Abstract
Mutations in the p63 gene (TP63) underlie several monogenic malformation syndromes manifesting cleft lip with or without cleft palate (CL/P). We investigated whether p63 mutations also result in non‐syndromic CL/P. Specifically, we performed mutation analysis of the 16 exons of the p63 gene for 100 Thai patients with non‐syndromic CL/P. In total, 21 variant sites were identified. All were single nucleotide changes, with six in coding regions, including three novel non‐synonymous changes: S90L, R313G, and D564H. The R313G was concluded to be pathogenic on the basis of its amino acid change, evolutionary conservation, its occurrence in a functionally important domain, its predicted damaging function, its de novo occurrence, and its absence in 500 control individuals. Our data strongly suggest, for the first time, a causative role of a heterozygous mutation in the p63 gene in non‐syndromic CL/P, highlighting the wide phenotypic spectrum of p63 gene mutations.
Keywords: non‐syndromic cleft lip, p63 , mutations
Cleft lip with or without cleft palate (CL/P), is the most common craniofacial anomaly, with an estimated incidence of 1 in 500–2500 live births. The frequency depends upon geographic origin, with populations of East Asian and Native American ancestry having the highest rates and African populations having the lowest.1 Approximately 70% of cases of CL/P occur as an isolated abnormality—that is, non‐syndromic CL/P.2 The majority of non‐syndromic CL/P cases are considered multifactorial in aetiology, with several genes and environmental factors involved.3,4,5,6 The remaining 30% of CL/P occur as part of more than 300 different malformation syndromes in which CL/P is only one manifestation.5 Many of these syndromes are monogenic disorders, and mutations in the responsible genes, such as MSX17,8 and IRF6,9,10,11,12 are associated with non‐syndromic CL/P. These findings led us to hypothesise that some patients with non‐syndromic CL/P might have mutations in genes known to cause “syndromic” CL/P.
Some examples of mendelian clefting syndromes include ectrodactyly‐ectodermal dysplasia‐clefting syndrome (EEC, MIM #129900), ankyloblepharon‐ectodermal dysplasia‐clefting syndrome (AEC, MIM #106260) and Rapp‐Hodgkin syndrome (RHS, MIM #129400). All three of these syndromes are allelic disorders caused by mutations in the p63 gene.13,14,15,16 This gene has several isoforms with two different transcription initiation sites, one giving rise to proteins containing the transactivating (TA) domain (the TA isotypes) and the other lacking the TA domain (the ▵N isotypes). Alternative splicing at the 3′ end of the gene results in three different C‐termini, α, β, and γ. The largest p63 isotype, TA‐p63α, has TA, DNA binding (DB), polymerisation, sterile α‐motif (SAM), and transactivation inhibitory (TI) domains.17 We performed mutation analysis of all the domains and exons of the p63 gene in Thai patients with non‐syndromic CL/P.
MATERIALS AND METHODS
Participants
The participants in the study were 88 sporadic cases of non‐syndromic CL/P and 12 additional cases with a positive family history. These two groups were further characterised by type, laterality, severity, and sex (table 1). All patients were studied under the auspices of the Thai Red Cross, a national charity organisation devoted to providing clinical care for the poor. Subjects were recruited between 2000 and 2004 from 10 centres in Thailand (Nakornratchaseema, Nan, Uthaithanee, Maehongsorn, Trang, Srakaew, Kalasin, Nongkhai, Mahasarakam, and Bangkok). All patients were screened for the presence of associated anomalies or syndromes by a geneticist (VS), and only those determined to have isolated cleft lip with or without cleft palate (normal growth, normal development, no other major anomalies, and no apparent visual or hearing deficits) were included in this study. The study was approved by the institutional review board of the Faculty of Medicine of Chulalongkorn University, and written informed consent was obtained from each person included in the study. Blood samples for DNA analysis were obtained at the time of blood typing and haematocrit determination. The control samples were Thai blood donors with no oral clefts, who denied history of oral clefts in other family members.
