Abstract
Background
High myopia is a common genetic variation in most cases, affecting 1–2% of people, and is the fourth most common disorder causing blindness worldwide. Six autosomal dominant loci and one X‐linked recessive locus have been reported, but no genes responsible for high myopia have been identified.
Objective
To report a Chinese family in which six males presented with high myopia consistent with an X linked recessive trait.
Results
Affected individuals shared three common features: high myopia, reduced visual acuity, and fundal changes of high myopia. Protan and deutan were observed in the family, but they did not co‐segregate with the high myopia phenotype. X‐chromosome‐wide linkage analysis mapped the high myopia locus to a 25 cM (14.9 Mb) region on Xq23–q25 between DXS1210 and DXS8057, with maximum two point lod scores at θ = 0 of 2.75 and 2.29 for DXS1001 and DXS8059, respectively.
Conclusions
This new myopia locus is outside the linked region of the first high myopia locus (MYP1). Refinement of the linkage region with additional families and screening candidate genes for mutation may lead to the identification of the defect gene.
Keywords: X linked recessive high myopia, linkage, cone dysfunction, MYP1
Myopia is the most common ocular health problem, affecting an average of about 30% (3% to 84%) of people throughout the world, usually for most of a person's life.1,2,3,4,5,6 The cost involved in correction of this problem is enormous,4,7 accounting for about one fourth of the entire expenditure in ophthalmology and optometry.
High myopia, the fourth most common cause of blindness, is an extreme form of myopia associated with retinal detachment and macular degeneration, occurring in 1–2% of the general population.8,9,10,11,12 It may be inherited as an autosomal dominant, autosomal recessive, X linked recessive, or complex trait. The genes responsible for high myopia have not been identified, although several chromosomal loci have been suggested.9,10,11,13,14,15,16,17 Most of these loci have not been confirmed by independent study, suggesting that the identified loci may be responsible for only a small portion of high myopia.18 A small fraction of high myopia is associated with other ocular or systemic diseases as part of a syndrome.19,20,21,22,23 The first locus (MYP1) for high myopia was mapped to Xq28 in 1990 but its genetic basis remains unknown.13,17
A four generation Chinese family with X linked recessive high myopia was collected in order to map the genetic locus as an initial step towards identifying the genetic cause of this condition. An X‐chromosome‐wide linkage study was undertaken and the myopia in the family was mapped to a novel locus on Xq23–q25, which is outside the linkage region of MYP1.
Methods
Family and clinical data
A Chinese family of Han ethnicity with X linked recessive high myopia was identified in a small town in southeast China. This family has six affected individuals in four generations. Sixteen individuals, including five affected and 11 unaffected, participated in the study. Informed consent conforming to the tenets of the Declaration of Helsinki and following the Guidance of Sample Collection of Human Genetic Diseases (863‐Plan) by the Ministry of Public Health of China was obtained from the participating individuals before the study. Medical and ophthalmic histories were obtained, and ophthalmological examination was carried out (by XG). Refractive error was measured by retinoscopy. A subject was considered to have high myopia if they met the following criteria:
the myopia was noted before school age;
there was bilateral cycloplegic refraction of −6.00 D or lower (spherical equivalent) in individuals <30 years of age, or manifest refraction of −6.00 D or lower (spherical equivalent) in individuals 30 years or more of age;
other known ocular or systemic diseases were excluded.
Electroretinogram (ERG) responses were recorded in the proband consistent with ISCEV standards.24 Colour vision was evaluated using an Ishihara plate and classified by analysing exon 5 of red‐green visual pigment genes, as previously described.25,26
Genotyping and linkage analysis
Genomic DNA was prepared from venous blood. An X‐chromosome‐wide linkage scan was carried out as previous described, except that only microsatellite markers for the X chromosome were analysed.15 High myopia in the family was analysed as an X linked recessive trait with full penetrance and a disease allele frequency of 0.0001. Haplotypes were generated using the Cyrillic 2.1 program and confirmed by inspection. Equal marker allele frequencies were arbitrarily assumed for the initial scan and were calculated from 15 unrelated unaffected individuals for markers in the linked region.
Results
The myopia in five affected individuals ranged from −6.00 D to −20.00 D. Refractive error for unaffected siblings, offspring, and individuals marrying into the family was between −3.00 D and +2.00 D. All affected individuals developed myopia before school age and the best corrected visual acuity was rather poor, between 0.15 and 0.5 (table 1). Progression of myopia was slow except in case 19. No patient had night blindness or photophobia. One affected individual (No 14) and one unaffected individual (No 37) had nystagmus. Age related macular degeneration (AMD) was observed in case 35.
