Abstract
Molecular definition at the BAC level of an 8p dicentric chromosome and an 8p deleted chromosome is reported in a patient with two different cell lines. The dicentric, which differed from that generating the recurrent inv dup del(8p) for the location of its break point, originated during the paternal meiosis on the background of the classical 8p23.1 inversion polymorphism. The breakage of this dicentric gave rise to the 8p deleted chromosome which, as a result of the inversion, had two non‐contiguous deletions. These findings confirm previous data on 1p distal deletions, showing that at least some of the deletions stem from the breakage of dicentric chromosomes. They suggest that non‐contiguous deletions may be frequent among distal deletions. This type of rearrangement can easily be overlooked when two contiguous clones, one absent and the other present by FISH analysis, are taken as boundaries of the deletion break point; in this case only high resolution array‐CGH will reveal their real frequency. The definition of such non‐contiguous distal deletions is relevant for phenotype/karyotype correlations. There are historical examples of blunders caused by overlooking a second non‐contiguous deletion. This paper shows how small scale structural variations, such as common polymorphic inversions, may cause complex rearrangements such as terminal deletions.
Keywords: inversion polymorphism, dicentric chromosomes, terminal deletions, non‐contiguous deletions
Mosaicisms with an aneuploid cell line often arise as a result of trisomy or monosomy rescue during the early phases of development in embryos with aneuploidies.1 It has been estimated that 10–30% of fertilised human eggs have a chromosome anomaly of number2 but the majority of these cannot be found among full term fetuses.3 The result of these phenomena can be a confined placental mosaicism, or a mosaicism involving both the embryo and the extrafetal tissue. These events are strictly correlated with the occurrence of uniparental disomy.4
Other kinds of mosaicism, such as those where different structural anomalies of the same chromosome can be detected in distinct cell lines, are described in some patients, if rarely. These conditions can take place, for example, in embryos which have inherited a dicentric chromosome (pter→q::q→pter or qter→p::p→qter). In fact a dicentric chromosome, because of the presence of two centromeres, can provoke the formation of an anaphase bridge during cell division, followed by breakage of the dicentric at an apparently random site. Asymmetric breakage of the dicentric chromosome can lead to the formation of an inverted duplicated chromosome and a terminally deleted chromosome. This phenomenon has been described in patients with 8p anomalies, where it has been shown that an isodicentric chromosome 8 is the unstable precursor of inv dup del(8p) and del(8pter) chromosomes.5,6,7 Initially it has been proposed that the breakage of the dicentric occurs during the second meiotic division, but recent data suggest that the dicentric could persist until the early stages of embryonic development. In fact, few cases have been described in which different products of the breakage of a dicentric chromosome 8 can be found in different cell lines.7,8,9
In some rare cases, however, one of the centromeres of a dic(8) becomes inactive; at least in some cells, the pseudodicentric chromosome 8 behaves like a normal chromosome and it can be inherited (from cell to cell) without any further rearrangement.10,11
We report a case of mosaic 46,XX,psu dic(8)(p23.2)/46,XX,del(8)(p23.1). The peculiarity of this case lies in the fact that the deleted 8p has two deleted regions that in respect to the reference genome sequence are non‐contiguous as a result of a polymorphic 8p inversion.6 Moreover, this work provides new insights into the formation and behaviour of dicentric chromosomes 8, both in recurrent and non‐recurrent 8p anomalies. We also emphasise the role of the polymorphic 8p inversion in the generation of uncommon chromosome rearrangements such as non‐contiguous deletions.
Methods
Clinical report
This case was previously published as case 1 by Digilio et al.10 At that time, the karyotype of the proposita was wrongly interpreted as a mosaic 46,XX/46,XX,‐8,+idic(8)(p23); in fact, the line without the dicentric 8 was interpreted as normal and the 8p deletion was not noted. At the age of 14 years (at the time of writing), the patient's height was 151 cm (3rd–10th centile), her weight was 61.5 kg (75th–90th centile), and her head circumference was 56.5 cm (97th centile). Dysmorphic features include an asymmetrical facies with the left eye lower than the right, left palpebral ptosis, dental malocclusion, zygomatic arch hypoplasia, low set ears, and a short neck with webbing. She has kyphoscoliosis, globous abdomen, short upper and lower limbs, premature grey hair, and severe mental retardation.