Table 1 Characteristics of the patients with nonsyndromic cleft lip with or without cleft palate.
| Cleft lip only | Cleft lip with cleft palate | Total | ||||
|---|---|---|---|---|---|---|
| No. of probands | 37 | 63 | 100 | |||
| Sporadic | 31 | 57 | 88 | |||
| Familial | 6 | 6 | 12 | |||
| Laterality | ||||||
| Right side cleft | 9 | 15 | 24 | |||
| Left side cleft | 26 | 32 | 58 | |||
| Bilateral cleft | 2 | 16 | 18 | |||
| Severity | ||||||
| Complete cleft | 19 | 61 | 80 | |||
| Incomplete cleft | 18 | 2 | 20 | |||
| Sex | ||||||
| Male | 16 | 42 | 58 | |||
| Female | 21 | 21 | 42 |
PCR
Genomic DNA was isolated from peripheral blood, according to established protocols. Intronic primers were used to specifically amplify fragments encompassing each of exons 1–15 and exon 3′ of the p63 gene (table 2). PCR reactions were carried out in a 20 µl volume containing 50 ng genomic DNA,1X PCR buffer, 1.5 mmol/l MgCl2, 0.2 mmol/l dNTPs, 0.2 µmol/l of each primers and 0.5 U Taq polymerase, using the following parameters: 35 cycles of 30 seconds at 94°C, 30 seconds at the appropriate annealing temperature (table 2), and 30 seconds at 72°C. PCR products were treated with ExoSAP‐IT (USP Corporation, Cleveland, OH, USA) according to the manufacturer's recommendations, and sent for direct sequencing (Macrogen Inc., Seoul, Korea). Analyses were performed using Sequencher software (version 4.2). When the results indicated a possible new variant, the sample was resequenced. All non‐synonymous coding variants were verified by restriction enzyme digestions of the PCR products of the patients. The parents (paternity and maternity confirmed by typing 15 microsatellite markers on 13 different chromosomes; data not shown) were also examined for the variants by sequencing and restriction enzyme analysis; and 500 Thai control individuals by restriction enzyme analysis. The position of mutations corresponds to the coding sequence for the original published TA‐p63α isotype (GenBank accession AF075430).
Table 2 Oligonucleotides and PCR conditions for p63 mutation analysis.
| Exon | Primer sequences for PCR (5′ to 3′) | Product size (bp) | AT (C°) | |||||
|---|---|---|---|---|---|---|---|---|
| Forward | Reverse | |||||||
| 1 | CCCTATTGCTTTTAGCCTCC | ACTGTGCTGACTAAACAAGG | 281 | 55 | ||||
| 2 | CTACATATATACCTGCATGG | AAAAACATGCCCTAGTAAGC | 344 | 52 | ||||
| 3 | AGCCTTGCTGACTTTGAAGC | CACATGACTGAAAAGACAGG | 317 | 55 | ||||
| 3′ | CATATTGTAAGGGTCTCAGAGG | GACCGAGAACCGCAAATACG | 223 | 62 | ||||
| 4 | ATGCATTCACCCATGGATGC | GAATCGCTAAACTGGGAAGG | 437 | 55 | ||||
| 5 | GTAAACAGGCAGCATGCAGC | AGTCTGAATCAGGTAGGTGG | 401 | 55 | ||||
| 6 | CACCAACATCCTGTTCATGC | GCTAGAAACATCCCTGTTGC | 296 | 55 | ||||
| 7 | AGAGGGAAGAACTGAGAAGG | CAGCCACGATTTCACTTTGC | 256 | 55 | ||||
| 8 | GGAAGTGGTAGATCTTCAGG | GCAGCTTCTCCAATATCACC | 294 | 55 | ||||
| 9 | GTGTTGCTGGTACTACTGTC | GACTAAGACACCTCCTTTCC | 334 | 55 | ||||
| 10 | ACTTCTAACAGTTCTACAGC | CTCATCAATCACCCTATTG | 275 | 52 | ||||
| 11 | CCATGTTTTAAACAGAGACC | CACAGAGTCTTGTCCTAAGC | 313 | 52 | ||||
| 12 | TTAACCAGACAAGATGGACC | CCCTTCCAACTGTTTTATGG | 321 | 52 | ||||
| 13 | CTTATCTCGCCAATGCAGTT | TACAAGGCGGTTGTCATCAG | 238 | 62 | ||||
| 14 | GGAATGATAGGATGCTGTGG | GCAGGAGTGCGCAGGAGTGC | 450 | 55 | ||||
| 15 | CAGGCACTCTATTCTGTCTA | GGAAATACAACACACACACT | 280 | 62 | ||||
AT, annealing temperature.