Table 1 Clinical data on the family members participating in the study.
| ID | Sex | Age | Age at first symptom | Visual acuity | Refraction | Others | Nystagmus | ||
|---|---|---|---|---|---|---|---|---|---|
| Unaided (corrected) | R eye | L eye | |||||||
| (y) | R eye | L eye | |||||||
| 3 | M | 43 | 1.0 | 1.0 | 0 | 0 | Protan | No | |
| 4 | F | 60 | 0.5 | 0.2 | Hyperopia and astigmatism | No | No | ||
| 6 | F | 49 | 1.0 | 0.8 | 0 | 0 | No | No | |
| 10 | F | 68 | 15 y | (0.5) | (0.5) | −3.00 D | −3.00 D | No | No |
| 12 | M | 35 | <7 y | (0.5) | (0.5) | −6.00 D | −7.00 D | Deutan | No |
| 14 | M | 32 | <7 y | (0.15) | (0.3) | −8.00 D | −8.00 D | Deutan | Yes |
| 15 | F | 33 | 1.0 | 1.0 | 0 | 0 | No | No | |
| 17 | F | 36 | 1.2 | 1.0 | 0 | 0 | No | No | |
| 18 | M | 36 | 1.2 | 1.2 | 0 | 0 | No | No | |
| 19 | M | 29 | <7 y | (0.15) | (0.15) | −22.00 D | −23.00 D | Protan | No |
| 25 | M | 7 | <5 y | (0.4) | (0.4) | −6.00 D | −7.00 D | No | No |
| 35 | F | 72 | 0.3 | 0.3 | +2.00 D | +2.00 D | AMD | No | |
| 37 | M | 70 | 0.2 | 0.08 | −1.00 DS to 1.00 DC | −2.00 DS to 1.5 DC | No | Yes | |
| 42 | M | 46 | 0.8 | 0.8 | −1.00 DS to 1.5 DC | −1.00 DS to 1.5 DC | No | No | |
| 43 | M | 40 | 1.0 | 0.8 | 0 | −0.50 DS | No | No | |
| 51 | M | 19 | <7 y | (0.4) | (0.4) | −5.50 DS to 2.00 DC | −6.00 DS to 1.50 DC | No | No |
AMD, age related macular degeneration; D, dioptre; DC, dioptres cylinder; DS, dioptres sphere; F, female; L, left; M, male; R, right; y, years.
Red‐green colour vision defects were found in three of the five affected individuals (Nos 12, 14, and 19) and were identified as having patterns of protan in individual 19 and deutan in individuals 12 and 14, based on Ishihara plate screening and heteroduplex‐SSCP analysis of the exon 5 of red‐green visual pigment genes (data supplied as supplemental material and can be seen on the journal website: www.jmedgenet.com/supplemental). Unaffected individual No 3 also had protan. It is thus obvious that the colour vision defects are not associated with high myopia in this family.