Cytogenetic investigations, array‐CGH, and genotyping
G‐banded chromosome analysis was undertaken on metaphases from peripheral blood lymphocytes of the patient and her parents at 400–550 band resolution. All BAC probes used in this study were selected according to the UCSC Human Genome Browser and were obtained from the Human Library RPCI‐11; chromosome 8 α‐satellite probe was extracted from the “chromosome 8 α‐satellite” clone.12 DNAs extraction and fluorescent in situ hybridisation (FISH) experiments were undertaken as described in Ciccone et al.13 The classical 8p23.1 paracentric inversion6 was determined by using clones RP11‐399J23 (biotin labelled) and RP11‐589N15 (digoxigenin labelled) in dual colour FISH experiments. Clones RP11‐5E15 and RP11‐589N15 were used to exclude the two deletions in the father.
Array‐CGH was carried out using the Spectral Genomics 1 Mb chip (Spectral Genomics Inc, Houston, Texas, USA), as already reported in Ciccone et al.13 Genotyping of polymorphic loci was undertaken by amplification with primers labelled with fluorescent probes (ABI 5‐Fam, Hex and Tet) followed by analysis on an ABI 310 genetic analyser (Applied Biosystems, Foster City, California, USA). The UCSC Genome Browser map and sequence were used as references (table 1).
Table 1 Results of microsatellite typing in the patient and her parents.
| Marker | Position (kb) | Proband | Mother | Father | Status |
|---|---|---|---|---|---|
| D8S504 | 1004 | 200.46 | 200.00 | 202.03/205.88 | del(P) |
| D8S264 | 2117 | 132.46 | 126.73/132.31 | 124.78/136.42 | del(P) |
| D8S201 | 3027 | 255.9 | 255.8 | 263.3 | del(P) |
| D8S1824 | 3540 | 242.38 | 242.50/246.29 | 228.84/238.49 | del(P) |
| D8S262 | 3664 | 111.89 | 111.79/113.67 | 107.92/113.72 | del(P) |
| D8S518 | 4475 | 226.60 | 226.50/242.04 | 226.58/242.03 | U |
| D8S277 | 6504 | 153.64 | 153.77 | 153.72/169.79 | U |
| D8S1819 | 6737 | 203.20/218.60 | 203.21/218.57 | 218.63 | N |
| D8S503 | 9270 | 213.85(*)/215.71 | 215.75/223.47 | 211.97/213.82 | ? |
| D8S1721 | 10178 | 167.55/200.57(*) | 167.68 | 190.98/200.57 | ? |
| D8S520 | 10593 | 190.76 | 190.79 | 188.88/190.77 | U |
| D8S550 | 10990 | 245.35/262.73 | 262.86/264.65 | 245.35/253.10 | ? |
| D8S265 | 11317 | 210.95(*)/212.92 | 204.15/212.84 | 210.5 | ? |
| D8S552 | 12752 | 164.82/172.76* | 164.82/172.67 | 164.84/172.72 | dup(P) |
| D8S511 | 14690 | 129.94 | 130.01/131.92 | 128.03/129.94 | U |
| D8S549 | 15660 | 164.76/166.75* | 164.77/168.67* | 162.69/166.71 | dup(P) |
| D8S254 | 16618 | 61.44/66.35 | 61.42/66.38 | 61.42/66.38 | U |
| D8S261 | 17836 | 123.58 | 123.62/133.52 | 123.40 | U |
| D8S258 | 20377 | 143.80/145.97* | 143.79/145.97 | 145.97 | dup(P) |
| D8S282 | 21425 | 262.27*/264.08 | 258.22/264.09 | 258.15/262.15 | dup(P) |
| D8S1734 | 22817 | 112.84*/114.68 | 114.65 | 112.80/114.67 | dup(P) |
| D8S1771 | 25463 | 224.32/226.22 | 224.32/226.14 | 224.12/233.84 | ? |
| D8S1809 | 28213 | 158.46 | 158.46 | 158.46/166.02 | U |
| D8S278 | 32606 | 231.37/233.39 | 231.34/237.24 | 233.16 | ? |
| D8S513 | 33727 | 198.33*/202.08 | 198.21/201.99* | 196.41/198.31 | dup(P) |
| D8S1750 | 35470 | 211.86/217.55* | 211.68/215.53 | 211.70/217.47 | dup(P) |
| D8S1821 | 38369 | 145.73/163.02* | 143.55/145.73 | 141.42/162.85 | dup(P) |
| D8S255 | 39902 | 117.25 | 117.26 | 117.17 | U |
| D8S268 | 41264 | 257.12*/259.06 | 256.93/258.92 | 256.86 | dup(P) |
| D8S1115 | 42554 | 160.45 | 157.09/160.34 | 160.23 | U |
| D8S531 | 49074 | 114.91 | 114.91 | 114.93/120.59 | U |
The asterisks indicate the paternal alleles whit an increased ratio of fluorescence intensity with respect to those of maternal origin.
del(P) and dup(P), paternal origin of the deletion and the duplication, respectively; U, uninformative results; ?, results are ambiguous, probably because of duplication in one cell line and deletion in the other.
