can be categorised into two groups. The first category comprises 26 cases that showed the typical 1.44–1.58 Mb deletion identified between pools 45p and 155p (fig 2A and table 1). This typical deletion is known to be mediated by segmentons known as NF1 LCR‐P and NF1 LCR‐D16 (fig 1). It is important to mention that PCR pool 45p spans the large 238 kb segmenton in the NF1 locus and is always scored as deleted for the typical deletions (fig 2A).
Table 3 Summary of previous mutation analysis and our array‐CGH results.
| Previous mutation analysis | No. | Deletions detected by array‐CGH |
|---|---|---|
| Previous point mutation or | 117* | |
| deletion analysis | ||
| Mutations detected | 67* | 33 |
| No mutations detected | 50 | 3 |
| Mutation analysis not performed | 44 | 3 |
*12 of these are MPNSTs.
The second category of deletions consists of 13 samples that showed atypical deletions, that is, heterozygous loss other than the typical 1.44–1.58 Mb (fig 2A and table 2). In two cases, namely DNA from NF1 patient 806 and tumour (MPNST) derived sample T165, the size of the deletion was greater than the span of the microarray. Thus neither the centromeric nor the telomeric breakpoints of these aberrations could be determined. In case 282775, the proximal breakpoint of the deletion was delimited to pool 80p. The distal breakpoint, however, was beyond the coverage of the array. Therefore, in this case the deletion is at least 1.33 Mb. Case 619 is intriguing as it displays two discontinuous deletions. The first 875 kb deletion is entirely embraced by the array and encompasses the NF1 gene, while the second one is at least 920 kb in size and extends beyond the coverage of the array towards the telomere of 17q. The latter deletion does not involve the NF1 gene as its centromeric breakpoint (138p) is outside the NF1 gene. In the remaining nine cases, the size of the atypical deletions ranged from 1.6 Mb to 6 kb and in each case both deletion breakpoints could be accurately determined (table 2, figs 2 and 4). An important point to mention is that we detected the loss of one copy of genomic segments in 17q11 in three samples in which no NF1 point mutations were previously detected. In two cases (2292 and 2337) we observed the typical 1.44–1.58 Mb deletion, whereas in sample 2338, a 66.4 kb atypical deletion, spanning intron 1 to intron 11 of the NF1 gene, was observed.
Figure 4 Detection of atypical deletions in NF1 patients using the NF1 locus specific microarray and confirmatory MLPA analysis. The array‐CGH profiles of DNA derived from four NF1 samples versus normal female reference DNA are shown in panels A, C, E, and G. The general layout of array‐CGH profiles follows the structure described in fig 3. Rectangles in the panels indicate the measurement points displaying fluorescence ratios consistent with a heterozygous deletion. Panels B, D, F, and H show the corresponding MLPA analysis for each patient. The x axis displays the 81 probes (ID 1 to 81) from the MLPA SALSA kit. The normalised peak area (NPA) value of each probe (y axis) indicates the relative copy number of the corresponding exon when compared to autosomal control loci. (A) An array‐CGH profile of the 223 kb heterozygous deletion (ID 80p to 113p, intron 1 to intron 56) in female NF1 patient sample 119688. The normalised fluorescence ratios for the deleted measurement points are 0.55±0.05 and for autosomal controls are 1.00±0.09. An identical array‐CGH profile was detected in another female NF1 sample (case F538, profile not shown, see table 2). (B) MLPA profile from analysis of case 119688. The heterozygous deletion of the NF1 gene exons 2 to 56 is detected by 51 probes (ID 4 to 54) with an NPA value of 0.53±0.05. (C) Detection of a 93 kb heterozygous deletion in female patient 193, who has been previously characterised as having a 90 kb deletion within the NF1 gene.23 Array‐CGH analysis of DNA from this patient displayed a heterozygous deletion (ID 88p to 96p; ANILFR 0.53±0.04) spanning 93 kb of the NF1 gene from intron 8 to intron 35. (D) Profile from MLPA analysis of case 193. The graph displays one copy loss of 25 probes (ID 9 to 33; NPA 0.52±0.04) which encompass the NF1 exons 8 to 35. (E) Detection of a small heterozygous deletion within the NF1 gene in case F548. The loss of one DNA copy is detected between intron 7 and intron 8 by a single PCR pool (88p) with a normalised fluorescence value (0.53); the genomic coverage of this pool is 7.1 kb. (F) Confirmatory MLPA analysis of case F548. We detected a heterozygous deletion spanning NF1 exons 6 to 8 (ID 8 to 9; NPA 0.56±0.01). (G) Array‐CGH profile of NF1 case F486. We detected heterozygous deletion of a single measurement point (74p; normalised ratio of fluorescence 0.60) spanning the 5′ UTR of the first exon of the NF1 gene. Empty space following pool 74p and indicating data points not scored is a result of a printing failure of the particular array used in the analysis of this patient. (H) Confirmatory MLPA analysis of case F486. Two probes (ID 2 and 3) within exon 1 of the NF1 gene displayed NPA values (0.52±0.05) consistent with one copy loss.