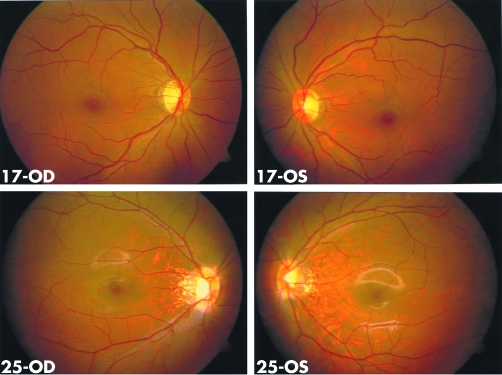
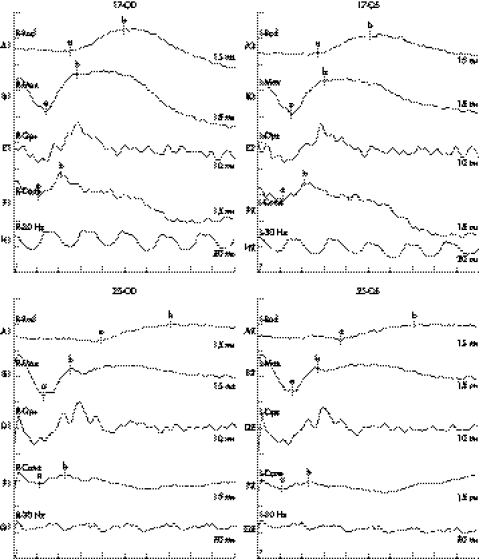
Ophthalmological examination excluded ocular diseases known to be associated with myopia. All affected individuals had a temporal crescent of the optic disc, thinning of the retinal pigment epithelium (“tigroid” appearance) between the fovea and the optic disc, and posterior extension of central macular region, as demonstrated in the proband (fig 1). Ocular A‐scan of the proband at age seven years recorded an axial length of 25.55 mm for the right eye and 25.90 mm for the left. Keratometry at the same time gave 42.00/40.00D for the right eye and 42.50/40.50D for the left. ERG in the proband showed severely reduced amplitude of the cone response. The implicit times of the rod response were delayed (fig 2). No systemic abnormalities were noted in any affected individual. Female carriers have normal visual acuity, normal fundal appearances, and normal responses on ERG recording (figs 1 and 2), except that reduced visual acuity has been recorded in individuals 10 and 35. Individual 10 had a normal fundal appearance and the reason for her reduced visual acuity is unknown. AMD may be contributing to the reduced visual acuity as she says she had better vision when she was young.
Figure 1 Fundus photographs of individuals 17 (carrier, top) and 25 (affected, bottom). The temporal crescent of the optic disc and thinning of the retinal pigment epithelium between the macula and optic disc are seen in individual 25 but not in the normal fundus of individual 17.
Figure 2 Severely reduced amplitude of the cone response and delayed implicit time of the rod response in the electroretinogram (ERG) recording of individual 25. His mother (individual 17) has normal cone‐rod response in the ERG recording.
The affected Chinese individuals share three common phenotypes: high myopia; reduced visual acuity which is stationary and could not be corrected; and typical high myopic fundal changes.
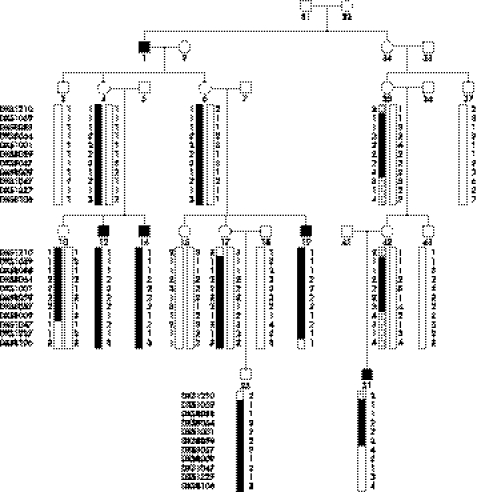
Upon linkage analysis of the X chromosome, maximum two point lod scores of 2.75 and 2.29 were obtained for DXS1001 and DXS8059 (table 2). Haplotype analysis showed a conserved haplotype between DXS1059 and DXS8059. This haplotype is present in all affected individuals as well as in unaffected carriers, but not in unaffected males (fig 3). An obligate recombinant at DXS1210 in affected individual 25 and confirmed in carrier individual 17 sets the proximal boundary. Recombination at DXS8057 in affected individual 51 and further recombination at DXS8106 in affected individual 19 sets the telomeric boundary for the linked region. Thus the linked interval for high myopia in this family is located in the 25 cM (14.9 Mb) region at Xq23–q25 between DXS1210 and DXS 8057.
Table 2 Two point linkage results between microsatellite markers of the X chromosome and high myopia in the Chinese family.