Results
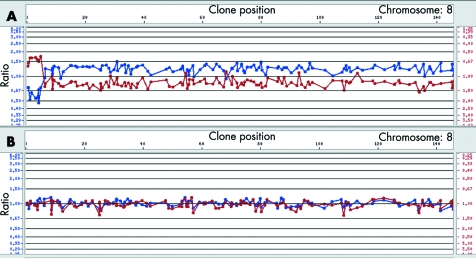
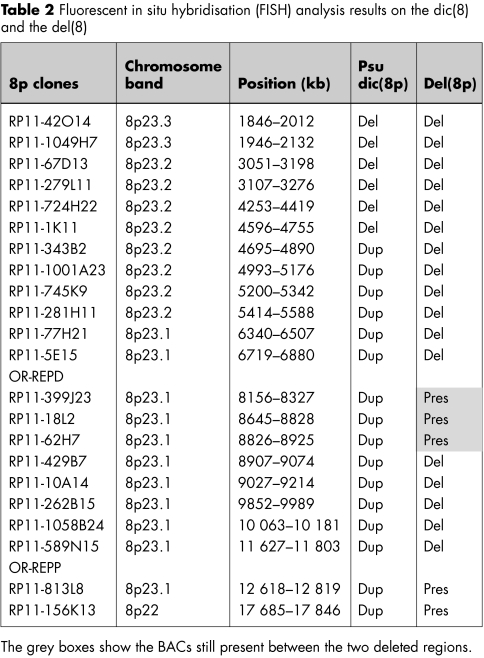
Cytogenetics investigations of the proband's metaphases revealed the presence of a dicentric chromosome 8 with a single primary constriction in 60% of the cells and an 8p‐ chromosome in 40% of the lymphocyte metaphases (100 analysed cells). Both parents had a normal karyotype. Microsatellite analysis showed the absence of paternal alleles at some polymorphic loci in the distal 3.6 Mb of 8p. Fluorescence intensities of several proximal paternal alleles showed an increased ratio with respect to the maternal alleles, showing that the rearrangement arose within the same paternal chromosome 8 and was caused by an intrachromosomal event (table 1). These results should clearly be interpreted as the mean of the allele dosage in both cell lines. Array‐CGH gave the first hint of the extension of the aneuploidy, demonstrating that the distal 8p region was deleted in both cell lines (fig 1). FISH experiments allowed the definition of the breakpoints in dicentric and deleted chromosomes (table 2) and excluded the presence of the two deletions in the father.
Figure 1 (A) Array‐CGH profile (1 Mb resolution) of the proposita's chromosome 8. As shown by the different ratio values the terminal deleted region is certainly at the hemizygous state in all the cell lines, whereas the rest of the duplicated chromosome is in mosaic (as confirmed by the FISH data). From array‐CGH analysis the last deleted clone is RP11‐1K11; this is deleted both in the dic(8) and in the del(8). Clones that are deleted in the del(8p) and duplicated in the dic(8) gave an ambiguous profile with values around 1. (B) Array‐CGH profile of a normal chromosome 8. CGH, comparative genomic hybridisation; FISH, fluorescent in situ hybridisation.
