Abstract
Objective
To screen cDNA for NLGN3 and NLGN4 from lymphoblastoid cells from autistic subjects.
Methods and results
10 young autistic females and 30 non‐autistic subjects were studied for alterations in two X linked genes, NLGN3 and NLGN4. A novel NLGN4 isoform lacking exon 4, which occurred de novo on the paternal allele, was identified in one of the autistic females. Monoallelic expression of NLGN4 was seen in this subject and in 11 of 14 informative autistic and non‐autistic females using a single nucleotide polymorphism found at 3′ UTR. Additionally, the NLGN3 transcript was present in two isoforms (with and without exon 7) in nine of 10 autistic females and in 30 non‐autistic subjects, including parents of the autistic female having only the complete transcript with exon 7, and from the whole brain of a control. The novel truncated NLGN3 product may have a regulatory role, as reported in other proteins (for example, vasopressin receptor) by attenuating the function of the full length isoform, resulting in a reduction of the mature protein. Three dimensional protein structures were characterised using comparative modelling, and significant changes were suggested in the protein cores for these two neuroligin isoforms.
Conclusions
Splice variants may lead to potentially abnormal neuroligins in the causation of autism spectrum disorders.
Keywords: NLGN3, NLGN4, autism, splicing, three dimensional structure
Autism (MIM 209850) is a genetically heterogeneous early onset neurodevelopmental disorder with developmental difficulties noted by three years of age. It belongs to a group of conditions known as autism spectrum disorders (ASD), including classical autism, pervasive developmental disorder–not otherwise specified (PDD‐NOS), and Asperger syndrome. Diagnostic features for classical autism include significant impairment in communication and social interaction accompanied by a pattern of repetitive or stereotypical behaviours and interests.1
To date, several candidate genes have been examined to evaluate their possible associations with autism. These have been selected on the basis of supporting linkage data, cytogenetic evidence, or clinical presentation. The main screening method for candidate genes is direct DNA sequencing of exons and their flanking intronic regions using genomic DNA from subjects with autism compared with controls. The proposed candidate genes for autism include the neuroligins which hold promise to uncover molecular causes of this complex neurological disorder.
Neuroligins are adhesion proteins that bind to β‐neurexin, a cell surface protein, to form functional synapses.2 Neuroligins 1, 3, and 4 are localised to excitatory glutamatergic axons, while neuroligin 2 is localised to inhibitory GABA axons.3 Neuroligins are composed of an extended N‐terminal extracellular region containing a large esterase‐homology domain necessary for the activity of neuroligins, and a short cytoplasmic domain.2
The neuroligin 3 gene (NLGN3), localised to Xq13, is composed of eight exons with the start codon in exon 2.4 Exons 7 and 8 are the largest exons, encoding about 65% of the NLGN3 protein. A point mutation of NLGN3 (R451C) was first reported in two affected brothers, one with typical autism and the other with Asperger syndrome.5 This mutation resulted in intracellular retention of the mutant protein, causing loss of synaptic function.6
The fourth member of the neuroligin gene family, NLGN4, localised to Xp22.3, is composed of six exons and has 63–73% amino acid identity with other human neuroligin genes.7 Jamain et al5 identified a 1 base pair (bp) insertion (1186insT) in the NLGN4 gene in two affected sons from a Swedish family (one with autism and the other with Asperger syndrome), resulting in a frame shift mutation causing a premature termination (D396X). Later, a 2 bp deletion in this neuroligin gene was reported in a large French family including several male members affected by non‐specific X linked mental retardation, with or without autism or PDD‐NOS.8 Functional analysis showed that the D396X frame shift mutation resulted in intracellular retention of the NLGN4 mutant protein and loss of synaptic function.6
Recently, Chih et al2 reported additional data using electrophysiological studies on mutant neuroligins carrying deletions in either the cytoplasmic tail or in the esterase homology domain which emphasised the critical role of the neuroligin genes in maintaining a functional balance between excitatory and inhibitory synapses in hippocampal neurones. They concluded that neuroligin defects led to selective loss of inhibitory function and abnormal excitatory/inhibitory balance in neurones. Such a defect is believed to play a role in autism.
Despite these positive findings, mutation screening of neuroligins using genomic DNA from subjects with ASD suggest that these mutations are not common or occur at a low frequency in the autistic population.9,10,11,12 To further investigate the role of neuroligins in autism, we screened the NLGN3 and NLGN4 genes using cDNA generated from actively growing lymphoblastoid cell lines from autistic females and non‐autistic males and females.