| Markers | Position | Lod score at θ = | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CM | Mb | 0 | 0.01 | 0.05 | 0.1 | 0.2 | 0.3 | 0.4 | |
| DXS1060 | 10.10 | 5.27 | −∞ | −7.31 | −3.91 | −2.51 | −1.24 | −0.61 | −0.23 |
| DXS8051 | 15.70 | 9.31 | −∞ | −4.27 | −2.16 | −1.28 | −0.50 | −0.17 | −0.03 |
| DXS987 | 25.50 | 14.47 | −∞ | −3.47 | −1.49 | −0.74 | −0.16 | 0.03 | 0.05 |
| DXS1226 | 36.80 | ? | −∞ | −1.92 | −0.65 | −0.20 | 0.08 | 0.12 | 0.08 |
| DXS1214 | 46.20 | 31.02 | −∞ | −5.43 | −2.71 | −1.60 | −0.63 | −0.20 | −0.01 |
| DXS1068 | 56.20 | 38.66 | −∞ | −4.35 | −2.28 | −1.43 | −0.66 | −0.29 | −0.10 |
| DXS993 | 66.10 | 40.90 | −∞ | −0.50 | 0.20 | 0.45 | 0.56 | 0.47 | 0.28 |
| DXS8080 | 71.60 | 44.00 | −∞ | −1.65 | −0.88 | −0.54 | −0.22 | −0.08 | −0.02 |
| DXS991 | 86.90 | 55.40 | −∞ | 1.11 | 1.62 | 1.67 | 1.44 | 1.04 | 0.56 |
| DXS1213 | 87.40 | 65.05 | −∞ | −0.77 | −0.05 | 0.22 | 0.38 | 0.35 | 0.22 |
| DXS8052 | 94.20 | 69.60 | −∞ | 0.47 | 1.01 | 1.11 | 0.99 | 0.71 | 0.36 |
| DXS986 | 95.90 | 79.19 | −∞ | −1.17 | 0.11 | 0.54 | 0.77 | 0.69 | 0.42 |
| DXS990 | 104.90 | 92.81 | −∞ | −1.17 | 0.11 | 0.54 | 0.77 | 0.69 | 0.42 |
| DXS1106 | 115.10 | 102.54 | −∞ | −0.77 | −0.11 | 0.12 | 0.26 | 0.24 | 0.14 |
| DXS1210 | 119.20 | 108.40 | −∞ | −2.17 | −0.83 | −0.32 | 0.07 | 0.16 | 0.13 |
| DXS1059 | 121.00 | 111.13 | 0.57 | 0.56 | 0.51 | 0.44 | 0.32 | 0.20 | 0.09 |
| DXS8088 | 122.80 | 113.16 | 0.50 | 0.49 | 0.45 | 0.41 | 0.31 | 0.20 | 0.09 |
| DXS8064 | 131.80 | 117.05 | 0.61 | 0.60 | 0.56 | 0.51 | 0.41 | 0.29 | 0.16 |
| DXS1001 | 139.40 | 119.62 | 2.75 | 2.70 | 2.51 | 2.27 | 1.75 | 1.20 | 0.61 |
| DXS8059 | 141.90 | 121.99 | 2.29 | 2.25 | 2.10 | 1.90 | 1.47 | 1.01 | 0.52 |
| DXS8057 | 144.20 | 123.30 | −∞ | −0.55 | 0.15 | 0.40 | 0.51 | 0.42 | 0.23 |
| DXS8009 | 148.40 | 125.90 | −∞ | −1.45 | −0.19 | 0.23 | 0.45 | 0.40 | 0.23 |
| DXS1047 | 150.30 | 128.80 | −∞ | −1.63 | −0.37 | 0.05 | 0.29 | 0.27 | 0.16 |
| DXS8074 | 152.70 | 133.81 | −∞ | −2.34 | −1.09 | −0.66 | −0.38 | −0.27 | −0.15 |
| DXS1062 | 153.70 | 137.03 | −∞ | −2.15 | −0.81 | −0.30 | 0.08 | 0.17 | 0.13 |
| DXS1205 | 163.70 | 139.99 | −∞ | 0.52 | 1.07 | 1.17 | 1.04 | 0.76 | 0.40 |
| DXS1227 | 164.70 | 140.53 | −∞ | −0.57 | 0.01 | 0.16 | 0.15 | 0.04 | −0.02 |
| DXS8106 | 173.60 | 141.91 | −∞ | −2.26 | −1.01 | −0.58 | −0.24 | −0.05 | 0.04 |
| DXS8043 | 176.70 | 143.73 | −∞ | −3.73 | −1.70 | −0.90 | −0.22 | 0.04 | 0.09 |
| DXS8091 | 186.30 | 147.31 | −∞ | −4.68 | −2.60 | −1.74 | −0.94 | −0.53 | −0.24 |
| DXS8069 | 190.40 | 149.31 | −∞ | −2.91 | −0.96 | −0.26 | 0.21 | 0.29 | 0.19 |
| DXS1073 | 196.50 | 153.39 | −∞ | −3.46 | −1.45 | −0.67 | −0.03 | 0.17 | 0.17 |
Figure 3 Pedigree and haplotype diagram of the family with X linked high myopia. Blackened bars indicate disease alleles. Filled squares represent individuals affected with high myopia.
