Abstract
Background
Angelman syndrome (AS) is a neurodevelopmental disorder characterised by severe mental retardation, dysmorphic features, ataxia, seizures, and typical behavioural characteristics, including a happy sociable disposition. AS is caused by maternal deficiency of UBE3A (E6 associated protein ubiquitin protein ligase 3A gene), located in an imprinted region on chromosome 15q11‐q13. Although there are four different molecular types of AS, deletions of the 15q11‐q13 region account for approximately 70% of the AS patients. These deletions are usually detected by fluorescence in situ hybridisation studies. The deletions can also be subclassified based on their size into class I and class II, with the former being larger and encompassing the latter.
Methods
We studied 22 patients with AS due to microdeletions using a microarray based comparative genomic hybridisation (array CGH) assay to define the deletions and analysed their phenotypic severity, especially expression of the autism phenotype, in order to establish clinical correlations.
Results
Overall, children with larger, class I deletions were significantly more likely to meet criteria for autism, had lower cognitive scores, and lower expressive language scores compared with children with smaller, class II deletions. Children with class I deletions also required more medications to control their seizures than did those in the class II group.
Conclusions
There are four known genes (NIPA1, NIPA2, CYFIP1, & GCP5) that are affected by class I but not class II deletions, thus raising the possibility of a role for these genes in autism as well as the development of expressive language skills.
Keywords: Angelman Syndrome, comparative genomic hybridization, autism, genotype‐phenotype correlation, chromosome microdeletion
Angelman syndrome (AS) was initially described in 1965 by the paediatrician Harry Angelman in three unrelated children presenting with severe motor and cognitive delays, microcephaly, absent speech, ataxic gait, and seizures, in association with distinctive facial features that include a large open mouth, widely spaced teeth, midface retrusion, and prognathism.1 Subsequent clinical reports described typical behaviour in individuals with AS, including a happy sociable disposition, inappropriate laughter, and flapping of the arms.2,3 The 15q11‐q13 region was implicated in the pathogenesis of AS, when some individuals were found to have deletions or rearrangements in the proximal long arm of chromosome 15.4,5,6 The same genomic segment on proximal 15q had previously been implicated in Prader‐Willi syndrome (PWS);7 subsequently it was shown that deletions of the maternal chromosome produce AS and deletions of the paternal chromosome produce PWS.8,9 AS may ensue from four different molecular mechanisms, all of which result in deficiency of the maternally inherited E6 associated protein ubiquitin protein ligase 3A gene (UBE3A). These include deletion of the maternal chromosome 15q11‐q13 region (70%), paternal uniparental disomy for chromosome 15 (2%),10,11 mutations of the UBE3A gene (10%),12,13 and mutations or deletions of the imprinting centre (5%).14,15 The genomic organisation of the 15q11‐q13 region is complex and is perhaps one of the most variable regions of the human genome.16,17 This region is known to harbour multiple low copy repeats that probably mediate these deletions, duplications, and inversions.17,18,19
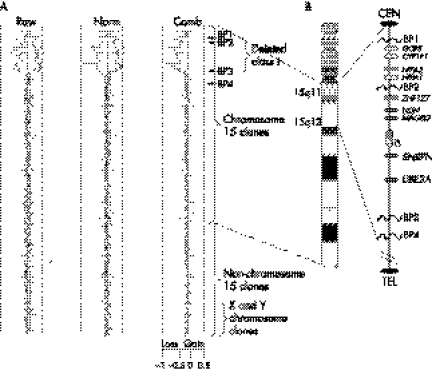
There are two types of deletions seen in AS, encompassing a region of over 6 Mb20 (Sahoo et al, 2005; personal communication). A common distal breakpoint (BP3) and two proximal breakpoints (BP1 and BP2) define the frequently occurring types of deletions.16,18,21,22,23 The deletions that extend from BP1 to BP3 are designated class I deletions, and those extending from BP2 to BP3 are designated class II deletions (fig 1A, B). The majority of deletions in AS and PWS are class II.22 Some well known genes, many of them imprinted, are located in the interval between BP2 and BP3, including the gene encoding small nuclear ribonucleoprotein polypeptide‐N (SNRPN), UBE3A, and the imprinting centre.