Methods
Subjects
Our autistic group consisted of 10 females diagnosed with classical autism (one from simplex and nine from multiplex families). These autistic females had skewed X chromosome inactivation (that is, a ratio of >80:20) using the androgen receptor gene,13 and were selected from a previous study on X chromosome inactivation patterns in females with autism.14 The control group consisted of 30 subjects (12 female and 18 male) without a history of autism or mental retardation. Autistic subjects were ascertained from the Autism Genetics Resource Exchange (AGRE), a publicly available biomaterials repository located in Los Angeles. The diagnosis of autism was established in the affected females with the use of the Autism Diagnostic Interview–Revised (ADI‐R).1 Chromosome analysis and fragile X testing were reportedly normal. DNA and clinical information (medical and pedigree data, diagnostic assessments and scores) were obtained on each subject from AGRE.
cDNA mutation screening
Total RNA was extracted from growing lymphoblastoid cell lines. One microgram of total RNA was used to synthesise cDNA using transcriptor reverse transcriptase kit from Roche (Indianapolis, Indiana, USA). The entire coding sequences of the NLGN3 and NLGN4 genes were amplified using three pairs of primers for each one of these candidate genes: NLGN3‐244F 5′‐TGATGCTGTCACCCTGGAGTC‐3′ and NLGN3‐1172R: 5′‐GGATGATGGCTCTCTGGAAAAG‐3′ generating a 929 bp polymerase chain reaction (PCR) product;NLGN3‐1049F 5′‐ATTGCCTTCTTCGGGGGAG‐3′ and NLGN3‐2037R: 5′‐TGGTCTCGGGGATTGTATTTG‐3′ generating a 989 bp PCR product; NLGN3‐1884F 5′‐ATGTTATGCTCAGTGCTGTCGTC‐3′ and NLGN3‐3005R: 5′‐GGGTTTGTTCAGGTTTACTTCCG‐3′ generating a 1122 bp PCR product; NLGN4‐184F 5′‐TTGTCCCTGGAGGTGTTGG‐3′ and NLGN4‐1458R 5′‐TGATGGTCTGCTGGATGAGC‐3′ generating a 1275 bp PCR product; NLGN4‐832F 5′‐TGCTGCCCATCTGGTTTACC‐3′ and NLGN4‐1664R 5′‐GGACACGGAGAAGTCAAAGTCG‐3′ generating a 833 bp PCR product; and NLGN4‐1603F 5′‐TGGACGGCATCGTGGATAAC‐3′ and NLGN4‐2946R 5′‐GCAGAGGGATAGGAAGGGAAATAG‐3′ generating a 1344 bp PCR product. In order to screen the intron/exon junction of exon 4 in subject HI 1920, we amplified a fragment from genomic DNA using intronic primers: exon 4‐F 5′‐TTCAGTGAGAAAGACAGGCG‐3′ and exon 4‐R 5′‐CAGAGCAAATGGAACAAAAGC‐3′. We then sequenced the amplified fragment with two internal primers (exon 4A‐R 5′‐CGAGCCAAAGATGGTCACTC‐3′ and exon 4B‐F 5′‐AAGCACTGCGGTGGATTG‐3′) to screen the splice junctions.
PCR was carried out in a total volume of 25 μl using FailSafe PCR premix selection kit according to the manufacturer's instructions (Epicentre, Madison, Wisconsin, USA). The amplified products were run on a 2% agarose gel, and the DNA bands were visualised by use of ethidium bromide staining under ultraviolet light. For sequence analysis, amplified products were purified using centrisep columns (Princeton Separations, Adelphia, New Jersey, USA). The purified PCR fragments were then sequenced with capillary electrophoresis ABI 3100 sequencer (Applied Biosystems, Foster City, California, USA). The primers for the NLGN3 (NM_018977) and NLGN4 (NM_020742) genes were designed using MacVector version 2.0 computer program (ACCELRYS, San Diego, California, USA).
Single nucleotide polymorphism analysis and allelic expression of NLGN4
PCR primers NLGN4‐2515F (5′‐CGCTCCTCTTCCTCAACATC‐3′) and NLGN4‐3507R: (5′‐TCAGGCTGGCAAAACACTC‐3′) were designed to amplify a 993 bp fragment containing the A/G polymorphism at position 3199 in the 3′UTR. The forward primer is located in the last coding exon and the reverse primer is inside the untranslated region. The same primer pair was used to amplify the corresponding region from both genomic DNA and cDNA. An internal primer upstream of the polymorphism position (NLGN4‐3060F: 5′‐AAAAGGCAGTCATCCCATCCCG‐3′) was used for DNA sequencing. For females having two different alleles in their genomic DNA (informative for the single nucleotide polymorphism (SNP)), the same region was amplified from cDNA to determine allelic expression. To evaluate whether or not exon 4 was present in the expressed allele for subject HI 1920, the cDNA was amplified with forward and reverse primers, NLGN4‐832F and NLGN4‐3507R, which span both the exon 4 and the SNP region. The amplified fragment of about 2.5 kb size was then sequenced with NLGN4‐832F primer (to verify the presence of exon 4) and NLGN4‐3060F (to screen for the SNP).