Discussion
We report X linked recessive high myopia with reduced visual acuity in a Chinese family. The lack of macular degeneration seen in other types of hereditary retinopathy, the stationary reduction in visual acuity in all the affected individuals, and the reduced ERG cone response shown in the proband all suggest cone dysfunction in the affected individuals. Such phenotypes were also found in a further four large Chinese families with X linked recessive high myopia (manuscript in preparation). In this study, the high myopia is assigned to a new locus on chromosome Xq23–q25 between DXS1210 and DXS8057. Exclusion of other regions in the X chromosome including the MYP1 region, a maximum two point lod score of 2.75, and haplotype observation all support a new locus for X linked recessive high myopia in this family. There are about 101 genes in the linked interval. Possible candidate genes include, but are not limited to, GUCY2F, GLUD2, GRIA3, BIRC4, STAG2, and LRCH2.
The first locus (MYP1) for high myopia,13,17 which was excluded in this study by linkage and haplotype analysis, is at least 16.66 Mb (42 cM) away from the telomeric boundary of the new locus. Individuals from both branches of the current family show obligate recombinations with this region. MYP1 was mapped to Xq28 by linkage analysis of a large Danish family with Bornholm eye disease, using three markers. Exclusion of other regions of the X chromosome has not been carried out and the diagnostic criteria were not meet by all affected individuals in the linkage analysis in that family.13,27 Another family with X linked high myopia associated with cone dysfunction, of Danish origin, was mapped to Xq27.3–q28.17 The disease in the Chinese family shares some features observed in those two families mapped to MYP1, including high myopia, impaired visual acuity, a myopic fundus, and abnormal cone responses on ERG recordings. Colour vision defects are present in all affected members in the previous two families mapped to MYP1 but in only three of the five affected individuals (two with deutan and one with protan) in the Chinese families, although variations in the visual pigment genes was not responsible for myopia.17 Apart from high myopia, common myopia has been shown to be linked to Xp,28,29 which is away from the novel locus identified in the Chinese family. Ocular refraction has been shown to have sex linked effect but its linked region on the X chromosome has not been identified.30 Different prevalences of refractive errors have been observed between males and females.31 All these indicate the importance of X linked genes in myopia development.
In addition, X linked high myopia has been well documented in association with congenital stationary night blindness (CSNB1/NYX)22 and retinitis pigmentosa (RP2).23 Mild to high myopia has been observed in other X linked ocular diseases includes cone‐rod dystrophy32,33,34 and Aland eye disease.35 The loci for these myopia associated syndromes are located outside the linked interval of the high myopia in the Chinese family.
In summary, a novel locus for X linked recessive high myopia in a Chinese family is mapped to Xq23–q25 with the highest lod score being 2.75 for DXS1001 at θ = 0. This locus is at least 16.66 Mb away from MYP1. Refinement of the linkage region with additional families and screening candidate genes for mutation may lead to the identification of the defect gene.
Supplementary data can be viewed on the journal website (www.jmedgenet.com/supplemental)
Acknowledgements
We thank all patients and family members for their participation. This study was supported in part by the National 863 Plan of China (Z19‐01‐04‐02 to QZ; 04AA104092 to XG), National Natural Science Foundation of China (30572006 to QZ), Department of Science and Technology of Guangdong Province (99M04805G to QZ), Guangdong Natural Science Foundation (04009335 to XG), Returnee Foundation from Sun Yat‐sen University (3030901010022 to QZ), and Zhongshan Ophthalmic Centre (3031002006 to QZ).
Abbreviations
AMD - age related macular degeneration
ERG - electroretinogram
ISCEV - International Society for Clinical Electrophysiology of Vision
Footnotes
Conflicts of interest: none declared.