Figure 1 Plot of the hybridisation results for representative case with class I deletion. (A) Raw, normalised (Norm) and combined (Comb) log2 ratio plots of array hybridisation are shown. The combined ratio plot provides a final estimate of gain, loss, and no change distribution for each clone. The dashed lines on the scale for the logarithmic plot (x axis) indicate the position of −1.0 and +0.5, which are the theoretical values for single copy loss or gain respectively. The plot is in clone by clone order starting (y axis: top to bottom) from 15 centromere to 15q telomere followed by chromosomes 1 to Y. The deletion in this case encompasses the segment between BP1 to BP3 typical of class I deletions. Fluorescent in situ hybridisation (FISH) using clones within and flanking the BP1‐BP3 interval confirmed the size and boundaries of the deletion. (B) On the right is a map of human chromosome 15 highlighting the 15q11‐q13 segment encompassing the PWS/AS imprinted domain. The common deletion breakpoints are indicated as BP1, BP2, and BP3. BP4 refers to the distal breakpoint commonly involved in isodicentric 15q.
Some recent studies aimed at identifying phenotypic differences in PWS and AS patients with different deletion sizes have revealed interesting genotype‐phenotype relationships. Adults with PWS harbouring class I deletions had a greater incidence of obsessive‐compulsive behaviour, and more deficits in adaptive skills as compared with individuals with smaller class II deletions.24 Similar studies in AS demonstrated that all individuals with class I deletions showed a complete lack of vocalisation, while those with class II deletions were able to produce at least some syllabic sounds.25 Neither of these studies examined symptoms of autism. Because (a) previous studies demonstrate significant overlap between AS and autism,26,27,28 and symptoms of autism persist over time in AS; (b) the obsessive‐compulsive behaviours, deficits in adaptive behaviour, and deficits in language skills found in previous studies comparing deletion types in AS and PWS are part of the broad autism phenotype, and (c) the AS/PWS critical region has been implicated in autism,29,30,31,32,33,34 we thought it important to examine the relationship between autism and deletion class in AS. As seizures occur in nearly 80% of children with AS,35 we also examined the relationship between the degree of seizure severity, EEG findings, and the number of medications required to achieve good seizure control across deletion classes.
Currently, the characterisation of deletion classes in the 15q11‐q13 region is performed by fluorescence in situ hybridisation studies (FISH) or microsatellite marker analysis. The advent of molecular tools such as array comparative genomic hybridisation (array CGH) allows us to define these rearrangements in a more detailed and comprehensive manner. A recent report has highlighted the usefulness of CGH arrays to characterise the AS/PWS region.16 For the current study, we analysed a group of 22 deletion bearing AS patients using a chromosome 15 specific array CGH to further characterise their deletion, and to examine genotype‐phenotype correlations.
The objectives of this study were threefold: (a) to characterise by array CGH a cohort of AS patients with deletions, (b) to examine the expression of autism spectrum disorder in this cohort, and (c) to analyse genotype‐phenotype differences between participants. It was hypothesised that accurate characterisation of the underlying molecular defect could help predict clinical outcomes, and therefore aid with early, appropriate interventions.
METHODS
Clinical samples
Research samples were collected in accordance with institutional review board approved protocols at Baylor College of Medicine and University of California, San Diego. In total, 22 patients with AS bearing deletions (13 boys and 9 girls; age range 17 months to 11 years) previously diagnosed by FISH were characterised using array CGH. The following clinical parameters were selected to assess genotype‐phenotype correlations: (a) developmental assessments using standardised testing for cognitive skills, language, and adaptive behaviour; (b) formal evaluation for autism; (c) seizure quality, frequency, and number of drugs required for seizure control; (d) routine 16 channel EEG recorded awake and asleep; and (e) head circumference.
Molecular studies
For CGH, a total of 106 genomic bacterial artificial chromosome (BAC) clones across the length of the long arm of chromosome 15 were used for the microarray. The highest density of clones is across the ∼10 Mb 15q11‐q14 interval encompassing the PWS/AS critical region, including the common deletion/duplication breakpoints. This particular microarray achieves a resolution greater than one clone per Mb for the entire chromosome 15. Additionally, over 50 clones (BACs and P1 derived artificial chromosomes) specific for the subtelomeric regions of all other chromosomes were included. The validation of genomic clones, and production and analysis of array CGH experiments were carried out as described previously (fig 1).36,37 The breakpoints defining the deletion class predicted by array CGH were confirmed by FISH analysis using clones within and flanking the deletion.