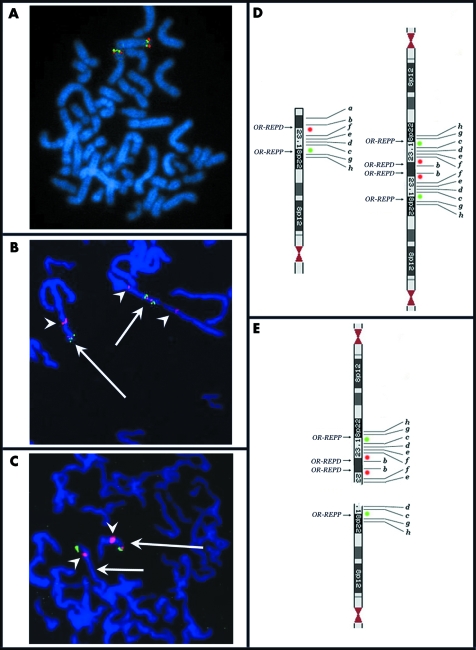
The psu dic(8) was deleted for the distal 4.7 Mb of 8p (from the telomere to clone RP11‐1K11), whereas the rest of the chromosome was duplicated. Thus its breakpoint does not coincide with the two clusters of homologous segmental duplications lying at 8p23.16 (8p‐OR‐REPD and 8p‐OR‐ REPP) originating the recurrent inv dup del(8p). As concerning the del(8p), FISH analysis showed that BAC clones from the telomere to 6.7 Mb (RP11‐5E15) were deleted, those from 8.15 Mb (RP11‐399J23) to 8.82 Mb (RP11‐62H7) were present, those from 8.82 Mb (RP11‐429B7) to 11.80 Mb (RP11‐589N15) were deleted, and those from 12.61 Mb (RP11‐813L8) to the centromere were present (table 2). Clones encompassing 8p‐OR‐REPD (from 6.7 to 8.15 Mb) and 8p‐OR‐REPP (from 11.80 to 12.61 Mb) could not be investigated owing the their low copy repeats content. Thus the del(8p) lacks two non‐contiguous regions. Dual colour FISH experiments, using clones RP11‐399J23 and RP11‐589N15, showed that the father was a heterozygous carrier of the 8p23.1 inversion encompassed between 8p‐OR‐REPD and 8p‐OR‐REPP (fig 2A). Indeed, the same experiment on proposita's metaphases showed that the dicentric chromosome had an inverted pattern of signals at 8p23.1 with respect to that expected from a dicentric originated from a normal chromosome 8 (green‐red‐red‐green instead of red‐green‐green‐red, see fig 2B). Moreover, probe RP11‐399J23 at 8.15 Mb was not deleted in the del(8p) chromosome, whereas probe RP11‐589N15 at 11.80 Mb was (fig 2C).
Figure 2 (A) metaphase from the proposita's father showing the inversion at 8p23,1. (B) Dual colour fluorescent in situ hybridisation (FISH) showing the inverted sequence of colours in the psu dic(8) (short arrow) with respect to the normal chromosome 8 (long arrow). (C) The del(8) (short arrow) shows only the green signal; note that this signal (RP11‐399J23 at 8.15 Mb) is distal to the red one (RP11‐589N15 at 11.80 Mb) present in the normal chromosome 8 (long arrow). In (B) and (C), arrowheads indicate the centromeres. (D) Ideogram of a chromosome 8 with the polymorphic inversion (left); this chromosome is the background for the formation of the dic(8) (right) found in the proposita that is deleted for a. (E) As a consequence of the dic(8) breakage, the del(8) (below) we found in the proposita has two non‐contiguous deletions (a–b and e–f) and its distal region is inverted (d–c instead of c–d). In (D) and (E) green and red spots indicate probes RP11‐399J23 and RP11‐589N15, respectively.
Discussion
In the last 5–10 years the mechanisms of several recurrent constitutional chromosome rearrangements such as interstitial duplications and deletions, translocations, and inversions have been elucidated. In most of these cases segmental duplications (also termed “low copy repeats”, LCRs) are located at the rearrangement's breakpoints predisposing to non‐allelic homologous recombination (NAHR).14 On the other hand, the molecular basis of the mechanisms leading to terminal deletions is poorly clarified, essentially for two reasons: first, in only a few cases have the breakpoints been determined at the base pair level; second, the observed breakpoints could be different from the original breakpoints as a result of the action of repair mechanisms activated by DNA double strand breakages.15 In any case, such broken chromosomes are stabilised by the addition of the telomeric sequence (TTAGGG)n through at least two mechanisms: telomere capture and telomere healing.16,17
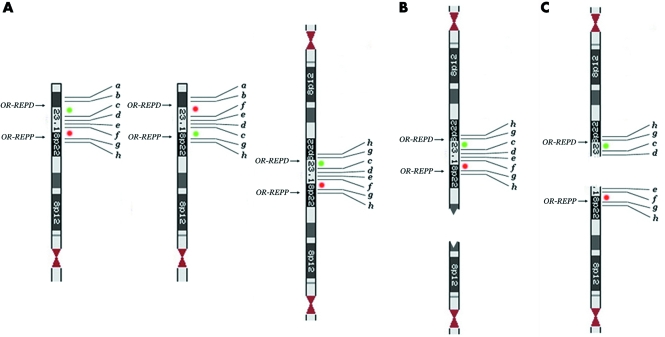
However, it should be borne in mind that, at least in some circumstances, terminal deletions arise from the breakage of dicentric chromosomes. The formation of dicentric chromosomes as the first product of abnormal meiotic recombination has been documented for chromosome 8p. In particular the presence of the polymorphic inversion mediated by LCRs (8p‐OR‐REPP and 8p‐OR‐REPD) renders this region prone to the formation of dicentric and acentric recombinant products.6,7 This dicentric contains two duplicated regions separated by a single copy region flanked by homologous LCRs (fig 3A). An asymmetrical breakage between the two centromeres but outside the single copy region leads to the formation of an inverted duplicated chromosome deleted in its distal