Structural modelling
Comparative modelling was carried out in order to generate approximate three dimensional structures for the esterase‐like domains of the NLGN3 (NCBI accession number NM_018977) and the X linked NLGN4 (NM_020742) through analogy to previously characterised human,15 mouse,16 and Pacific electric ray (Torpedo californica)17 acetylcholinesterase crystal structures. To establish the underlying structural relations, we aligned the neuroligin sequences to those of human, mouse, and Pacific electric ray acetylcholinesterases with the Clustal‐W program,18 using the BLOSUM‐30 substitution matrix,19 and standard gap penalties of 10 for opening and 0.1 for extension. Portions of the NLGN3 and NLGN4 sequences that lay outside the esterase‐like domain (NLGN3 residues 1–37, 616–828; NLGN4 residues 1–41, 602–816) were omitted from consideration. Based on the resulting multiple sequence alignment, we used the Modeller program20 to predict the corresponding three dimensional neuroligin structures by fitting the aligned NLGN3 and NLGN4 sequences to a consensus structural template derived from the three aforementioned crystal structures. In the modelling process we applied default restraint settings, and effected a rigorous relaxation protocol involving five simulated annealing relaxation cycles (4.4 ps stepwise warming from 0–1000 K, followed by 19.2 ps stepwise cooling back down to 300 K, all done through Charmm force field and charges).21 The resulting structures were visualised and rendered in SYBYL 7.0 (The Tripos Associates, St Louis, Missouri, USA).
Results
cDNA screening
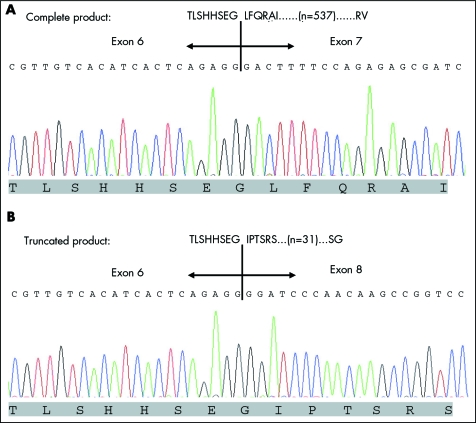
The entire coding sequence of both NLGN3 and NLGN4 genes were screened in 10 autistic females and 30 non‐autistic controls. In nine of the 10 autistic females, the NLGN3 transcript was present in two isoforms (with and without exon 7), shown in fig 1. This novel isoform was predicted to result in premature termination shortly after exon 6, producing a truncated product. We identified these two transcripts in both males and females using lymphoblastoid cDNA from 30 non‐autistic subjects including parents of the autistic female not producing a transcript without exon 7 (subject ID: HI 0959) and a control female whole brain.
Figure 1 A novel NLGN3 transcript lacking exon 7. Partial translation and nucleotide sequences of (A) the intact NLGN3 gene and (B) the truncated NLGN3‐exon 7 missing isoform. Amino acids encoded/ predicted by the representative nucleotides are shown below each electropherograms. The intervening region contains 537 and 31 residues in the complete and truncated products, respectively.
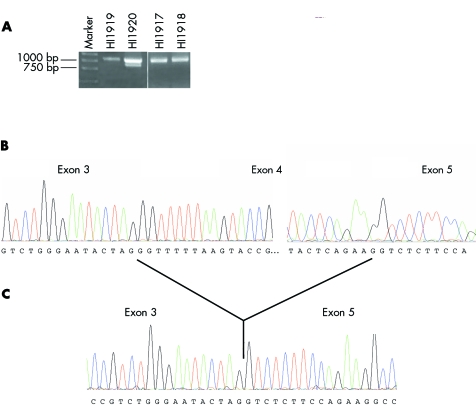
No mutations or polymorphisms were identified in the coding sequences of the NLGN4 gene in the 10 autistic females. However, when amplifying a fragment from cDNA encompassing the middle section of the gene (from exon 2 to exon 5), in one autistic female (subject ID: HI 1920) there was evidence for the presence of a shorter fragment in addition to the fragment of expected size (that is, a faint band was seen below the band of expected size on the agarose gel). To evaluate the presence of the second unexpected band, this PCR product was reamplified (fig 2A). The two fragments were excised from the gel and their DNA isolated and sequenced separately. DNA sequencing generated from both bands matched to NLGN4 but exon 4 was missing in the lower (smaller) band (fig 2, panels B and C). The size difference between these two bands corresponded to exon 4 (that is, 185 nt). When exon 4 and its flanking intronic region (about 200 nt from each side) was amplified and sequenced from the subject's genomic DNA, no changes were detected in the splice junctions. No other tissue was available from subject HI 1920 to examine for the presence of the NLGN4 transcript without exon 4.