Analysis
For protein sequence comparisons, p63 orthologues were first identified through a BLAST search of the non‐redundant database using Homo sapiens p63, accession NP_003713, as the reference sequence. All known and complete p63 sequences were included from the vertebrate lineage. These files in FASTA format were then analysed by ClustalX software (version 1.81). The human p63 was aligned with Norway rat (accession NP_062094), house mouse (accession no.AAP87982), chicken (NP_989682), African clawed frog (AAK15622), and zebrafish (NP_694518). The program classified amino acids by the variation in polarity, assessing both amino acid class conservation and evolutionary conservation at any given site.
PolyPhen (http://www.bork.embl‐heidelberg.de/PolyPhen) was used to predict the effect of the non‐synonymous mutations. ESEfinder software (http://exon.cshl.org/ESE/index.html) was used to predict potential exonic splicing enhancers (ESEs).18
RESULTS
In 100 DNA samples from subjects with non‐syndromic CL/P, 21 variant sites were identified. All were single nucleotide changes, comprising 14 transitions (five in coding regions) and seven transversions (one in a coding region) (table 3).
Table 3 Variant sites of P63 found in 100 Thai patients with nonsyndromic CL/P .
| Nucleotide position | Exon/intron | Nucleotide change | Expected amino acid change | Frequencies of : | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hetero‐ zygotes | Homo‐ zygotes | |||||||||
| 261 | Exon 4 | C→T | N87N | 1 | 0 | |||||
| 269 | Exon 4 | C→T | S90L | 1 | 0 | |||||
| 462+39 | Intron 4 | T→A | – | 6 | 0 | |||||
| 463−49 | Intron 4 | T→A | – | 1 | 0 | |||||
| 463−42 | Intron 4 | G→A | – | 1 | 0 | |||||
| 463−41 | Intron 4 | G→A | – | 1 | 0 | |||||
| 463−40 | Intron 4 | T→A | – | 1 | 0 | |||||
| 649+42 | Intron 5 | G→A | – | 9 | 0 | |||||
| 742 | Exon 6 | C→T | L248L | 7 | 0 | |||||
| 875+51 | Intron 7 | G→A | – | 1 | 0 | |||||
| 937 | Exon 8 | A→G | R313G | 1 | 0 | |||||
| 1013−22 | Intron 8 | A→G | – | 23 | 1 | |||||
| 1095+79 | Intron 9 | A→G | – | 41 | 11 | |||||
| 1218 | Exon 10 | C→T | H406H | 1 | 0 | |||||
| 1232+25 | Intron 10 | A→T | – | 1 | 0 | |||||
| 1232+26 | Intron 10 | G→C | – | 1 | 0 | |||||
| 1232+40 | Intron 10 | G→C | – | 39 | 10 | |||||
| 1232+41 | Intron 10 | G→A | – | 24 | 2 | |||||
| 1233−23 | Intron 10 | T→C | – | 42 | 35 | |||||
| 1629+22 | Intron 13 | C→T | – | 1 | 0 | |||||
| 1690 | Exon 14 | G→C | D564H | 1 | 0 | |||||
The coding regions of p63 contained six different variants, three synonymous and three non‐synonymous. The three non‐synonymous variants, 269C→T, 937A→G, and 1690G→C, have not been reported previously. One non‐synonymous variant was found in each of three patients; all were sporadic cases, with no anomalies besides the oral clefts, normal radiographs of hands and feet (data not shown), no consanguinity, normal development, and different geographic origins.