It is also interesting to mention the results obtained from one patient among those with the typical 1.44–1.58 Mb deletions (case F110). This case was, prior to the start of this study, known to be a mosaic case for the total gene deletion. This patient was studied by FISH analysis and approximately 90% of the scored metaphases displayed a single FISH signal (Messiaen et al, unpublished) (data not shown). This information was, however, not known to the array‐CGH team prior to their analysis of sample F110. This case was scored as a typical deletion and the levels of fluorescence ratios for deleted data points on the array‐CGH profile (not shown) were not particularly different from other samples with the typical deletion. This illustrates that array‐CGH is suitable for detection of aberrations using mosaic samples and confirms previous conclusions from tumour related array‐CGH analyses. Indeed, the ability to accurately assess DNA copy number alterations in a mixed cell population is one of the major assets of array‐CGH. A number of recent array‐CGH papers have also reported the detection of DNA copy number imbalances on a heterogeneous cell background in the context of mouse tumour cell lines24 and also in human primary tumours such as schwannoma, ovarian cancer, and meningioma.25,26,27 The current series of samples from NF1 patients also included those from 12 MPNSTs, along with matching constitutional DNA available for nine of these. We detected deletions in three MPNSTs (one typical and two atypical, fig 2 and tables 1 and 2) and none of these were present in their respective constitutional DNA samples (not shown). We therefore assume that these deletions represent the second hit mutation, the first most likely being a point mutation in the NF1 gene.
Confirmation of array‐CGH findings using MLPA
We applied MLPA analysis as an independent methodology to confirm the NF1 gene deletions detected by array‐CGH, as it provides exon‐level resolution in profiling the gene of interest for DNA copy number changes.20 We used the commercially available SALSA P081/082 NF1 MLPA kit (MRC Holland) for deletion detection across the NF1 gene. In total, 81 probes are included in the kit, of which 56 (ID 1 to 56) span the NF1 exons (except exons 7 and 9) and 25 (ID 57 to 81) cover autosomal control loci. The normalised peak area (NPA) for each probe is indicative of the relative copy number of the corresponding exon when compared to the control loci. In total, DNA from 29 NF1 samples, including six typical deletion cases, all 13 atypical deletion cases, and 10 non‐deleted cases were profiled by MLPA. For the typical deletion cases, in which the entire NF1 gene was deleted, a heterozygous deletion of exons 1–57 was detected (data not shown). The samples which were negative for deletions by array‐CGH, were also negative for copy number alterations within the NF1 gene by MLPA (data not shown). Upon analysis by MLPA of the 13 atypical deletion cases, we observed that there was concordance in all but one (case F548) with the data derived from array‐CGH. The data from patient F548 are in partial disagreement, owing to the array design for the NF1 gene exons deleted in this case (see Discussion below). Four representative profiles from these experiments are shown in fig 4 (panels B, D, F, and H). Figure 4B displays the detection of a 223 kb deletion encompassing NF1 exons 2 to 56 in sample 119688 by MLPA. For comparison, the array‐CGH data for the same sample (fig 4A) indicated a heterozygous deletion from intron 1 to intron 56. Similarly, the two 7.1 and 6 kb microdeletions detected by NF1 locus array in samples F548 and F486, respectively, were also confirmed by MLPA analysis (fig 4F,H). In summary, in all the analysed cases, the data obtained from MLPA independently verified all the analysed deletions detected by array‐CGH.