portion (inv dup del(8p)) and a simply deleted chromosome (del(8p)) (fig 3B); the two cell lines may even persist in the same subject.7,8,9 A breakage between the two centromeres but within the single copy region would produce two 8p deleted chromosomes (fig 3C). Owing to the presence of the polymorphic inversion, one of these should have two non‐contiguous deletions and should be inverted in the region between them (fig 3C). In the present case, the mechanism of the deletion is different; first, the abnormal recombination generating it occurred about 2 Mb distal to 8p‐OR‐REPD, and second, the dicentric carries the heterozygous inversion. The breakage of the dicentric within one of the two inverted regions encompassed by 8p‐OR‐REPP and 8p‐OR‐REPD produced a del(8p) with two non‐contiguous deletions and an inversion (fig 2E: d–c instead of c–d). The main question is how often this situation occurs; that is, how often will a terminally deleted chromosome be the result of non‐contiguous deletions? This situation seems to be extremely rare as only one case has been reported relating to 1p36.18 This case was detected through array‐CGH analysis, using a large contig of BAC clones. The following considerations suggest that non‐contiguous deletions are frequent (although overlooked when FISH is used, and two contiguous clones—one absent and the other present—are taken as the deletion breakpoint), and are always associated with an inversion:
Figure 3 (A) The classical dicentric chromosome 8 deleted for a and b (right) generated by non‐allelic homologous recombination (NAHR) between the 8p‐OR‐REPP and 8p‐OR‐REPD the background of the polymorphic inversion6; the chromosome 8 on the left is normal, while the one in the middle is inverted. (B) Recurrent inv dup del(8p)s and del(8p)s are originated by the breakage of this dicentric chromosome.7 (C) Hypothetical breakage of the dicentric within the single copy region (8p23.1)6: two del(8p)s are generated, one of which (the upper one) having two non‐contiguous deletions (a–b and e–f) and an inverted distal region (d–c instead of c–d). Green and red spots indicate probes RP11‐399J23 and RP11‐589N15, respectively.
(1) Rearrangements of the inv dup del type are more frequent than deduced by the early cytogenetics. Many inv dup del rearrangements have now been reported19,20 and several cases interpreted at first as terminal duplications21 and even terminal deletions22 are in fact inverted duplications with a terminal deletion. All these cases should have originated as dicentric chromosomes, possibly as the result of NAHR between inverted LCRs. As we have already stressed, the breakage of these dicentric chromosomes may lead to deletion of chromosomes for two non‐contiguous regions.
(2) Cryptic inversions are frequent in the human genome and it seems very likely that those reported up to now are only the tip of the iceberg. They have been documented as causing several recurrent rearrangements such as the t(X,Y)23 and t(4;8)24 translocations and the interstitial deletions associated with Williams and Sotos syndromes25,26,27 but also as being at the basis of linkage disequilibrium generating specific haplotypes.28
A few cases are also known in which the cryptic recurrent inversion is associated with a Mendelian disease such as haemophilia A29 and Hunter disease30 as a result of NAHR breaking the disease gene. The comparison of the human genome reference sequence with a second genome represented by fosmid paired end sequences to detect intermediate sized structural variants revealed 297 sites of these variants, including 56 inversion breakpoints.31 Thus, cryptic inversions, whatever their significance, are unexpectedly frequent. Most of them may be mediated by LCRs and it is well known that not only the pericentromeric regions but also the subtelomeric ones are enriched in LCRs.32 It seems very likely that polymorphic inversions should be particularly frequent at the level of subtelomeric regions and that this situation predisposes to the formation of dicentric chromosomes with the duplicated regions interrupted by a single copy region. As a result, several deletions not only should result from the loss of two non‐contiguous regions (a, b, e, f), but the region interrupting the deletion (c, d) should also be inverted (fig 2E).