Patient 1
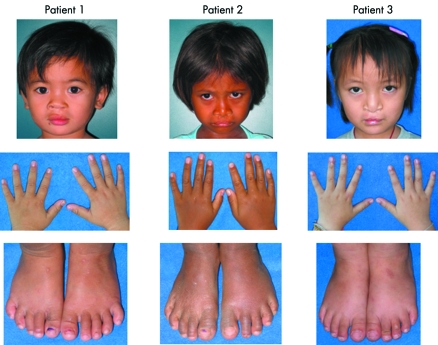
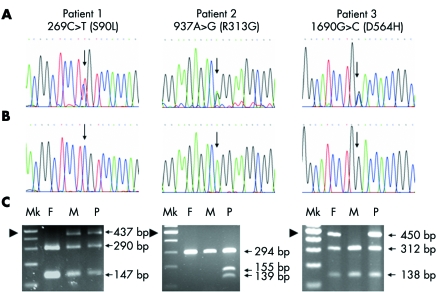
A 26 month old girl from Kalasin province was the second child of a 23 year old mother and a 28 year old father. The mother had had only the one previous pregnancy. The child had a surgically repaired left complete cleft lip (fig 1, left panel). Direct sequencing of the PCR products of the exons of her p63 gene revealed that she was heterozygous for a C→T point mutation at nucleotide position 269 (269C→T) in exon 4 (fig 2A, left panel). The mutation was confirmed by digestion of the PCR products with the restriction enzyme BseRI, the recognition site is removed by the mutation (fig 2C, left panel). The patient's clinically normal mother was also heterozygous for the 269C→T mutation (fig 2C, left panel). Her father had only the normal C alleles (fig 2C, left panel). The mutation 269C→T was expected to result in conversion of a serine to a leucine (S90L). Serine is polar and uncharged, while leucine is non‐polar. The serine at codon 90 is evolutionarily conserved in rat, mouse, chicken, and frog (data not shown). Codon 90 of zebrafish is threonine, which is a polar, uncharged amino acid, in the same group as serine. Codon 90 of p63 is 52 amino acids 5′ to the DB domain and is present in all p63 isotypes. PolyPhen predicted this variant to be benign. The nucleotide 269C is in the ESE motifs of SC35 and SRp55 serine/arginine‐rich (SR) proteins with scores of 3.96 and 3.00, respectively. The 269C→T variant is in the ESE motif of the SC35 SR protein, also with a score of 3.96, but is eliminated as a potential ESE for the SRp55 SR protein. S90L was absent from 1000 control chromosomes.
Figure 1 Clinical features of patients with non‐synonymous variants. The left, middle, and right panels relate to patients 1, 2, and 3, respectively. Note that there were no other dysmorphic features besides oral clefts in all three patients. Written consents were obtained from the patients' legal guardians for publication of the images.
Figure 2 Mutation analysis. The left, middle, and right panels relate to patients 1, 2, and 3, respectively. Electropherograms of (A) patients, showing 269C→T, 937A→G, and 1690G→C (arrows) in patients 1, 2, and 3, respectively; and (B) controls, showing normal genotypes at codons 269 as CC, 937 AA and 1690 GG (arrows). (C) Restriction enzyme digestion analysis. F, father; M, mother; Mk, 100 bp marker; P, patient. The arrowhead indicates the 500 bp marker. In the left panel, BseRI digested the 437 bp product of the father into two products of 147 and 290 bp. The 269C→T mutation in patient 1 and her mother removes the BseRI site, leaving the undigested 437 bp products present with the digested 147 and 290 bp products of the normal alleles, indicating that patient 1 and her mother are heterozygous for the 269C→T mutation. In the middle panel, Esp3I does not have a restriction site in the normal alleles of the patient's parents, leaving the 294 bp PCR product intact. The 937A→G mutation in patient 2 creates an Esp3I restriction site. Therefore, the 294 bp PCR product of the mutant allele of the patient is digested into two products of 139 and 155 bp, present along with the undigested 294 bp product of the normal allele, indicating that patient 2 is heterozygous for the 937A→G mutation. In the right panel, BstNI digested the 450 bp PCR product of the mother to two products of 138 and 312 bp. The 1690G→C mutation in patient 3 and her father removes the BstNI site, leaving the undigested 450 bp product present with the digested 138 and 312 bp products of the normal allele, indicating that patient 3 and her father are heterozygous for the 1690G→C mutation.