Instrumentation for clinical evaluations
Psychologists were blinded to a child's deletion class when conducting clinical evaluations. All participants were given the Autism Diagnostic Observation Schedule (ADOS‐G), generic, module 1 and the Autism Diagnostic Interview, revised (ADI‐R). The rationale for this procedure is described in detail in a previous publication.27 The Bayley Scale of Infant Development, second edition (BSID‐II) was used to assess cognitive and motor skills. Parents were interviewed using the standardised administration of the Vineland Adaptive Behavior Scale (VABS), interview edition. The Preschool Language Scale, third edition (PLS‐III) was used to assess communication skills.38
Statistical analysis
Differences among deletion classes (class I versus class II) on categorical variables (sex, autism) were analysed using a χ2 analysis. The scores from dependent measures (mental scores, adaptive behaviour scores, language scores) were normally distributed. Analyses of variance were conducted to compare differences between the deletion classes for all dependent variables. Chronological age was used as a covariate in all analyses. Means and standard deviation scores for these measures are expressed as age equivalents. All reported p values are two sided; p values ⩽0.05 were considered to indicate statistical significance, and p values <0.10 were considered to indicate a statistical trend.
RESULTS
Table 1 shows the sex of patients in each deletion class. There were comparable numbers of boys and girls within each deletion subgroup. A representative CGH profile for a child with a typical class I deletion is shown (fig 1A), showing loss (log2 ratio <0.18–0.2) of clones located within the BP1–BP3 interval. One male patient had an extended class II deletion with a breakpoint extending from the proximal BP2 to an unusual distal breakpoint (BP3A) that encompassed an additional ∼1 Mb genomic loss.
Table 1 Deletion class by sex.
| Deletion | Male | Female |
|---|---|---|
| Class I | 6 | 4 |
| Class II | 6 | 5 |
| Atypical class II | 1 | 0 |
Developmental evaluation
Table 2 shows the age equivalent means and standard deviations for cognitive, language, and adaptive behaviour measures, broken down by deletion subtype. For the purpose of this analysis, the child with the unique extended class II deletion was excluded.
Table 2 Developmental scores by deletion class.
| Scales | Deletion class | Mean | SD | |||
|---|---|---|---|---|---|---|
| Bayley mental, age equivalent | I | 8.42 | 0.67 | |||
| II | 10.35 | 0.63 | ||||
| Bayley motor, age equivalent | I | 9.5 | 0.80 | |||
| II | 10.7 | 0.84 | ||||
| PLS auditory comprehension, | I | 6.39 | 0.67 | |||
| age equivalent | II | 7.83 | 0.64 | |||
| PLS expressive communication, | I | 3.73 | 0.76 | |||
| age equivalent | II | 5.74 | 0.72 | |||
| PLS composite, age equivalent | I | 5.05 | 0.76 | |||
| II | 7.23 | 0.72 | ||||
| VABS communication, age | I | 9.94 | 0.82 | |||
| equivalent | II | 11.6 | 0.77 | |||
| VABS daily living, age | I | 12.24 | 1.44 | |||
| equivalent | II | 13.87 | 1.37 | |||
| VABS socialization, age | I | 10.96 | 0.70 | |||
| equivalent | II | 12.49 | 0.66 |
Age equivalent scores are reported in months.
Broadly, on all measures of development, children with class II deletions scored higher than children with class I deletions. Statistically significant differences were noted between deletion class groups for Bayley mental scores (F(1,20) = 3.50; p = 0.05; adjusted R2 = 0.042). Trends toward significance were noted for expressive communication (p = 0.09) and for total language scores (p = 0.066) on the PLS. It is important to note that these differences are independent of chronological age. There were no significant differences between children with class I and class II deletions for Bayley Motor scale scores, auditory comprehension scores (receptive language), or for any of the parent report measures on the VABS.