Ballif et al22 explored the causes of terminal deletions using the del(1p)s as a model. In their series of 1p36 deletions they found two in which the deleted region was followed by two duplicated regions that were separated by a single copy region of 1.7 and 1.8 kb, respectively. They explained this situation assuming that an initial break is repaired by non‐homologous end joining, thus leading to the formation of a dicentric chromosome. An asymmetric breakage of the dicentric would lead to a breakage‐fusion‐bridge cycle that might explain the finding of concurrent deletions and duplications (fig 5 in Ballif et al, 200322). As assumed by the same investigators, this model does not explain the initial double strand break. In our model, the formation of the dicentric chromosome is not explained by NAHR between LCRs, as demonstrated for the classical inv dup del(8p) but two possible scenarios can be envisaged: either the mechanism is the same as that described by Ballif et al,22 or the presence of the polymorphic inversion also disrupts the synapsis regularity above the inverted region itself, thus favouring the occurrence of the dicentric (fig 2E).
The phenotypic consequences of two non‐contiguous deletions are noteworthy. Usually, the phenotype–genotype correlations are based on a strict association between deleted genes and malformation signs. A typical example is the Xp22 contiguous gene syndrome: males with Xp deletions of different size, most being consequent to t(X;Y)(p22;q11.2), are affected by one or more X linked diseases according to the number of deleted genes. This well consolidated procedure would totally collapse when applied to non‐contiguous deletions. In fact, genes supposed to be deleted may be present and genes supposed to be present may be deleted. A significant example of how this situation may indeed lead to a blunder is provided by the history of the identification of the testis determining factor (TDF). Initially, the TDF was identified as the ZFY gene thanks to a 45,X,t(Y;22) female patient with a submicroscopic deletion of the Yp region containing the ZFY gene.33 Subsequently, this hypothesis was refuted because the TDF was really identified as the SRY gene.34,35 The paradox of the 45,X,t(Y;22) female patient was explained by the discovery of a second more distal Yp deletion involving the SRY gene, non‐contiguous to that previously identified.36
Conclusions
The case we have studied suggests a mechanism that may explain part of the distal deletions. This mechanism is based on the evidence that several rearrangements interpreted as deletions and duplications are in fact inverted duplications associated with terminal deletions, and on the unexpected frequency of paracentric inversion polymorphisms.31 It is obvious that non‐contiguous deletions arise because the genome map reports inversion polymorphisms in only one of their possible orientations. Array‐CGH studies with high resolution platforms will make it possible to prove our hypothesis.
Electronic database information
Resources for Molecular Cytogenetics (http://www.biologia.uniba.it/rmc)
UCSC Genome Bioinformatics Site (http://genome.ucsc.edu/index)
Spectral Genomics (www.spectralgenomics.com).
Acknowledgements
This work was supported by cofin03‐MIUR (to OZ), cofin04‐MIUR, the FIRB 2001(to OZ), the Italian Telethon Foundation (GP0247Y01 to OZ), and the Cariplo Foundation (to OZ).
Abbreviations
BAC - bacterial artificial chromosome
CGH - comparative genomic hybridisation
FISH - fluorescent in situ hybridisation
LCR - low copy repeat
NAHR - non‐allelic homologous recombination
UCSC - University of California Santa Cruz
Footnotes
Conflicts of interest: none declared
References
- 1.Youssoufian H, Pyeritz R E. Mechanisms and consequences of somatic mosaicism in humans. Nat Rev Genet 20023748–758. [DOI] [PubMed] [Google Scholar]
- 2.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 20012280–291. [DOI] [PubMed] [Google Scholar]
- 3.Daniel A, Wu Z, Darmanian A, Malafiej P, Tembe V, Peters G, Kennedy C, Ades L. Issues arising from the prenatal diagnosis of some rare trisomy mosaics—the importance of cryptic fetal mosaicism. Prenat Diagn 200424524–536. [DOI] [PubMed] [Google Scholar]