Identification of novel segmentons within 17q11
From the above analysis of DNA dosage alterations we identified 13 atypical deletions. We also observed that 10 of these 13 deletions shared a common breakpoint either on the proximal or distal end of the deletion. In five samples (282775, F538, 119688, 2338, and 162) we identified a common proximal breakpoint within intron 1 of the NF1 gene delimited by the data point 80p. This pool contained 1.7 kb of sequence in four PCR fragments and it spaned 17.9 kb of genomic sequence (fig 2B). In two additional samples (cases 193 and F548) displaying intragenic microdeletions, we observed a common breakpoint on the proximal end of the deletions. This was encompassed in array data point 88p, which contained 1.2 kb of sequence in two PCR fragments and spanned 7.1 kb of the locus (fig 2B). We also observed that some of the atypical deletions apparently had the same centromeric or telomeric breakpoints, when compared with the breakpoints for the typical 1.44–1.58 Mb deletion. In two samples (619 and ASB4) the centromeric breakpoint was defined in pool 45p and in case T145 (MPNST derived DNA) the telomeric breakpoint was located in pool 155p. As described before, the proximal and distal breakpoints of the typical 1.44–1.58 Mb deletion are located in the vicinity of pools 45p and 155p, respectively. These observations have prompted us to perform a refined bioinformatic analysis across the 2.24 Mb with the aim of localising novel segmentons in the vicinity of breakpoints for atypical deletions. Identification of all types of redundant sequences is a by‐product of the Allocator program. We therefore utilised this part of the output and further processed it to identify new segmentons.
Contiguous pieces of redundant sequence with a minimum size of 3 kb, identified by Allocator, were regarded as potentially containing segmental duplications. For a more detailed analysis, these regions were blasted against the reference sequence of the human genome (http://www.ncbi.nlm.nih.gov/genome/seq/HsBlast.html), using default parameters. Sequences showing at least 90% similarity over 1000 bp to another location in the human genome were considered to be segmental duplications. The 33 segmental duplications resulting from this analysis are shown in fig 2 and in table 4. Special attention was paid to the segmentons that were present in pools associated with the recurrent breakpoints for atypical deletions (80p and 88p) (fig 2B and table 2). Pool 80p, which was the common proximal breakpoint in five atypical deletions, displays high similarity to 4.7 kb on chromosome 13 (18569.3 to 18574 kb). Pool 88p was observed to be the common breakpoint in two atypical deletion cases. Blast analysis revealed that the genomic sequence of this pool displayed high sequence similarity to regions on chromosomes 18 and 21 of similar size, which means that there are three copies of this segmenton within the haploid genome. In summary, we identified two novel segmentons bordering proximal breakpoints in atypical deletions; these should be studied further for their role in mediating deletions in NF1 patients.