Autism
The results of evaluations for autism revealed that eight of 10 children with larger class I deletions met criteria for autism, compared with two of 11 children with class II deletions, which was statistically significant (χ2 = 6.60; p = 0.01). There were no differences in autism diagnosis according to chronological age. The patient with the unique extended class II deletion also met criteria for comorbid autism. Nine of the 11 children diagnosed with autism were male. The preponderance of males with autism is similar to that seen in the general population. In contrast to their peers with AS alone, the children with comorbid diagnoses of autism and AS rarely directed vocalisations to others, were not responsive to their names being called, and, although many of them exhibited the excessive laughter commonly associated with AS, they did not exhibit shared enjoyment in interactions with others. They were typically more focused on objects (and the repetitive use of objects), as opposed to interactions with other people, and made very few social overtures. Although all children with AS exhibited severe language delays, and deficits in their play skills, children without comorbid autism had developed the use of nonverbal gestures and nearly all of their vocalisations were socially directed.
Although it would be desirable to examine any interactions between deletion subtype and autism for all of the developmental outcomes, the fact that only two children with class II deletions met criteria for autism, and only two children with class I deletions did not meet criteria for autism prevented these analyses from being conducted. Nevertheless, as concluded from the scores described in table 2, differences between the two classes of patients with respect to cognitive impairments and developmental delay is significant and most likely determines whether they do or do not express the autism phenotype.
Head circumference
Fronto‐occipital circumference (FOC) measurements were obtained at clinic visits. For those subjects who were seen on more than one occasion, the last measurement was the one selected for this analysis. All measurements were compared with standard growth curves according to sex and age. Any FOC measurement below 2.5 SD was considered microcephalic. Of the 22 children studied, 14 were microcephalic. Seven of 10 patients with class I deletions were microcephalic; six of the 11 patients with class II deletions were microcephalic (non‐significant, p = 0.6).
Seizures and medications
Of the 22 children, 20 (91%) had experienced or were experiencing seizures. Some children had more than one type of seizure. Of the 10 children with class I deletions, eight (80%) had generalised seizures including tonic and atypical absence seizures; three (30%) had partial (complex) seizures. Eight (80%) of these children were receiving antiepileptic drug(s) (AEDs) at the time of the index visit; none of these children had uncontrolled seizures. Of the 11 children with class II deletions, six (55%) had generalised seizures including tonic, myoclonic, and atypical absence seizures; four (36%) had partial (complex) seizures. Five (45%) of these children were receiving AEDs; none had uncontrolled seizures. Two children from the class I group and one child from the class II group had febrile seizures. The child with atypical class II deletion has severe generalised tonic‐clonic seizures and was initially diagnosed with infantile spasms. The number of seizure medications required to achieve good control in class I was significantly increased (χ2: F = 4.571; p = 0.046) compared with class II patients. Patients with class I deletions (n = 10) required a mean of 1.60 drugs in comparison to 0.73 drugs in the class II group (n = 11), with a mean of 0.73.
The severity of EEG findings was estimated by evaluating the presence of expected developmental features (such as occipital dominant), degree and character of slowing (such as high voltage delta activity), and presence of epileptiform abnormalities (such as generalised spike and slow wave activity) or recording of EEG seizure discharges. Of the children with class I deletions, 90% had epileptiform abnormalities compared with 91% of those with class II deletions. In all cases, the epileptiform abnormalities were generalised. In addition to the generalised anomalies, 20% of focal changes were seen in class I versus 36% in class II. Multifocal findings were also seen in 10% of class I versus 18% in class II. Lastly, 20% of the children in class I had a hypsarrhythmia pattern not seen in class II. Overall, the children with class I deletions appeared to have a more severe degree of EEG abnormalities. Using a scale of 0 (normal EEG) to 20 (severely abnormal EEG), the mean for the scores in the class I group (n = 10) was 11.78, and for class II 10.87 (n = 11) (p = 0.540, non‐significant).
DISCUSSION
Phenotypic profiling of patients with accurate molecular genotypes provides an opportunity to make crucial genotype‐phenotype correlations. Our array CGH based analysis of the deletion type in AS helped accurately define the nature and size of the deletions in these 22 patients. This study demonstrates that: children with AS who had larger class I deletions (a) were significantly more likely to meet criteria for comorbid autism, (b) have significantly lower cognitive scores, and (c) were significantly more likely to require more seizure medications than their class I counterparts. There was a trend for class I individuals to have lower expressive and total language abilities.