- 4.Robinson W P. Mechanisms leading to uniparental disomy and their clinical consequences. Bioessays 200022452–459. [DOI] [PubMed] [Google Scholar]
- 5.Floridia G, Piantanida M, Minelli A, Dellavecchia C, Bonaglia C, Rossi E, Gimelli G, Croci G, Franchi F, Gilgenkrantz S, Grammatico P, Dalpra L, Wood S, Danesino C, Zuffardi O. The same molecular mechanism at the maternal meiosis I produces mono‐ and dicentric 8p duplications. Am J Hum Genet 199658785–796. [PMC free article] [PubMed] [Google Scholar]
- 6.Giglio S, Broman K W, Matsumoto N, Calvari V, Gimelli G, Neumann T, Ohashi H, Voullaire L, Larizza D, Giorda R, Weber J L, Ledbetter D H, Zuffardi O. Olfactory receptor‐gene clusters, genomic‐inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet 200168874–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pramparo T, Giglio S, Gregato G, de Gregori M, Patricelli M G, Ciccone R, Scappaticci S, Mannino G, Lombardi C, Pirola B, Giorda R, Rocchi M, Zuffardi O. Inverted duplications: how many of them are mosaic? Eur J Hum Genet 200412713–717. [DOI] [PubMed] [Google Scholar]
- 8.Soler A, Sanchez A, Carrio A, Badenas C, Mila M, Borrell A. Fetoplacental discrepancy involving structural abnormalities of chromosome 8 detected by prenatal diagnosis. Prenat Diagn 200323319–322. [DOI] [PubMed] [Google Scholar]
- 9.Vermeesch J R, Thoelen R, Salden I, Raes M, Matthijs G, Fryns J P. Mosaicism del(8p)/inv dup(8p) in a dysmorphic female infant: a mosaic formed by a meiotic error at the 8p OR gene and an independent terminal deletion event. J Med Genet 200340e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Digilio M C, Giannotti A, Floridia G, Uccellatore F, Mingarelli R, Danesino C, Dallapiccola B, Zuffardi O. Trisomy 8 syndrome owing to isodicentric 8p chromosomes: regional assignment of a presumptive gene involved in corpus callosum development. J Med Genet 199431238–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piantanida M, Dellavecchia C, Floridia G, Giglio S, Hoeller H, Dordi B, Danesino C, Schinzel A, Zuffardi O. Ataxic gait and mental retardation with absence of the paternal chromosome 8 and an idic(8)(p23.3): imprinting effect or nullisomy for distal 8p genes. Hum Genet 199799766–771. [DOI] [PubMed] [Google Scholar]
- 12.Archidiacono N, Antonacci R, Marzella R, Finelli P, Lonoce A, Rocchi M. Comparative mapping of human alphoid sequences in great apes, using fluorescence in situ hybridization. Genomics 199525477–484. [DOI] [PubMed] [Google Scholar]
- 13.Ciccone R, Giorda R, Gregato G, Guerrini R, Giglio S, Carrozzo R, Bonaglia M C, Priolo E, Lagana C, Tenconi R, Rocchi M, Pramparo T, Zuffardi O, Rossi E. Reciprocal translocations: a trap for cytogenetists? Hum Genet 2005117571–582. [DOI] [PubMed] [Google Scholar]
- 14.Shaw C J, Lupski J R. Implications of human genome architecture for rearrangement‐based disorders: the genomic basis of disease. Hum Mol Genet 2004(suppl 13)R57–R64. [DOI] [PubMed]
- 15.Ballif B C, Gajecka M, Shaffer L G. Monosomy 1p36 breakpoints indicate repetitive DNA sequence elements may be involved in generating and/or stabilizing some terminal deletions. Chromosome Res 200412133–141. [DOI] [PubMed] [Google Scholar]
- 16.Wilkie A O M, Lamb J, Harris P C, Finney R D, Higgs D R. A truncated human chromosome 16 associated with a thalassaemia is stabilized by addition of telomeric repeat (TTAGGG)n. Nature 1990346868–871. [DOI] [PubMed] [Google Scholar]
- 17.Meltzer P S, Guan X‐Y, Trent J M. Telomere capture stabilizes chromosome breakage. Nat Genet 19934252–255. [DOI] [PubMed] [Google Scholar]
- 18.Heilstedt H A, Ballif B C, Howard L A, Lewis R A, Stal S, Kashork C D, Bacino C A, Shapira S K, Shaffer L G. Physical map of 1p36, placement of breakpoints in monosomy 1p36, and clinical characterization of the syndrome. Am J Hum Genet 2003721200–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonaglia M C, Giorda R, Poggi G, Raggi M E, Rossi E, Baroncini A, Giglio S, Borgatti R, Zuffardi O. Inverted duplications are recurrent rearrangements always associated with a distal deletion: description of a new case involving 2q. Eur J Hum Genet 20008597–603. [DOI] [PubMed] [Google Scholar]