Patient 2
A 4 year old girl from Srakaew province was the second child of a 20 year old mother and a 30 year old father. The mother had had only the one previous pregnancy. The child had a surgically repaired bilateral complete cleft lip (fig 1, middle panel). She was heterozygous for an A→G point mutation at nucleotide 937 in exon 8 of p63 (fig 2A, middle panel). The mutation was confirmed by digestion of the PCR products with the restriction enzyme Esp3I; the patient's point mutation adds a recognition site (fig 2C, middle panel). Both parents had only the normal A alleles, by sequencing (data not shown) and restriction enzyme analysis (fig 2C, middle panel). The mutation 937A→G is predicted to result in conversion of an arginine to glycine (R313G). Arginine is a polar, positively charged amino acid while glycine is non‐polar. The arginine at codon 313 is conserved in rat, mouse, chicken, frog, and zebrafish (data not shown). It is in the DB domain and is present in all isotypes of p63. PolyPhen predicted this variant to be probably damaging. The nucleotide 937A is in the ESE motifs of SF2/ASF and SRp40 SR proteins with scores of 4.41 and 4.45, respectively. The 937A→G mutation reduces the score of the site being a potential ESE for the SF2/ASF SR protein to 4.04, and eliminates it as a potential ESE for SRp40 SR protein. R313G was absent from 1000 control chromosomes.
Patient 3
A 4 year old girl from Nongkhai province was the product of the first pregancy of a 26 year old woman. The child's father was aged 26 years. The child had a surgically repaired bilateral complete cleft lip and palate (fig 1, right panel). Direct sequencing revealed that she was heterozygous for a G→C mutation at position 1690 of exon 14 of p63 (fig 2A, right panel). The mutation was confirmed by digestion of the PCR products with the restriction enzyme BstNI; the point mutation removes a recognition site (fig 2C, right panel). The patient's father was also heterozygous for the 1690G→C mutation (fig 2C, right panel) but had no dysmorphic features. Her mother had only the normal G alleles (fig 2C, right panel). The mutation 1690G→C is predicted to result in conversion of an aspartic acid, a polar, negatively charged amino acid, to histidine, a polar, positively charged amino acid (D564H). The aspartic acid at codon 564 is conserved in rat, mouse, and chicken (data not shown). Codon 564 of frog was not available, while that of zebrafish is glutamic acid, which is a polar, negatively charge amino acid, like aspartic acid. Codon 564 is in the SAM domain, just four amino acids from its end. The mutation is expected to alter α isotypes, whereas the β and γ isotypes are unaffected. PolyPhen predicted this variant to be possibly damaging. The nucleotide 1690G is in the ESE motifs of SC35 SR protein with a score of 4.97. The 1690G→C mutation reduces the score of the site being a potential ESE for the SC35 SR protein to 2.94, and makes it a potential ESE for the SRp55 SR protein with a score of 2.92. D564H was found in one of 1000 control chromosomes.
DISCUSSION
Evidence that the 937A→G variant in patient 2 is indeed causative for non‐syndromic CL/P was provided by six observations:
The variant is a nonconservative substitution, predicted to result in conversion of an arginine to glycine (R313G). Arginine is a polar, positively charged amino acid while glycine is non‐polar.
The arginine at codon 313 is evolutionarily conserved in all examined vertebrates, including rat, mouse, chicken, frog, and zebrafish.
It is in the DB domain, a functionally important area, present in all isotypes of p63. A previously reported mutation in p63, D312H, has been found in patients with EEC syndrome and occurs just one amino acid N‐terminal to the mutation found in our patient.19
PolyPhen predicted this variant to be probably damaging.
The variant apparently arose de novo (fig 2C, middle panel).
It was not identified in a cohort of 500 control individuals.
The roles of the 269C→T and 1690G→C variants were inconclusive. They were inherited from one of the patients' clinically normal parents. S90L is not in any functionally important domain and was predicted by PolyPhen to be benign. D564H was found in only one of 1000 control chromosomes. These variants, therefore, could be non‐pathogenic. It may, however, be that they are pathogenic, being deleterious either by causing amino acid changes or from disrupting potential ESEs causing splicing defects, and inherited in a dominant but incompletely penetrant fashion in these families, similar to a previously reported MSX1 P147Q mutation, which also caused non‐syndromic CL/P in Vietnamese families.20 Their causative roles, therefore, need further investigation. Two of the three synonymous changes and eight of the 15 variants in intronic regions occurred only once in 200 chromosomes of our non‐syndromic CL/P patients (table 3); they could contribute to the disease phenotype and also need further investigation.