There are four genes in the interval that differentiates individuals with class I from those with class II deletions: NIPA‐1, NIPA‐2, CYFIP1, and GCP5.17 The predicted function of the NIPA genes and their encoded polypeptides suggests a transporter or receptor function. NIPA‐1 is also strongly expressed in brain. Mutations in conserved regions of NIPA‐1 have been found in selected families with an autosomal dominant form of spastic paraplegia: SPG6.39,40,41 This condition is characterised by progressive spasticity in the lower extremities. Patients with AS or PWS with deletions of this gene region do not have progressive spasticity, therefore it has been hypothesised that NIPA‐1 mutations may cause disease by gain of function rather than by haploinsufficiency.39CYFIP1 has been shown to interact with fragile X mental retardation protein (FMRP).42 In addition, the CYFIP1 protein has been shown to interact with the small GTPase RAC1, which is involved the development and maintenance of neuronal structures.42 It has been suggested that RAC1 and CYFIP1 are part of the pathway regulating certain functions of FMRP.42 The considerable overlap between fragile X syndrome and autism (33% of children with fragile X syndrome were found to meet criteria for autism in a recent study43), gives additional support for a role of CYFIP1 in autism. Lastly, GCP5 encodes a human γ‐globulin complex required for microtubule nucleation at the centrosome. It is unclear whether these genes may play any role in AS given that none of them are imprinted. It should be mentioned that NIPA‐1, NIPA‐2, and CYFIP1 show asynchronous replication, a phenomenon often seen for imprinted genes that have monoallelic expression.17 Given that the replication asynchrony in this case is independent of the parental origin, a parental imprinting effect is less likely.
CONCLUSION
Individuals with AS with larger class I deletions appear to have a more severe phenotype associated with a comorbid diagnosis of autism, lower developmental scores, and lower verbal skills, and are more likely to require a larger number of medications for seizure control. Children with class I deletions may therefore require more intensive interventions including: (a) behavioural therapy (such as applied behavioural analysis) to address skills more specific to autism such as eye gaze, sitting, imitation, and self stimulatory/repetitive behaviours; (b) more intensive speech therapy, including the teaching of nonverbal gestures; and (c) occupational and physical therapy. Given our findings, children with AS with class I deletions may benefit from close monitoring of seizures as well as the antiepileptic drugs they require. This study extends previous work examining differences in deletion classes in AS and PWS, and provides further evidence that the four genes located between BP1 to BP2 (NIPA‐1, NIPA‐2, CYFIP1, and GCP5) may have a role in the increased severity and/or a role in communication and social development. We hypothesise that one or more of these genes in the class I region, or their downstream regulators, may have further implications in autism spectrum disorders.
ACKNOWLEDGEMENTS
This study was funded in part by the National Association for Autism Research, pilot research award (NAAR 704/TS/01‐201‐004‐00‐00 to TS) and by the March of Dimes (MOD 6FY03‐73 to CAB). We also like to thank the support from the MRDDRC core at Baylor College of Medicine.