Figure 2 A novel NLGN4 transcript lacking exon 4. The gel image illustrating the amplified product of the NLGN4 gene for an autistic subject (HI 1920) and other family members is shown in (A). The corresponding partial DNA sequencing for this autistic subject (HI 1920) generated from (B) the top band and (C) the lower band is demonstrated. The smaller band reveals exon 4 skipping which results in an in‐frame deletion of 62 amino acids.
The NLGN4 allele with the exon 4 skipping is predicted to result in an in‐frame exclusion of 62 amino acids. Protein sequence alignment of the four members of the neuroligin gene family (NLGN1–4) has demonstrated that amino acids encoded by NLGN4 exon 4 are highly conserved among different neuroligin proteins (fig 3). The NLGN4 transcript lacking exon 4 was not found in lymphoblastoid cDNA from any of the 30 non‐autistic subjects including parents of HI1920 screened in our study.
Figure 3 Multiple protein alignment. Partial amino acid sequence alignment of the four members of the human neuroligin gene family (NLGN4, NLGN3, NLGN2, NLGN1) and their homologous gene in mouse, rat, and chimpanzee corresponding to exon 4 in NLGN4. The human Y chromosome homologue (NLGN4Y) sequence is also shown. Amino acids that differ from the human X linked form of NLGN4 (NLGN4) sequence are highlighted. The conserved cysteine residue7 is marked by asterisks. The conserved glycine residue, thought to be aligned with the triad's serine in cholinesterases,7 is marked by a vertical arrow.
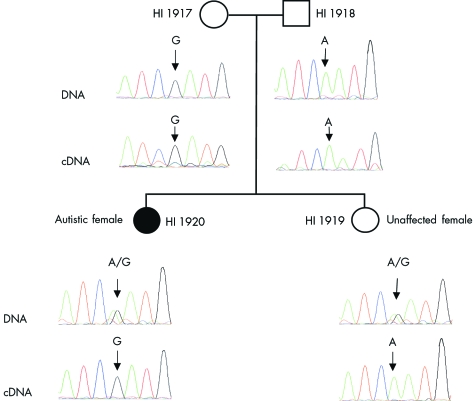
To investigate the parental origin of the allele with exon 4 skipping in autistic subject HI 1920, the 3′ untranslated region (3′UTR) was screened for the presence of a polymorphism. One SNP (A/G) in NLGN4 was identified in the genomic DNA at position 3199 in this autistic female with exon 4 skipping. However, only one of the two alleles (G) was detected when the same region was amplified from her cDNA (fig 4). DNA sequencing using a primer upstream of exon 4 indicated the presence of this exon in the expressed G allele in subject HI 1920. Monoallelic expression was also observed in her unaffected sister except for the A allele. When other family members were screened for this SNP, it was shown that the maternal allele (G) was expressed in the autistic female while only the paternal allele (A) was detected in her unaffected sister (fig 4). Two other autistic families were screened for this SNP but no consistent pattern was observed for the parental origin of the expressed allele. The monoallelic expression of NLGN4 appears to be random and not caused by imprinting.
Figure 4 Single nucleotide polymorphism (SNP) analysis and monoallelic expression of the NLGN4 gene. Inheritance of the SNP found in the 3′ untranslated (3′UTR) region of the NLGN4 gene is illustrated for the autistic female (HI 1920) with exon 4 missing allele, along with her parents and unaffected sibling. DNA sequencing from genomic DNA shows the presence of two alleles in HI 1920 and her sister (HI 1919), while only one of the two alleles (maternal and paternal allele, respectively) was detected in their cDNA. As shown in fig 2A, two products were polymerase chain reaction (PCR) amplified for subject HI 1920 after second amplification. A weak signal representing a smaller band was present in the first PCR amplification (data not shown). However, when a 2.5 kb size fragment encompassing both exon 4 and the SNP was sequenced, only one product was seen for subject HI 1920. It should be noted that if a second product (lacking exon 4) is present but has an intensity of less than 10% it could not be observed in DNA sequencing.
Genomic DNA from a total of 25 females (affected and unaffected) was screened for the SNP at the 3′UTR of the NLGN4 gene and 14 females (56%) were found heterozygous. Eleven of 14 informative females (79%) displayed monoallelic expression in their lymphoblastoid cDNA while the remaining showed expression from both alleles. Furthermore, cDNA from the frontal cortex, visual cortex, and pons were also available for one of these informative non‐autistic females for this SNP (A/G). Monoallelic expression of the A allele was seen in her frontal cortex and pons while both alleles were expressed in her lymphoblastoid cells and visual cortex.