CL/P is a multifactorial disorder caused by a combination of genes and environmental interactions. These factors may contribute differently to CL/P in different populations.10,20 In the Thai population, a significantly higher frequency of the MTHFR 677CT/1298AC genotype occurs in the mothers of CL/P patients compared with controls (odds ratio 4.43; 95% confidence interval 1.33 to 15.10).6 This observation was reinforced by some studies of certain populations21,22 but not by other studies.23 Therefore, the failure of a previous study to find mutations in p63 among 31 white and 31 Filipino cases with non‐syndromic orofacial clefts24 may be related to small sample size or different populations providing different susceptibility alleles in other genes.
Although at least 48 mutations in p63 have been reported in patients with EEC, AEC, RHS, acro‐dermato‐ungual‐lachrymal‐tooth syndrome, or split hand/split foot malformation (Human Genome Mutation Database, http://archive.uwcm.ac.uk/uwcm/mg/hgmd0.html, searched 10 June 2005), the R313G mutation is novel. This could suggest a genotype‐phenotype correlation. A mutation in p63 causing non‐syndromic CL/P may occur at a site different from those underlying mendelian malformation syndromes. Alternatively, identical mutations might cause either non‐syndromic CL/P or syndromes with clefts, depending on variability of expressivity.
This report demonstrates that the p63 R313G mutation is associated with non‐syndromic CL/P highlights the wide phenotypic spectrum of p63 mutations.
ACKNOWLEDGEMENTS
We wish to thank the patients and their families for their participation in the study, and the medical staff of the Thai Red Cross, the Craniofacial Center of King Chulalongkorn Memorial Hospital, and the Provincial Hospitals of Nakornratchaseema, Nan, Uthaithanee, Maehongsorn, Trang, Srakaew, Kalasin, Nongkhai and Mahasarakam for the excellent care of their patients. We are grateful to Ms I Bernardini of the National Institutes of Health, USA for reviewing the manuscript and to P Stanyasuwan for performing the paternity and maternity testing. This study was supported by the Research Unit Grant from Chulalongkorn University, the National Center for Genetic Engineering and Biotechnology, and the Thailand Research Fund.
Abbreviations
AEC - ankyloblepharon‐ectodermal dysplasia‐clefting syndrome
CL/P - cleft lip with or without cleft palate
DB - DNA binding
EEC - ectrodactyly‐ectodermal dysplasia‐clefting syndrome
ESE - exonic splicing enhancer
RHS - Rapp‐Hodgkin syndrome
SAM - sterile α‐motif, SR, serine/arginine‐rich
TA - transactivating
and TI - transactivation inhibitory
Footnotes
Competing interests: there are no competing interests
We received written consents from the patients' legal guardians for publication of the images.
References
- 1.Mossey P A, Little J. Epidemiology of oral clefts: an international perspective. In: Wyszynski DF, ed. Cleft lip and palate: from origin to treatment. Oxford: Oxford University Press, 2002127–158.