Table 4 Segmental duplications identified in the analysed 17q11 segment using a 3 kb threshold setting for minimum redundant sequence length.
| Segmenton number | Start on chr. 17 sequence | End on chr. 17 sequence | Size of the segmenton |
|---|---|---|---|
| 1 | 25840801 | 25841920 | 1119 |
| 2 | 25846358 | 25850382 | 4024 |
| 3 | 25852953 | 25854201 | 1248 |
| 4 | 25904240 | 25936929 | 32689 |
| 5 | 25940406 | 25941527 | 1121 |
| 6 | 25952455 | 26023997 | 71542 |
| 7 | 26027961 | 26036380 | 8419 |
| 8 | 26038649 | 26050367 | 11718 |
| 9 | 26053161 | 26061266 | 8105 |
| 10 | 26063421 | 26130956 | 67535 |
| 11 | 26329732 | 26332515 | 2783 |
| 12 | 26360373 | 26384016 | 23643 |
| 13 | 26387064 | 26402110 | 15046 |
| 14 | 26412845 | 26413874 | 1029 |
| 15 | 26422217 | 26423581 | 1364 |
| 16 | 26496020 | 26497571 | 1551 |
| 17 | 26541266 | 26542658 | 1392 |
| 18 | 26546257 | 26547392 | 1135 |
| 19 | 26550729 | 26552976 | 2247 |
| 20 | 26572600 | 26587209 | 14609 |
| 21 | 26608916 | 26611300 | 2384 |
| 22 | 26615910 | 26619020 | 3110 |
| 23 | 26696521 | 26699863 | 3342 |
| 24 | 27288579 | 27313290 | 24711 |
| 25 | 27315594 | 27337683 | 22089 |
| 26 | 27352384 | 27356101 | 3717 |
| 27 | 27360773 | 27362123 | 1350 |
| 28 | 27364177 | 27386904 | 22727 |
| 29 | 27389104 | 27452780 | 63676 |
| 30 | 27457013 | 27459848 | 2835 |
| 31 | 27466425 | 27470656 | 4231 |
| 32 | 27479299 | 27482802 | 3503 |
| 33 | 27530551 | 27531651 | 1100 |
Discussion
We report a new tool for accurate, high throughput, and cost effective analysis of disease causing deletions for a very common genetic disorder. The average resolution of analysis for the whole 2.24 Mb locus is ∼12 kb per measurement point. However, the average resolution for the NF1 gene is twice as high (∼6 kb per array data point), that is, 57 NF1 gene exons are resolved by 44 measurement points, which is close to exon level resolution. Indeed, in one case we detected a deletion involving only a single exon (case F486, exon 1, fig 2). We also show that the same array allows, in a single experiment, detection and simultaneous sizing of deletions from 6 kb up to the megabase range, including deletions as large as the entire extent of the array. Furthermore, we analysed a large series of patient samples (161 in number) and observed 39 deletions affecting the NF1 tumour suppressor gene, of which 26 were the typical 1.44–1.58 Mb deletions and 13 were atypical deletions. If three cases of somatic deletions in MPNST samples are excluded from the series of deletion cases, then we detected 25 typical and 11 atypical deletions occurring in the constitutional DNA of NF1 patients. These results are in agreement with previous studies6,8,9 reporting a high frequency of the typical deletions, which are mediated by the large segmentons flanking the NF1 gene on both sides. Moreover, our study clearly indicates that there exists considerable heterogeneity of deletion breakpoints among patients with NF1 gene microdeletions. One disadvantage of the current version of the NF1 locus microarray is that with the chosen settings for the Allocator program and the criteria for the construction of the array, it is not possible to differentiate between the 1.44–1.58 Mb and 1.2 Mb deletions mediated by NF1 LCRs (proximal and distal) and JJAZ1 gene elements, respectively. The 237.8 kb genomic sequence, spanning NF1 LCR‐P and JJAZ1‐P, is identified as redundant by Allocator and hence, no measurement points could be created to resolve these two regions.