Using the most recent assembly (May 2004) from the UCSC genome browser (http://genome.ucsc.edu), two RefSeq accessions (NM_020742 and NM_181332) were identified for NLGN4, known as variant 1 and variant 2, respectively. These two NLGN4 variants encode the same protein as they differ only in the 5′UTR region. Furthermore, sequences for two mRNA with alternative splice exons, AY358562 and AX773938, have been found to match with NLGN4. Sequence alignment between these mRNA and NM_020742 shows inclusion of 60 nt (before exon 3) and 112 nt (before exon 4) in AY358562 and AX773938, respectively. The primers used in our study flanked this region but no alternative splicing was seen, as suggested by the UCSC genome browser. For NLGN3, one RefSeq accession, NM_018977, was found in the UCSC browser; however, Philibert et al4 reported two transcripts for this gene. The longer transcript (AF217411) consisted of sequences from all exons except exon 3 and a shorter transcript (AF217412) using exon 3 terminated in the poly A tail shortly before the end of exon 7. We found transcripts with and without exon 3 in our study but our primers would not have picked up products with premature termination in the poly A tail.
Structural modelling
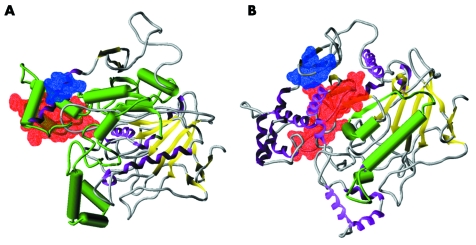
Exploiting a reasonable level of sequence similarity between the neuroligin esterase‐like domains and acetylcholinesterase, a prior comparative modelling study was carried out to predict the structure of human neuroligin 1.22,23 Strong homology with human acetylcholinesterase also make NLGN3 (33% identical, 62% similar) and NLGN4 (33% identical, 63% similar) good candidates for such analysis, offering reasonable expectations that the basic underlying fold structure will be well represented, and that predicted intra‐chain interactions, if approximate, should generally be qualitatively correct. There are two gaps of note in the neuroligin/acetylcholinesterase alignments that should be addressed: segments of 9 and 13 residues in NLGN4 and of eight and 24 residues in NLGN3 for which there is no analogous complement in the acetylcholinesterase templates. In our predicted models, these gaps appear in surface regions and, based on the simulated annealing structural relaxation, appear to correspond to unstructured segments that should have little effect on the fold structure. Taking all of this into account, it is reasonable to assert that both NLGN3 and NLGN4, depicted in fig 5, panels A and B, respectively, correspond to three‐layer (αβα) sandwiches with the basic Rossman fold topology characteristic of cholinesterases.
Figure 5 Predicted three dimensional structure for the esterase‐like domains of the NLGN3 and NLGN4 proteins. (A) Comparative three dimensional model of human NLGN3. Protein segment corresponding to exon 7 is rendered in green, with helices shown as cylinders and sheets as flat arrows. Spatial extents of the putative EF‐hand domain and important mutagenesis residues (K620, V621, H626, L627) are shown as red and blue line surfaces, respectively. Remainder of the protein is rendered according to secondary structure features with magenta helices, yellow sheets, and grey coils. (B) Comparative three dimensional model of human NLGN4. The protein segment corresponding to exon 4 is rendered in green, with helices shown as cylinders and sheets as flat arrows. Spatial extents of the putative EF‐hand domain and important mutagenesis residues (K586, V587, E592, L593) are shown as red and blue line surfaces, respectively. The remainder of the protein is rendered according to secondary structure features with magenta helices, yellow sheets, and grey coils.
Discussion
Except for the one autistic female, who only produced the complete NLGN3 transcript (HI 0959), the truncated isoform of NLGN3 was present in all subjects screened including the parents of this autistic female. It therefore appears that the truncated isoform is a normal product. However, this truncated product may not have normal neuroligin activity owing to the absence of an intact esterase domain involving exons 7 and 8. The novel isoform lacking exon 7 would not have been detected in the original description of NLGN3 which used a probe corresponding to exons 7 and 8.4
We suggest that this novel truncated NLGN3 product may have a regulatory role in the activity of neuroligins by attenuating the function of the full length isoform resulting in a reduction of the mature protein. This potential regulatory impact on the function of the complete transcript might be a necessary factor in maintaining balance between excitatory/inhibitory neurones. Similar attenuating regulatory mechanisms using splicing variants have been shown for other proteins such as the vasopressin V2 receptor,24 whereby two alternatively spliced transcripts are generated. The protein encoded by the small transcript (V2b splice variant) is retained inside the cell while the longer wild type variant (V2a) is found at the cell surface. Expression of the splice variant V2b downregulates the surface expression of the V2a receptor by forming V2aV2b heterodimers inside the cell.24 Such a mechanism may occur in the regulation of NLGN3 function. We suggest that the absence of the truncated product in our affected female resulting in only a full length NLGN3 isoform may have contributed to her autism.