Supplementary data can be viewed on the journal website (www.jmedgenet.com/supplemental)
References
- 1.Feldkamper M, Schaeffel F. Interactions of genes and environment in myopia. Dev Ophthalmol 20033734–49. [DOI] [PubMed] [Google Scholar]
- 2.Saw S M. A synopsis of the prevalence rates and environmental risk factors for myopia. Clin Exp Optom 200386289–294. [DOI] [PubMed] [Google Scholar]
- 3.Sperduto R D, Seigel D, Roberts J, Rowland M. Prevalence of myopia in the United States. Arch Ophthalmol 1983101405–407. [DOI] [PubMed] [Google Scholar]
- 4.Choo V. A look at slowing progression of myopia. Lancet 20033611622–1623. [DOI] [PubMed] [Google Scholar]
- 5.He M, Zeng J, Liu Y, Xu J, Pokharel G P, Ellwein L B. Refractive error and visual impairment in urban children in southern China. Invest Ophthalmol Vis Sci 200445793–799. [DOI] [PubMed] [Google Scholar]
- 6.Fredrick D R. Myopia. BMJ 20023241195–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellwein L B. Updating the Hu 1981 estimates of the economic costs of visual disorders and disabilities ( http://www.nei.nih.gov/eyedata/hu_estimates.asp#note ) 2004
- 8.Hu D N. Prevalence and mode of inheritance of major genetic eye diseases in China. J Med Genet 198724584–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young T L, Ronan S M, Drahozal L A, Wildenberg S C, Alvear A B, Oetting W S, Atwood L D, Wilkin D J, King R A. Evidence that a locus for familial high myopia maps to chromosome 18p. Am J Hum Genet 199863109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naiglin L, Gazagne C, Dallongeville F, Thalamas C, Idder A, Rascol O, Malecaze F, Calvas P. A genome wide scan for familial high myopia suggests a novel locus on chromosome 7q36. J Med Genet 200239118–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paluru P, Ronan S M, Heon E, Devoto M, Wildenberg S C, Scavello G, Holleschau A, Makitie O, Cole W G, King R A, Young T L. New locus for autosomal dominant high myopia maps to the long arm of chromosome 17. Invest Ophthalmol Vis Sci 2003441830–1836. [DOI] [PubMed] [Google Scholar]
- 12.Pararajasegaram R. VISION 2020‐the right to sight: from strategies to action. Am J Ophthalmol 1999128359–360. [DOI] [PubMed] [Google Scholar]
- 13.Schwartz M, Haim M, Skarsholm D. X‐linked myopia: Bornholm eye disease. Linkage to DNA markers on the distal part of Xq. Clin Genet 199038281–286. [PubMed] [Google Scholar]
- 14.Young T L, Ronan S M, Alvear A B, Wildenberg S C, Oetting W S, Atwood L D, Wilkin D J, King R A. A second locus for familial high myopia maps to chromosome 12q. Am J Hum Genet 1998631419–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik J F. A new locus for autosomal dominant high myopia maps to 4q22–q27 between D4S1578 and D4S1612. Mol Vis 200511554–560. [PubMed] [Google Scholar]
- 16.Paluru P C, Nallasamy S, Devoto M, Rappaport E F, Young T L. Identification of a novel locus on 2q for autosomal dominant high‐grade myopia. Invest Ophthalmol Vis Sci 2005462300–2307. [DOI] [PubMed] [Google Scholar]
- 17.Young T L, Deeb S S, Ronan S M, Dewan A T, Alvear A B, Scavello G S, Paluru P C, Brott M S, Hayashi T, Holleschau A M, Benegas N, Schwartz M, Atwood L D, Oetting W S, Rosenberg T, Motulsky A G, King R A. X‐linked high myopia associated with cone dysfunction. Arch Ophthalmol 2004122897–908. [DOI] [PubMed] [Google Scholar]
- 18.Farbrother J E, Kirov G, Owen M J, Pong‐Wong R, Haley C S, Guggenheim J A. Linkage analysis of the genetic loci for high myopia on 18p, 12q, and 17q in 51 U.K. families. Invest Ophthalmol Vis Sci 2004452879–2885. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, Guo X, Xiao X, Yi J, Jia X, Hejtmancik J F. Clinical description and genome wide linkage study of Y‐sutural cataract and myopia in a Chinese family. Mol Vis 200410890–900. [PubMed] [Google Scholar]
- 20.Dietz H C, Cutting G R, Pyeritz R E, Maslen C L, Sakai L Y, Corson G M, Puffenberger E G, Hamosh A, Nanthakumar E J, Curristin S M.et al Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature 1991352337–339. [DOI] [PubMed] [Google Scholar]