No clear genotype‐phenotype relationship has been found in NF1.12 This may be either due to the influence of additional modifier loci in different individuals or the variable nature, location, and developmental timing of the somatic mutations of the NF1 gene which determine the severity of disease. Although a correlation between the presence of large NF1 gene deletions and clinical features (including dysmorphic features, learning disability, and developmental delay) has been described,13 we find that not all patients with NF1 deletions have this specific phenotype.2 In this study, we did not observe any obvious relationship between the size of NF1 deletion and specific clinical features. This indicates that both unlinked modifying genes and the normal allele may be involved in the development of particular clinical features in NF1, and that the relative contributions may vary for different features. We also identified new segmentons bordering breakpoints of atypical deletions and these should be further studied to determine their role in causing these rearrangements. In the case of at least one of these novel segmentons (located within intron 1 of the NF1 gene and contained within the 80p array data point) we found an association with five separate deletion breakpoints. In each of these five cases, the deletions extended towards the telomere of 17q but varied considerably in size from 25.8 kb to >1.33 Mb. However, two of the deletion profiles derived from different patients (cases 119688 and F538, fig 2) were apparently identical. Hence, it is tempting to hypothesise that the segmenton, contained within the 80p array data point, represents a new hot spot for NF1 gene deletions, which warrants further study. It should be stressed in this context that this segmenton is present at least twice per haploid genome and on two different autosomes (that is, chromosomes 13 and 17). Assuming that our hypothesis regarding the involvement of “segmenton 80p” is correct, this indicates an as yet uncharacterised mechanism mediating deletions within the NF1 gene.
Our current series of NF1 derived samples included 12 samples from MPNSTs. Three of these disclosed deletions within the studied 2.24 Mb locus and these were not present in the constitutional DNA of affected patients. Interestingly, two of these deletions were entirely contained within the 2.24 Mb locus, including one sample (T127, table 1) that showed the typical deletion breakpoints. Previous studies analysing the NF1 gene in MPNSTs have shown a varying percentage (55–100%) of loss of heterozygosity at 17q.28,29,30 A recent FISH based study of MPNSTs also identified deletions of the NF1 gene in all eight analysed MPNST cases.30 However, as far as we are aware, this is the first report disclosing the typical NF1 locus deletion (located between the two major segmentons flanking the NF1 gene) in an MPNST sample. The absence of large deletions in the constitutional DNA of these patients indicates that small genetic lesions, most probably point mutations, are sufficient to cause malignancy in some NF1 patients. This result also suggests that abnormal intra‐chromosomal recombination, mediated by these flanking segmentons, is also causing deletions of the NF1 gene in somatic cells, via an as yet unknown mechanism that likely occurs during mitosis.
In only one case (F548) were the data obtained by array‐CGH not in complete agreement with our MLPA analysis (fig 4E,F) and previous data. The deletion in F548 was previously mapped from exon 6 to exon 8 using long range PCR and MLPA and was confirmed independently by our MLPA analysis. However, array‐CGH analysis detected the deletion starting in intron 7 and ending in intron 8. This discordance between previous data and our array‐CGH data can be explained by the design of the NF1 locus microarray. Pool 87p spans a 8.5 kb genomic sequence between intron 5 and intron 7. Most of the sequence (7.75 kb out of 8.5 kb; 92%) covered by pool 87p is within intron 5. Pool 88p also consisted of two PCR products and spans a 7.1 kb genomic sequence between intron 7 and intron 8. The deletion in F548, which spans intron 5 to intron 8, was detected by pool 88p but not by 87p. This can be explained by assuming that the proximal breakpoint of the deletion in F548 is situated closer to exon 6 than exon 5 and consequently only a small fraction of intron 5 is deleted. This would result in a situation where most of pool 87p is not deleted and hence the fluorescence ratio for this measurement point was consistent with two copies.
The array‐CGH methodology has emerged as a revolutionary and versatile platform over the past few years, allowing a number of different assays to be performed.31 Rapid development of this technology is likely to continue over the next few years, with improvements in coverage and resolution as the main focus. This will have a major impact on genetic research and the routine diagnostics of genetic disorders. From our NF1 locus array‐CGH data, we show that the shortest stretch of unique sequence which allows reliable detection of single copy number changes is 490 bp, using fluorescently labelled total human DNA in the hybridisation experiments. In conclusion, even at the current resolution, our array constitutes a novel and reliable tool offering significantly improved diagnostics for this common disorder.