The recent reports of allelic variation in gene expression for an unexpectedly large number of genes have shown that preferential expression of one allele is common in the human genome.25 Thus, unequal expression of two alleles may contribute to human variability and may have implications for disease susceptibility.25,26 For example, study of allelic variation in gene expression for 602 genes in kidney and liver tissues from seven individuals showed preferential expression of one allele (greater than twofold difference) in at least one individual for 326 genes (54%). Furthermore, a significant difference (greater than fourfold) between the expression level of the two alleles was also observed for 170 of these studied genes (28%).26
In a recent paper published in Nature, a comprehensive X inactivation profile of the human X chromosome genes was generated using nine rodent/human somatic cell hybrid fibroblast lines.27 In this hybrid system, expression of each transcript was examined using PCR with reverse transcription to determine if expression was detected only from hybrid lines retaining the active X (that is, the gene is subject to inactivation) or from both the active and inactive X hybrid lines (that is, the gene escapes from inactivation). This study showed that about 15% of X linked genes escaped inactivation, including NLGN4 which was expressed in all inactive X hybrids (9/9). A quantitative assay based on expressed single nucleotide polymorphisms was then used to evaluate allelic expression of 94 genes in skin cells from 40 women with complete skewing of X inactivation. This quantitative assay showed a variable expression level from the inactive X (from >75% to ∼25% of the active X expression) even for genes that escape from inactivation. Interestingly, only 12 of 20 genes expressed from all the inactive X hybrids (9/9) were biallelically expressed in all samples. However, some of these genes (5/20) that were assumed to escape inactivation were biallelic in only some fibroblast samples, and three genes were monoallelic in all samples. These data suggested a significant allelic variation in levels of X linked gene expression in females. Allelic expression was measured only for a subset of genes that escape from inactivation, not including NLGN4. Our allelic expression based on direct sequencing of a SNP at 3′UTR was not quantitative but clearly suggested that, for the NLGN4 gene which is reported to escape from inactivation,27 monoallelic expression can be seen in lymphoblastoid cell lines as well as in brain tissue in some females.
In our study, a novel splice variant of the NLGN4 gene lacking exon 4 was detected in one autistic female and none of the 30 non‐autistic subjects. DNA sequencing of the flanking intronic region of exon 4 in this autistic female did not reveal any sequence changes. However, it is possible that changes in a functionally important intronic region farther away from the splice sites might be responsible for the observed exon skipping. To evaluate this possibility, we propose to examine the entire intronic sequence upstream of this exon.
The possible functional implication of this novel NLGN4 isoform remains to be determined. However, findings in our study suggest that this NLGN4 isoform may be contributing to the autism in this affected female. First, NLGN4 exon 4 encodes for 62 residues containing two conserved amino acids: G254 (a residue in the sequence position corresponding to the catalytic serine in cholinesterases)7 and C259 (conserved cysteine).7 Furthermore, exon 4 encodes part of the highly conserved esterase‐like domain which is necessary for the activity of the neuroligins.2 Second, this isoform was not seen in 30 non‐autistic subjects including her unaffected parents and sister. Third, SNP analysis indicated monoallelic expression of maternal and paternal NLGN4 allele for this autistic female and her unaffected sister, respectively, in lymphoblastoid cells. DNA sequencing of the preferentially expressed allele (maternal) in this autistic subject showed the presence of exon 4. The intact paternal allele was expressed in the unaffected sister. Thus it appears that exon 4 skipping occurred de novo in the paternal allele in this autistic female. Fourth, the inconsistent expression pattern of NLGN4 was seen in four different tissues including frontal cortex, lymphoblastoid cells, pons, and visual cortex available from one control female without autism. Therefore, we suggest that if the NLGN4 allele lacking exon 4 is preferentially expressed in the brain of this autistic subject it may reduce functional activity of the encoded NLGN4 protein and contribute to her autism.
Neuroligins are cell surface proteins with high binding affinity to β‐neurexins. A striking feature of the neurexin genes is that thousands of neurexin proteins may be synthesised from only three genes through the use of two alternative promoters and alternative splicing.28,29 The extensive alternative splicing of the neurexins may be functionally important and play a regulatory role in controlling their interactions with ligands.29 Thus it is not surprising to see another layer of gene expression regulation (that is, monoallelic expression) existing for NLGN4, a member of the neuroligin gene family.