Abbreviations
ADI‐R - Autism Diagnostic Interview, revised
ADOS‐G - Autism Diagnostic Observation Schedule, Generic
AED - antiepileptic drug
AS - Angelman Syndrome
BAC - bacterial artificial chromosome
BSID‐II - Bayley Scale of Infant Development, second edition
CGH - comparative genomic hybridization
FISH - fluorescent in situ hybridisation
FMRP - fragile X mental retardation protein
FOC - fronto‐occipital circumference
PLS‐III - Preschool Language Scale, third edition
PWS - Prader‐Willi Syndrome
SNRPN - small nuclear ribonucleoprotein polypeptide‐N
UBE3A - E6 associated protein ubiquitin protein ligase 3A gene
VABS - Vineland Adaptive Behavior Scale
Footnotes
Competing interests: there are no competing interests
References
- 1.Angelman H. “Puppet children”: a report of three cases. Dev Med Child Neurol 19657681–688. [DOI] [PubMed] [Google Scholar]
- 2.Berg J M, Pakula Z. Angelman's (“happy puppet”) syndrome. Am J Dis Child 197212372–74. [DOI] [PubMed] [Google Scholar]
- 3.Williams C A, Angelman H, Clayton‐Smith J, Driscoll D J, Hendrickson J E, Knoll J H, Magenis R E, Schinzel A, Wagstaff J, Whidden E M, Zori R T. Angelman syndrome: consensus for diagnostic criteria. Angelman Syndrome Foundation. Am J Med Genet 199556237–238. [DOI] [PubMed] [Google Scholar]
- 4.Fryburg J S, Breg W R, Lindgren V. Diagnosis of Angelman syndrome in infants. Am J Med Genet 19913858–64. [DOI] [PubMed] [Google Scholar]
- 5.Magenis R E, Brown M G, Lacy D A, Budden S, LaFranchi S. Is Angelman syndrome an alternate result of del(15)(q11q13)? Am J Med Genet 198728829–838. [DOI] [PubMed] [Google Scholar]
- 6.Magenis R E, Toth‐Fejel S, Allen L J, Black M, Brown M G, Budden S, Cohen R, Friedman J M, Kalousek D, Zonana J, Lace D, La Franchi S, Lahr M, Macfarlane J, Williams C P S. Comparison of the 15q deletions in Prader‐Willi and Angelman syndromes: specific regions, extent of deletions, parental origin, and clinical consequences. Am J Med Genet 199035333–349. [DOI] [PubMed] [Google Scholar]
- 7.Donlon T A. Similar molecular deletions on chromosome 15q11.2 are encountered in both the Prader‐Willi and Angelman syndromes. Hum Genet 198880322–328. [DOI] [PubMed] [Google Scholar]
- 8.Knoll J H, Nicholls R D, Magenis R E, Graham J M, Jr, Lalande M, Latt S A. Angelman and Prader‐Willi syndromes share a common chromosome 15 deletion but differ in parental origin of the deletion. Am J Med Genet 19893285–290. [DOI] [PubMed] [Google Scholar]
- 9.Knoll J H, Nicholls R D, Lalande M. On the parental origin of the deletion in Angelman syndrome. Hum Genet 198983205–207. [DOI] [PubMed] [Google Scholar]
- 10.Engel E. Uniparental disomy (UPD). Genomic imprinting and a case for new genetics (prenatal and clinical implications: the “Likon” concept). Ann Genet 19974024–34. [PubMed] [Google Scholar]
- 11.Prasad C, Wagstaff J. Genotype and phenotype in Angelman syndrome caused by paternal UPD 15. Am J Med Genet 199770328–329. [DOI] [PubMed] [Google Scholar]
- 12.Matsuura T, Sutcliffe J S, Fang P, Galjaard R J, Jiang Y H, Benton C S, Rommens J M, Beaudet A L. De novo truncating mutations in E6‐AP ubiquitin‐protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet 19971574–77. [DOI] [PubMed] [Google Scholar]
- 13.Kishino T, Lalande M, Wagstaff J. UBE3A/E6‐AP mutations cause Angelman syndrome. Nat Genet 19971570–73. [DOI] [PubMed] [Google Scholar]