Electronic‐database information
The supplementary tables are available at http://www.jmedgenet.com/supplemental. Other URLs mentioned in this study are as follows: RepeatMasker, http://www.repeatmasker.org; BLAST, http://www.ncbi.nlm.nih.gov/blast; Primer3, http://frodo. wi.mit.edu/cgi‐bin/primer3/primer3_www.cgi; and Human Blast Sequences, http://www.ncbi.nlm.nih. gov/genome/seq/HsBlast.html.
Acknowledgements
We would like to thank Birgitta Sinder Wilen and Anne‐Marie Westlund for excellent technical assistance.
Abbreviations
ANILFR - average normalised inter‐locus fluorescence ratio
LCR - low copy repeat
LCR‐D - LCR‐distal
LCR‐P - LCR‐proximal
MLPA - multiplex ligation‐dependent probe amplification
MPNST - malignant peripheral nerve sheath tumours
NF1 - neurofibromatosis type 1
NPA - normalised peak area
UTR - untranslated region
Footnotes
This work was supported by grants from the U.S. Army Medical Research and Materiel Command, award no. W81XWH‐04‐1‐0269, the Swedish Cancer Foundation, Wallenberg Consortium North, The Swedish Research Council, and Uppsala University to JPD, and the Danish Platform for Integrated Biology of the Danish Basic Research Fund to LB
Competing interests: none declared
The supplementary tables are available at http://www.jmedgenet.com/supplemental. Other URLs mentioned in this study are as follows: RepeatMasker, http://www.repeatmasker.org; BLAST, http://www.ncbi.nlm.nih.gov/blast; Primer3, http://frodo. wi.mit.edu/cgi‐bin/primer3/primer3_www.cgi; and Human Blast Sequences, http://www.ncbi.nlm.nih. gov/genome/seq/HsBlast.html.
References
- 1.Marchuk D A, Collins F S. Molecular genetics of neurofibromatosis 1. In: Huson SM, Hughes RAC, eds. The neurofibromatoses: a pathogenetic and clinical overview. London: Chapman and Hall, 199423–49.
- 2.Upadhyaya M, Ruggieri M, Maynard J, Osborn M, Hartog C, Mudd S, Penttinen M, Cordeiro I, Ponder M, Ponder B A, Krawczak M, Cooper D N. Gross deletions of the neurofibromatosis type 1 (NF1) gene are predominantly of maternal origin and commonly associated with a learning disability, dysmorphic features and developmental delay. Hum Genet 1998102(5)591–597. [DOI] [PubMed] [Google Scholar]
- 3.Ars E, Serra E, Garcia J, Kruyer H, Gaona A, Lazaro C, Estivill X. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum Mol Genet 20009(2)237–247. [DOI] [PubMed] [Google Scholar]
- 4.Messiaen L M, Callens T, Mortier G, Beysen D, Vandenbroucke I, Van Roy N, Speleman F, Paepe A D. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat 200015(6)541–555. [DOI] [PubMed] [Google Scholar]
- 5.Cnossen M H, van der Est M N, Breuning M H, van Asperen C J, Breslau‐Siderius E J, van der Ploeg A T, de Goede‐Bolder A, van den Ouweland A M, Halley D J, Niermeijer M F. Deletions spanning the neurofibromatosis type 1 gene: implications for genotype‐phenotype correlations in neurofibromatosis type 1? Hum Mutat 19979(5)458–464. [DOI] [PubMed] [Google Scholar]
- 6.Lopez‐Correa C, Dorschner M, Brems H, Lazaro C, Clementi M, Upadhyaya M, Dooijes D, Moog U, Kehrer‐Sawatzki H, Rutkowski J L, Fryns J P, Marynen P, Stephens K, Legius E. Recombination hotspot in NF1 microdeletion patients. Hum Mol Genet 200110(13)1387–1392. [DOI] [PubMed] [Google Scholar]
- 7.Kluwe L, Siebert R, Gesk S, Friedrich R E, Tinschert S, Kehrer‐Sawatzki H, Mautner V F. Screening 500 unselected neurofibromatosis 1 patients for deletions of the NF1 gene. Hum Mutat 200423(2)111–116. [DOI] [PubMed] [Google Scholar]