In the cases of both NLGN3 and NLGN4, our structural models predict that the segments corresponding to the alternate splices of interest to this work (exon 7 in NLGN3 and exon 4 in NLGN4) participate in both surface and core structural roles. In the core region, the splice segments comprise a part of the central β sheet and also entail at least one embedded helix. From modelling alone, it is difficult to quantify precisely the impact of an alternate splice on the overall protein structure; however, the presence of splice segment elements within the protein core effectively guarantees that an alternate splice missing these elements would show an adjusted fold as required for structural equilibrium in a polar medium. Furthermore, it appears that the splice segments in both NLGN3 and NLGN4 impinge on regions important to neuroligin synaptogenic function. We find in both cases that the splice segment borders directly on a cluster of residues (blue line surface, corresponding to K620 + V621 + H626 + L627 for NLGN3; and K586 + V587 + E592 + L593 for NLGN4) identified through mutagenesis studies as being critical to synaptogenesis.30 The splice segments also appear to affect a sequence motif that has been identified as a putative EF‐hand structure (red line surface)—a prevalent Ca2+ binding motif in cytoplasmic proteins that has also been identified in extracellular species31 and has thus been suggested as a possible site for Ca2+ mediated protein–protein binding in neuroligins.22 These EF‐hand domains appear to manifest themselves in our predicted neuroligin structures as highly electronegative patches arising from a convergence of hydroxy and carboxy side chains: D362, T370, D373, S377, and D381 in the case of NLGN3; and D344, D353, D355, S357, and S359 for NLGN4. In the case of NLGN4, the splice segment closely borders the putative EF‐hand, and in the NLGN3 case, the entire motif is incorporated as a subset of the splice segment. The cumulative structural evidence therefore suggests that the absence of exon 4 segment of NLGN4 would have a substantial effect on the protein's basic function and that an even greater effect is likely for NLGN3 in the absence of its exon 7.
Conclusions
Our study suggests that other forms of genetic alteration (other than mutations) including splice variants may lead to potentially abnormal function of neuroligins in autism spectrum disorder. Using different screening strategies with the evaluation of transcripts instead of genomic sequencing alone may shed light on changes in neuroligin function. Based on preliminary examination of clinical data available from AGRE, no unusual features were found in the two autistic females with the specific neuroligin splice variants compared with the other subjects we examined. Additional studies are under way to characterise these splicing variants further, along with the frequency with which they are found in ASD subjects.
Acknowledgements
Lymphoblastoid cell lines from autistic subjects were provided by the Autism Genetics Research Exchange (AGRE) (Los Angeles). Partial funding support for this study was from CMH Special Gift Funds (GL 01.2650) and CMH Physician Scientist Award.
Abbreviations
AGRE - Autism Genetics Resource Exchange
ASD - autism spectrum disorder
PDD‐NOS - pervasive developmental disorder–not otherwise specified
SNP - single nucleotide polymorphism
UTR - untranslated region
Footnotes
Conflicts of interest: none declared.
References
- 1.Lord C, Rutter M, Le Couteur A. Autism diagnostic interview–revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 199424659–685. [DOI] [PubMed] [Google Scholar]
- 2.Chih B, Engelman H, Scheiffele P. Control of excitatory and inhibitory synapse formation by neuroligins. Science 20053071324–1328. [DOI] [PubMed] [Google Scholar]
- 3.Graf E R, Zhang X, Jin S X, Linhoff M W, Craig A M. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell 20041191013–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Philibert R A, Winfield S L, Sandhu H K, Martin B M, Ginns E I. The structure and expression of the human neuroligin‐3 gene. Gene 2000246303–310. [DOI] [PubMed] [Google Scholar]
- 5.Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg I C, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T. Mutations of the X‐linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet 20033427–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chih B, Afridi S K, Clark L, Scheiffele P. Disorder‐associated mutations lead to functional inactivation of neuroligins. Hum Mol Genet 2004131471–1477. [DOI] [PubMed] [Google Scholar]
- 7.Bolliger M F, Frei K, Winterhalter K H, Gloor S M. Identification of a novel neuroligin in humans which binds to PSD‐95 and has a widespread expression. Biochem J 2001356581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laumonnier F, Bonnet‐Brilhault F, Gomot M, Blanc R, David A, Moizard M P, Raynaud M, Ronce N, Lemonnier E, Calvas P, Laudier B, Chelly J, Fryns J P, Ropers H H, Hamel B C, Andres C, Barthelemy C, Moraine C, Briault S. X‐linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet 200474552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vincent J B, Kolozsvari D, Roberts W S, Bolton P F, Gurling H M, Scherer S W. Mutation screening of X‐chromosomal neuroligin genes: no mutations in 196 autism probands. Am J Med Genet 200412982–84. [DOI] [PubMed] [Google Scholar]