- 21.Ahmad N N, Ala‐Kokko L, Knowlton R G, Jimenez S A, Weaver E J, Maguire J I, Tasman W, Prockop D J. Stop codon in the procollagen II gene (COL2A1) in a family with the Stickler syndrome (arthro‐ophthalmopathy). Proc Natl Acad Sci USA 1991886624–6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bech‐Hansen N T, Naylor M J, Maybaum T A, Sparkes R L, Koop B, Birch D G, Bergen A A, Prinsen C F, Polomeno R C, Gal A, Drack A V, Musarella M A, Jacobson S G, Young R S, Weleber R G. Mutations in NYX, encoding the leucine‐rich proteoglycan nyctalopin, cause X‐linked complete congenital stationary night blindness. Nat Genet 200026319–323. [DOI] [PubMed] [Google Scholar]
- 23.Schwahn U, Lenzner S, Dong J, Feil S, Hinzmann B, van Duijnhoven G, Kirschner R, Hemberger M, Bergen A A, Rosenberg T, Pinckers A J, Fundele R, Rosenthal A, Cremers F P, Ropers H H, Berger W. Positional cloning of the gene for X‐linked retinitis pigmentosa 2. Nat Genet 199819327–332. [DOI] [PubMed] [Google Scholar]
- 24.International Standardization Committee Standard for clinical electroretinography. Arch Ophthalmol 1989107816–819. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Q, Xiao X, Shen H, Li S, Jiang F. Correlation of gene structure and psychophysical measurement in red‐green color vision deficiency in Chinese. Jpn J Ophthalmol 200044596–600. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Q, Minoda K. Detection of congenital color vision defects using heteroduplex‐SSCP analysis. Jpn J Ophthalmol 19964079–85. [PubMed] [Google Scholar]
- 27.Haim M, Fledelius H C, Skarsholm X‐linked myopia in Danish family. Acta Ophthalmol (Copenh) 198866450–456. [DOI] [PubMed] [Google Scholar]
- 28.Stambolian D, Ibay G, Reider L, Dana D, Moy C, Schlifka M, Holmes T, Ciner E, Bailey‐Wilson J E. Genomewide linkage scan for myopia susceptibility loci among Ashkenazi Jewish families shows evidence of linkage on chromosome 22q12. Am J Hum Genet 200475448–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stambolian D, Ciner E B, Reider L C, Moy C, Dana D, Owens R, Schlifka M, Holmes T, Ibay G, Bailey‐Wilson J E. Genome‐wide scan for myopia in the Old Order Amish. Am J Ophthalmol 2005140469–476. [DOI] [PubMed] [Google Scholar]
- 30.Biino G, Palmas M A, Corona C, Prodi D, Fanciulli M, Sulis R, Serra A, Fossarello M, Pirastu M. Ocular refraction: heritability and genome‐wide search for eye morphometry traits in an isolated Sardinian population. Hum Genet 2005116152–159. [DOI] [PubMed] [Google Scholar]
- 31.Zhao J, Mao J, Luo R, Li F, Munoz S R, Ellwein L B. The progression of refractive error in school‐age children: Shunyi district, China. Am J Ophthalmol 2002134735–743. [DOI] [PubMed] [Google Scholar]
- 32.Mantyjarvi M, Nurmenniemi P, Partanen J, Myohanen T, Peippo M, Alitalo T. Clinical features and a follow‐up study in a family with X‐linked progressive cone‐rod dystrophy. Acta Ophthalmol Scand 200179359–365. [DOI] [PubMed] [Google Scholar]
- 33.Jalkanen R, Demirci F Y, Tyynismaa H, Bech‐Hansen T, Meindl A, Peippo M, Mantyjarvi M, Gorin M B, Alitalo T. A new genetic locus for X linked progressive cone‐rod dystrophy. J Med Genet 200340418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebenezer N D, Michaelides M, Jenkins S A, Audo I, Webster A R, Cheetham M E, Stockman A, Maher E R, Ainsworth J R, Yates J R, Bradshaw K, Holder G E, Moore A T, Hardcastle A J. Identification of novel RPGR ORF15 mutations in X‐linked progressive cone‐rod dystrophy (XLCORD) families. Invest Ophthalmol Vis Sci 2005461891–1898. [DOI] [PubMed] [Google Scholar]
- 35.Alitalo T, Kruse T A, Forsius H, Eriksson E W, de la Chapelle A. Localization of the Aland Island eye disease locus to the pericentromeric region of the X chromosome by linkage analysis. Am J Hum Genet 19914831–38. [PMC free article] [PubMed] [Google Scholar]