- 14.Buiting K, Barnicoat A, Lich C, Pembrey M, Malcolm S, Horsthemke B. Disruption of the bipartite imprinting center in a family with Angelman syndrome. Am J Hum Genet 2001681290–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buiting K, Dittrich B, Gross S, Lich C, Farber C, Buchholz T, Smith E, Reis A, Burger J, Nothen M M, Barth‐Witte U, Janssen B, Abeliovich D, Lerer I, van den Ouweland A M, Halley D J, Schrander‐Stumpel C, Smeets H, Meinecke P, Malcolm S, Gardner A, Lalande M, Nicholls R D, Friend K, Schulze A, Matthijs G, Kokkonen H, Hilbert P, Van Maldergem L, Glover G, Carbonell P, Willems P, Gillessen‐Kaesbach G, Horsthemke B. Sporadic imprinting defects in Prader‐Willi syndrome and Angelman syndrome: implications for imprint‐switch models, genetic counseling, and prenatal diagnosis. Am J Hum Genet 19986170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Locke D P, Segraves R, Nicholls R D, Schwartz S, Pinkel D, Albertson D G, Eichler E E. BAC microarray analysis of 15q11‐q13 rearrangements and the impact of segmental duplications. J Med Genet 200441175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chai J H, Locke D P, Greally J M, Knoll J H, Ohta T, Dunai J, Yavor A, Eichler E E, Nicholls R D. Identification of four highly conserved genes between breakpoint hotspots BP1 and BP2 of the Prader‐Willi/Angelman syndromes deletion region that have undergone evolutionary transposition mediated by flanking duplicons. Am J Hum Genet 200373898–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amos‐Landgraf J M, Ji Y, Gottlieb W, Depinet T, Wandstrat A E, Cassidy S B, Driscoll D J, Rogan P K, Schwartz S, Nicholls R D. Chromosome breakage in the Prader‐Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet 199965370–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gimelli G, Pujana M A, Patricelli M G, Russo S, Giardino D, Larizza L, Cheung J, Armengol L, Schinzel A, Estivill X, Zuffardi O. Genomic inversions of human chromosome 15q11‐q13 in mothers of Angelman syndrome patients with class II (BP2/3) deletions. Hum Mol Genet 200312849–858. [DOI] [PubMed] [Google Scholar]
- 20.Sahoo T, Shaw C A, Young A S, Whitehouse N L, Schroer R J, Stevenson R E, Beaudet A L. Genomic array‐based comparative genomic hybridization analysis of recurrent chromosme 15q rearrangements. Am J Med Genet 2005139106–113. [DOI] [PubMed] [Google Scholar]
- 21.Knoll J H, Nicholls R D, Magenis R E, Glatt K, Graham J M, Jr, Kaplan L, Lalande M. Angelman syndrome: three molecular classes identified with chromosome 15q11q13‐specific DNA markers. Am J Hum Genet 199047149–155. [PMC free article] [PubMed] [Google Scholar]
- 22.Christian S L, Robinson W P, Huang B, Mutirangura A, Line M R, Nakao M, Surti U, Chakravarti A, Ledbetter D H. Molecular characterization of two proximal deletion breakpoint regions in both Prader‐Willi and Angelman syndrome patients. Am J Hum Genet 19955740–48. [PMC free article] [PubMed] [Google Scholar]
- 23.Christian S L, Fantes J A, Mewborn S K, Huang B, Ledbetter D H. Large genomic duplicons map to sites of instability in the Prader‐Willi/Angelman syndrome chromosome region (15q11‐q13). Hum Mol Genet 199981025–1037. [DOI] [PubMed] [Google Scholar]
- 24.Varela M C, Kok F, Otto P A, Koiffmann C P. Phenotypic variability in Angelman syndrome: comparison among different deletion classes and between deletion and UPD subjects. Eur J Hum Genet 200412987–992. [DOI] [PubMed] [Google Scholar]
- 25.Butler M G, Bittel D C, Kibiryeva N, Talebizadeh Z, Thompson T. Behavioral differences among subjects with Prader‐Willi syndrome and type I or type II deletion and maternal disomy. Pediatrics 2004113565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steffenburg S, Gillberg C L, Steffenburg U, Kyllerman M. Autism in Angelman syndrome: a population‐based study. Pediatr Neurol 199614131–136. [DOI] [PubMed] [Google Scholar]
- 27.Peters S U, Beaudet A L, Madduri N, Bacino C A. Autism in Angelman syndrome: implications for autism research. Clin Genet 200466530–536. [DOI] [PubMed] [Google Scholar]
- 28.Bolton P F, Dennis N R, Browne C E, Thomas N S, Veltman M W, Thompson R J, Jacobs P. The phenotypic manifestations of interstitial duplications of proximal 15q with special reference to the autistic spectrum disorders. Am J Med Genet 2001105675–685. [DOI] [PubMed] [Google Scholar]