- 10.Talebizadeh Z, Bittel D C, Veatch O J, Butler M G, Takahashi T N, Miles J H. Do known mutations in neuroligin genes (NLGN3 and NLGN4) cause autism? J Autism Dev Disord 200434735–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan J, Oliveira G, Coutinho A, Yang C, Feng J, Katz C, Sram J, Bockholt A, Jones I R, Craddock N, Cook E H, Vicente A, Sommer S S. Analysis of the neuroligin 3 and 4 genes in autism and other neuropsychiatric patients. Mol Psychiatry 200510329–332. [DOI] [PubMed] [Google Scholar]
- 12.Gauthier J, Bonnel A, St‐Onge J, Karemera L, Laurent S, Mottron L, Fombonne E, Joober R, Rouleau G A. NLGN3/NLGN4 gene mutations are not responsible for autism in the Quebec population. Am J Med Genet B Neuropsychiatr Genet 200513274–75. [DOI] [PubMed] [Google Scholar]
- 13.Allen R C, Zoghbi H Y, Moseley A B, Rosenblatt H M, Belmont J W. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen‐receptor gene correlates with X chromosome inactivation. Am J Hum Genet 1992511229–1239. [PMC free article] [PubMed] [Google Scholar]
- 14.Talebizadeh Z, Bittel D C, Veatch O J, Kibiryeva N, Butler M G. Non‐random X chromosome inactivation in females with autism. J Autism Dev Disord 2005161–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kryger G, Harel M, Giles K, Toker L, Velan B, Lazar A, Kronman C, Barak D, Ariel N, Shafferman A, Silman I, Sussman J L. Structures of recombinant native and E202Q mutant human acetylcholinesterase complexed with the snake‐venom toxin fasciculin‐II. Acta Crystallogr D Biol Crystallogr 2000561385–1394. [DOI] [PubMed] [Google Scholar]
- 16.Bourne Y, Taylor P, Bougis P E, Marchot P. Crystal structure of mouse acetylcholinesterase. A peripheral site‐occluding loop in a tetrameric assembly. J Biol Chem 19992742963–2970. [DOI] [PubMed] [Google Scholar]
- 17.Dvir H, Wong D M, Harel M, Barril X, Orozco M, Luque F J, Munoz‐Torrero D, Camps P, Rosenberry T L, Silman I, Sussman J L. 3D structure of Torpedo californica acetylcholinesterase complexed with huprine X at 2. 1 A resolution: kinetic and molecular dynamic correlates, Biochemistry 2002412970–2981. [DOI] [PubMed] [Google Scholar]
- 18.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position‐specific gap penalties and weight matrix choice. Nucleic Acids Res 1994224673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henikoff S, Henikoff J G. Performance evaluation of amino acid substitution matrices. Proteins 19931749–61. [DOI] [PubMed] [Google Scholar]
- 20.Marti‐Renom M A, Stuart A, Fiser A, Sánchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 200029291–325. [DOI] [PubMed] [Google Scholar]
- 21.MacKerell A D, Brooks B, Brooks C L, Nilsson L, Roux B, Won Y, Karplus M. CHARMM: The energy function and its parameterization with an overview of the program. In: Schleyer P, Schreiner P, Allinger N, Clark T, Gasteiger J, Kollman P, Schaefer H, eds. The encyclopedia of computational chemistry . Chichester: John Wiley & Sons, 1998, 1271–277.
- 22.Tsigelny I, Shindyalov I N, Bourne P E, Südhof T C, Taylor P. Common EF hand motifs in cholinesterases and neuroligins suggest a role for Ca2+ binding in cell surface associations. Protein Sci 20009180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffman R C, Jennings L L, Tsigelny I, Comoletti D, Flynn R E, Sudhof T C, Taylor P. Structural characterization of recombinant soluble rat neuroligin 1: mapping of secondary structure and glycosylation by mass spectrometry. Biochemistry 2004431496–1506. [DOI] [PubMed] [Google Scholar]
- 24.Sarmiento J M, Anazco C C, Campos D M, Prado G N, Navarro J, Gonzalez C B. Novel down‐regulatory mechanism of the surface expression of the vasopressin V2 receptor by an alternative splice receptor variant. J Biol Chem 200427947017–47023. [DOI] [PubMed] [Google Scholar]
- 25.Yan H, Yuan W, Velculescu V E, Vogelstein B, Kinzler K W. Allelic variation in human gene expression. Science 20022971143. [DOI] [PubMed] [Google Scholar]
- 26.Lo H S, Wang Z, Hu Y, Yang H H, Gere S, Buetow K H, Lee M P. Allelic variation in gene expression is common in the human genome. Genome Res 2003131855–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carrel L, Willard H F. X‐inactivation profile reveals extensive variability in X‐linked gene expression in females. Nature 2005434400–404. [DOI] [PubMed] [Google Scholar]
- 28.Ushkaryov Y A, Petrenko A G, Geppert M, Sudhof T C. Neurexins: synaptic cell surface proteins related to the alpha‐latrotoxin receptor and laminin. Science 199225750–56. [DOI] [PubMed] [Google Scholar]
- 29.Missler M, Sudhof T C. Neurexins: three genes and 1001 products. Trends Genet 19981420–26. [DOI] [PubMed] [Google Scholar]
- 30.Dean C, Scholl F G, Choih J, De Maria S, Berger J, Isacoff E, Scheiffele P. Neurexin mediates the assembly of presynaptic terminals. Nat Neurosci 20036708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pottgiesser J, Maurer P, Mayer U, Nischt R, Mann K, Timpl R, Krieg T, Engel J. Changes in calcium and collagen IV binding caused by mutation in the EF‐hand and other domains of extracellular matrix protein BM‐40 (SPARC, osteonectin). J Mol Biol 1994238563–574. [DOI] [PubMed] [Google Scholar]