- 8.Dorschner M O, Sybert V P, Weaver M, Pletcher B A, Stephens K. NF1 microdeletion breakpoints are clustered at flanking repetitive sequences. Hum Mol Genet 20009(1)35–46. [DOI] [PubMed] [Google Scholar]
- 9.Jenne D E, Tinschert S, Reimann H, Lasinger W, Thiel G, Hameister H, Kehrer‐Sawatzki H. Molecular characterization and gene content of breakpoint boundaries in patients with neurofibromatosis type 1 with 17q11.2 microdeletions. Am J Hum Genet 200169(3)516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenne D E, Tinschert S, Dorschner M O, Hameister H, Stephens K, Kehrer‐Sawatzki H. Complete physical map and gene content of the human NF1 tumor suppressor region in human and mouse. Genes Chromosomes Cancer 200337(2)111–120. [DOI] [PubMed] [Google Scholar]
- 11.De Raedt T, Brems H, Lopez‐Correa C, Vermeesch J R, Marynen P, Legius E. Genomic organization and evolution of the NF1 microdeletion region. Genomics 200484(2)346–360. [DOI] [PubMed] [Google Scholar]
- 12.Castle B, Baser M E, Huson S M, Cooper D N, Upadhyaya M. Evaluation of genotype‐phenotype correlations in neurofibromatosis type 1. J Med Genet 200340(10)e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kayes L M, Burke W, Riccardi V M, Bennett R, Ehrlich P, Rubenstein A, Stephens K. Deletions spanning the neurofibromatosis 1 gene: identification and phenotype of five patients. Am J Hum Genet 199454(3)424–436. [PMC free article] [PubMed] [Google Scholar]
- 14.Tonsgard J H, Yelavarthi K K, Cushner S, Short M P, Lindgren V. Do NF1 gene deletions result in a characteristic phenotype? Am J Med Genet 199773(1)80–86. [DOI] [PubMed] [Google Scholar]
- 15.De Raedt T, Brems H, Wolkenstein P, Vidaud D, Pilotti S, Perrone F, Mautner V, Frahm S, Sciot R, Legius E. Elevated risk for MPNST in NF1 microdeletion patients. Am J Hum Genet 200372(5)1288–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kehrer‐Sawatzki H, Kluwe L, Sandig C, Kohn M, Wimmer K, Krammer U, Peyrl A, Jenne D E, Hansmann I, Mautner V F. High frequency of mosaicism among patients with neurofibromatosis type 1 (NF1) with microdeletions caused by somatic recombination of the JJAZ1 gene. Am J Hum Genet 200475(3)410–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kehrer‐Sawatzki H, Tinschert S, Jenne D E. Heterogeneity of breakpoints in non‐LCR‐mediated large constitutional deletions of the 17q11.2 NF1 tumour suppressor region. J Med Genet 200340(10)e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petek E, Jenne D E, Smolle J, Binder B, Lasinger W, Windpassinger C, Wagner K, Kroisel P M, Kehrer‐Sawatzki H. Mitotic recombination mediated by the JJAZF1 (KIAA0160) gene causing somatic mosaicism and a new type of constitutional NF1 microdeletion in two children of a mosaic female with only few manifestations. J Med Genet 200340(7)520–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mantripragada K K, Buckley P G, Jarbo C, Menzel U, Dumanski J P. Development of NF2 gene specific, strictly sequence defined diagnostic microarray for deletion detection. J Mol Med 200381(7)443–451. [DOI] [PubMed] [Google Scholar]