- 29.Shao Y, Cuccaro M L, Hauser E R, Raiford K L, Menold M M, Wolpert C M, Ravan S A, Elston L, Decena K, Donnelly S L, Abramson R K, Wright H H, DeLong G R, Gilbert J R, Pericak‐Vance M A. Fine mapping of autistic disorder to chromosome 15q11‐q13 by use of phenotypic subtypes. Am J Hum Genet 200372539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philippe A, Martinez M, Guilloud‐Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, Van Maldergem L, Penet C, Feingold J, Brice A, Leboyer M. Genome‐wide scan for autism susceptibility genes. Paris Autism Research International Sibpair Study. Hum Mol Genet 19998805–812. [DOI] [PubMed] [Google Scholar]
- 31.Nurmi E L, Bradford Y, Chen Y, Hall J, Arnone B, Gardiner M B, Hutcheson H B, Gilbert J R, Pericak‐Vance M A, Copeland‐Yates S A, Michaelis R C, Wassink T H, Santangelo S L, Sheffield V C, Piven J, Folstein S E, Haines J L, Sutcliffe J S. Linkage disequilibrium at the Angelman syndrome gene UBE3A in autism families. Genomics 200177105–113. [DOI] [PubMed] [Google Scholar]
- 32.Thomas N S, Browne C E, Oley C, Healey S, Crolla J A. Investigation of a cryptic interstitial duplication involving the Prader‐Willi/Angelman syndrome critical region. Hum Genet 1999105384–387. [DOI] [PubMed] [Google Scholar]
- 33.Schroer R J, Phelan M C, Michaelis R C, Crawford E C, Skinner S A, Cuccaro M, Simensen R J, Bishop J, Skinner C, Fender D, Stevenson R E. Autism and maternally derived aberrations of chromosome 15q. Am J Med Genet 199876(327–336. [DOI] [PubMed] [Google Scholar]
- 34.Bass M P, Menold M M, Wolpert C M, Donnelly S L, Ravan S A, Hauser E R, Maddox L O, Vance J M, Abramson R K, Wright H H, Gilbert J R, Cuccaro M L, DeLong G R, Pericak‐Vance M A. Genetic studies in autistic disorder and chromosome 15. Neurogenetics 20002219–226. [DOI] [PubMed] [Google Scholar]
- 35.Clayton‐Smith J, Laan L. Angelman syndrome: a review of the clinical and genetic aspects. J Med Genet 20034087–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai W W, Mao J H, Chow C W, Damani S, Balmain A, Bradley A. Genome‐wide detection of chromosomal imbalances in tumors using BAC microarrays. Nat Biotechnol 200220393–396. [DOI] [PubMed] [Google Scholar]
- 37.Yu W, Ballif B C, Kashork C D, Heilstedt H A, Howard L A, Cai W W, White L D, Liu W, Beaudet A L, Bejjani B A, Shaw C A, Shaffer L G. Development of a comparative genomic hybridization microarray and demonstration of its utility with 25 well‐characterized 1p36 deletions. Hum Mol Genet 2003122145–2152. [DOI] [PubMed] [Google Scholar]
- 38.Peters S U, Goddard‐Finegold J, Beaudet A L, Madduri N, Turcich M, Bacino C A. Cognitive and adaptive behavior profiles of children with Angelman syndrome. Am J Med Genet A 2004128110–113. [DOI] [PubMed] [Google Scholar]
- 39.Rainier S, Chai J H, Tokarz D, Nicholls R D, Fink J K. NIPA1 gene mutations cause autosomal dominant hereditary spastic paraplegia (SPG6). Am J Hum Genet 200373967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reed J A, Wilkinson P A, Patel H, Simpson M A, Chatonnet A, Robay D, Patton M A, Crosby A H, Warner T T. A novel NIPA1 mutation associated with a pure form of autosomal dominant hereditary spastic paraplegia. Neurogenetics 2005679–84. [DOI] [PubMed] [Google Scholar]
- 41.Chen S, Song C, Guo H, Xu P, Huang W, Zhou Y, Sun J, Li C X, Du Y, Li X, Liu Z, Geng D, Maxwell P H, Zhang C, Wang Y. Distinct novel mutations affecting the same base in the NIPA1 gene cause autosomal dominant hereditary spastic paraplegia in two Chinese families. Hum Mutat 200525135–141. [DOI] [PubMed] [Google Scholar]
- 42.Schenck A, Bardoni B, Moro A, Bagni C, Mandel J L. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP‐related proteins FXR1P and FXR2P. Proc Natl Acad Sci USA 2001988844–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogers S J, Wehner D E, Hagerman R. The behavioral phenotype in fragile X: symptoms of autism in very young children with fragile X syndrome, idiopathic autism, and other developmental disorders. J Dev Behav Pediatr 20012409–417. [DOI] [PubMed] [Google Scholar]