- 20.Beaujard M P, Jouannic J M, Bessieres B, Borie C, Martin‐Luis I, Fallet‐Bianco C, Portnoi M F. Prenatal detection of a de novo terminal inverted duplication 4p in a fetus with the Wolf‐Hirschhorn syndrome phenotype. Prenat Diagn 200525451–455. [DOI] [PubMed] [Google Scholar]
- 21.De Brasi D, Rossi E, Giglio S, D'Agostino A, Titomanlio L, Farina V, Andria G, Sebastio G. Inv dup del (1)(pter→q44 q44→q42:) with the classical phenotype of trisomy 1q42‐qter. Am J Med Genet 2001104127–130. [DOI] [PubMed] [Google Scholar]
- 22.Ballif B C, Yu W, Shaw C A, Kashork C D, Shaffer L G. Monosomy 1p36 breakpoint junctions suggest pre‐meiotic breakage‐fusion‐bridge cycles are involved in generating terminal deletions. Hum Mol Genet 2003122153–2165. [DOI] [PubMed] [Google Scholar]
- 23.Jobling M A, Williams G A, Schiebel G A, Pandya G A, McElreavey G A, Salas G A, Rappold G A, Affara N A, Tyler‐Smith C. A selective difference between human Y‐chromosomal DNA haplotypes. Curr Biol 199881391–1394. [DOI] [PubMed] [Google Scholar]
- 24.Giglio S, Calvari V, Gregato G, Gimelli G, Camanini S, Giorda R, Ragusa A, Guerneri S, Selicorni A, Stumm M, Tonnies H, Ventura M, Zollino M, Neri G, Barber J, Wieczorek D, Rocchi M, Zuffardi O. Heterozygous submicroscopic inversions involving olfactory receptor–gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am J Hum Genet 200271276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osborne L R, Li M, Pober B, Chitayat D, Bodurtha J, Mandel A, Costa T, Grebe T, Cox S, Tsui L C, Scherer S W. A 1.5 million‐base pair inversion polymorphism in families with Williams‐Beuren syndrome. Nat Genet 200129321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bayes M, Magano L F, Rivera N, Flores R, Perez Jurado L A. Mutational mechanisms of Williams‐Beuren syndrome deletions. Am J Hum Genet 200373131–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visser R, Shimokawa O, Harada N, Kinoshita A, Ohta T, Niikawa N, Matsumoto N. Identification of a 3.0‐kb major recombination hotspot in patients with Sotos syndrome who carry a common 1.9‐Mb microdeletion. Am J Hum Genet 20057652–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, Baker A, Jonasdottir A, Ingason A, Gudnadottir V G, Desnica N, Hicks A, Gylfason A, Gudbjartsson D F, Jonsdottir G M, Sainz J, Agnarsson K, Birgisdottir B, Ghosh S, Olafsdottir A, Cazier J B, Kristjansson K, Frigge M L, Thorgeirsson T E, Gulcher J R, Kong A, Stefansson K. A common inversion under selection in Europeans. Nat Genet 200537129–137. [DOI] [PubMed] [Google Scholar]
- 29.Lakich D, Kazazian H H, Antonarakis S E, Gitschier J. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia A. Nat Genet 19935236–241. [DOI] [PubMed] [Google Scholar]
- 30.Bondeson M L, Dahl N, Malmgren H, Kleijer W J, Tonnesen T, Carlberg B M, Pettersson U. Inversion of the IDS gene resulting from recombination with IDS‐related sequences is a common cause of the Hunter syndrome. Hum Mol Genet 19954615–621. [DOI] [PubMed] [Google Scholar]
- 31.Tuzun E, Sharp A J, Bailey J A, Kaul R, Morrison V A, Pertz L M, Haugen E, Hayden H, Albertson D, Pinkel D, Olson M V, Eichler E E. Fine‐scale structural variation of the human genome. Nat Genet 200537727–732. [DOI] [PubMed] [Google Scholar]
- 32.International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature 2004431931–945. [DOI] [PubMed] [Google Scholar]
- 33.Page D C, Mosher R, Simpson E M, Fisher E M, Mardon G, Pollack J, McGillivray B, de la Chapelle A, Brown L G. The sex‐determining region of the human Y chromosome encodes a finger protein. Cell 1987511091–1104. [DOI] [PubMed] [Google Scholar]
- 34.Palmer M S, Sinclair A H, Berta P, Ellis N A, Goodfellow P N, Abbas N E, Fellous M. Genetic evidence that ZFY is not the testis‐determining factor. Nature 1989342937–939. [DOI] [PubMed] [Google Scholar]
- 35.Berta P, Hawkins J R, Sinclair A H, Taylor A, Griffiths B L, Goodfellow P N, Fellous M. Genetic evidence equating SRY and the testis‐determining factor. Nature 1990348448–450. [DOI] [PubMed] [Google Scholar]
- 36.Page D C, Fisher E M, McGillivray B, Brown L G. Additional deletion in sex‐determining region of human Y chromosome resolves paradox of X,t(Y;22) female. Nature 1990346279–281. [DOI] [PubMed] [Google Scholar]