- 20.Schouten J P, McElgunn C J, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation‐dependent probe amplification. Nucleic Acids Res 200230(12)e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhami P, Coffey A J, Abbs S, Vermeesch J R, Dumanski J P, Woodward K J, Andrews R M, Langford C, Vetrie D. Exon array‐CGH: detection of copy number changes at the resolution of individual exons in the human genome. Am J Hum Genet 200576(5)750–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Upadhyaya M, Roberts S H, Maynard J, Sorour E, Thompson P W, Vaughan M, Wilkie A O, Hughes H E. A cytogenetic deletion, del(17)(q11.22q21.1), in a patient with sporadic neurofibromatosis type 1 (NF1) associated with dysmorphism and developmental delay. J Med Genet 199633(2)148–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Upadhyaya M, Cheryson A, Broadhead W, Fryer A, Shaw D J, Huson S, Wallace M R, Andersen L B, Marchuk D A, Viskochil D.et al A 90 kb DNA deletion associated with neurofibromatosis type 1. J Med Genet 199027(12)738–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodgson G, Hager J H, Volik S, Hariono S, Wernick M, Moore D, Nowak N, Albertson D G, Pinkel D, Collins C, Hanahan D, Gray J W. Genome scanning with array CGH delineates regional alterations in mouse islet carcinomas. Nat Genet 200129(4)459–464. [DOI] [PubMed] [Google Scholar]
- 25.Mantripragada K K, Buckley P G, Benetkiewicz M, De Bustos C, Hirvela C, Jarbo C, Bruder C E, Wensman H, Mathiesen T, Nyberg G, Papi L, Collins V P, Ichimura K, Evans G, Dumanski J P. High‐resolution profiling of an 11 Mb segment of human chromosome 22 in sporadic schwannoma using array‐CGH. Int J Oncol 200322(3)615–622. [PubMed] [Google Scholar]
- 26.Benetkiewicz M, Wang Y, Schaner M, Wang P, Mantripragada K K, Buckley P G, Kristensen G, Borresen‐Dale A L, Dumanski J P. High‐resolution gene copy number and expression profiling of human chromosome 22 in ovarian carcinomas. Genes Chromosomes Cancer 200542(3)228–237. [DOI] [PubMed] [Google Scholar]
- 27.Buckley P G, Jarbo C, Menzel U, Mathiesen T, Scott C, Gregory S G, Langford C F, Dumanski J P. Comprehensive DNA copy number profiling of meningioma using a chromosome 1 tiling‐path microarray identifies novel candidate tumor suppressor loci. Cancer Res 2005652653–2661. [DOI] [PubMed] [Google Scholar]
- 28.Legius E, Marchuk D A, Collins F S, Glover T W. Somatic deletion of the neurofibromatosis type 1 gene in a neurofibrosarcoma supports a tumour suppressor gene hypothesis. Nat Genet 19933(2)122–126. [DOI] [PubMed] [Google Scholar]
- 29.Lothe R A, Slettan A, Saeter G, Brogger A, Borresen A L, Nesland J M. Alterations at chromosome 17 loci in peripheral nerve sheath tumors. J Neuropathol Exp Neurol 199554(1)65–73. [DOI] [PubMed] [Google Scholar]
- 30.Perry A, Roth K A, Banerjee R, Fuller C E, Gutmann D H. NF1 deletions in S‐100 protein‐positive and negative cells of sporadic and neurofibromatosis 1 (NF1)‐associated plexiform neurofibromas and malignant peripheral nerve sheath tumors. Am J Pathol 2001159(1)57–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mantripragada K K, Buckley P G, de Stahl T D, Dumanski J P. Genomic microarrays in the spotlight. Trends Genet 200420(2)87–94. [DOI] [PubMed] [Google Scholar]
- 32.Upadhyaya M, Majounie E, Thompson P, Han S, Consoli C, Krawczak M, Cordeiro I, Cooper D N. Three different pathological lesions in the NF1 gene originating de novo in a family with neurofibromatosis type 1. Hum Genet 2003112(1)12–17. [DOI] [PubMed] [Google Scholar]