- 2.Schutte B C, Murray J C. The many faces and factors of orofacial clefts. Hum Mol Genet 199981853–1859. [DOI] [PubMed] [Google Scholar]
- 3.Jugessur A, Murray J C. Orofacial clefting: recent insights into a complex trait. Curr Opin Genet Dev 200515270–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marazita M L, Murray J C, Lidral A C, Arcos‐Burgos M, Cooper M E, Goldstein T, Maher B S, Daack‐Hirsch S, Schultz R, Mansilla M A, Field L L, Liu Y E, Prescott N, Malcolm S, Winter R, Ray A, Moreno L, Valencia C, Neiswanger K, Wyszynski D F, Bailey‐Wilson J E, Albacha‐Hejazi H, Beaty T H, McIntosh I, Hetmanski J B, Tuncbilek G, Edwards M, Harkin L, Scott R, Roddick L G. Meta‐analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32–35. Am J Hum Genet 200475161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spritz R A. The genetics and epigenetics of orofacial clefts. Curr Opin Pediatr 200113556–560. [DOI] [PubMed] [Google Scholar]
- 6.Shotelersuk V, Ittiwut C, Siriwan P, Angspatt A. Maternal 677CT/1298AC genotype of the MTHFR gene as a risk factor for cleft lip. J Med Genet 200340e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van den Boogaard M J, Dorland M, Beemer F A, van Amstel H K. MSX1 mutation is associated with orofacial clefting and tooth agenesis in humans. Nat Genet 200024342–343. [DOI] [PubMed] [Google Scholar]
- 8.Jezewski P A, Vieira A R, Nishimura C, Ludwig B, Johnson M, O'Brien S E, Daack‐Hirsch S, Schultz R E, Weber A, Nepomucena B, Romitti P A, Christensen K, Orioli I M, Castilla E E, Machida J, Natsume N, Murray J C. Complete sequencing shows a role for MSX1 in non‐syndromic cleft lip and palate. J Med Genet 200340399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo S, Schutte B C, Richardson R J, Bjork B C, Knight A S, Watanabe Y, Howard E, de Lima R L, Daack‐Hirsch S, Sander A, McDonald‐McGinn D M, Zackai E H, Lammer E J, Aylsworth A S, Ardinger H H, Lidral A C, Pober B R, Moreno L, Arcos‐Burgos M, Valencia C, Houdayer C, Bahuau M, Moretti‐Ferreira D, Richieri‐Costa A, Dixon M J, Murray J C. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet 200232285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zucchero T M, Cooper M E, Maher B S, Daack‐Hirsch S, Nepomuceno B, Ribeiro L, Caprau D, Christensen K, Suzuki Y, Machida J, Natsume N, Yoshiura K, Vieira A R, Orioli I M, Castilla E E, Moreno L, Arcos‐Burgos M, Lidral A C, Field L L, Liu Y E, Ray A, Goldstein T H, Schultz R E, Shi M, Johnson M K, Kondo S, Schutte B C, Marazita M L, Murray J C. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med 2004351769–780. [DOI] [PubMed] [Google Scholar]
- 11.Shotelersuk V, Srichomthong C, Yoshiura K, Niikawa N. A novel mutation, 1234del(C), of the IRF6 in a Thai family with Van der Woude syndrome. Int J Mol Med 200311505–507. [PubMed] [Google Scholar]
- 12.Srichomthong C, Siriwan P, Shotelersuk V. Significant association between IRF6 820G>A and non‐syndromic cleft lip with or without cleft palate in the Thai population. J Med Genet 200542e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Bokhoven H, Hamel B C, Bamshad M, Sangiorgi E, Gurrieri F, Duijf P H, Vanmolkot K R, van Beusekom E, van Beersum S E, Celli J, Merkx G F, Tenconi R, Fryns J P, Verloes A, Newbury‐Ecob R A, Raas‐Rotschild A, Majewski F, Beemer F A, Janecke A, Chitayat D, Crisponi G, Kayserili H, Yates J R, Neri G, Brunner H G. Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell 199999143–153. [DOI] [PubMed] [Google Scholar]
- 14.McGrath J A, Duijf P H, Doetsch V, Irvine A D, de Waal R, Vanmolkot K R, Wessagowit V, Kelly A, Atherton D J, Griffiths W A, Orlow S J, van Haeringen A, Ausems M G, Yang A, McKeon F, Bamshad M A, Brunner H G, Hamel B C, van Bokhoven H. Hay‐Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum Mol Genet 200110221–229. [DOI] [PubMed] [Google Scholar]
- 15.Bougeard G, Hadj‐Rabia S, Faivre L, Sarafan‐Vasseur N, Frebourg T. The Rapp‐Hodgkin syndrome results from mutations of the TP63 gene. Eur J Hum Genet 200311700–704. [DOI] [PubMed] [Google Scholar]
- 16.Shotelersuk V, Janklat S, Siriwan P, Tongkobpetch S. De novo missense mutation, S541Y, in the p63 gene underlying Rapp‐Hodgkin ectodermal dysplasia syndrome. Clin Exp Dermatol 200530282–285. [DOI] [PubMed] [Google Scholar]
- 17.van Bokhoven H, Brunner H G. Splitting p63. Am J Hum Genet 2002711–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shotelersuk V, Desudchit T, Tongkobpetch S. ASA E382K disrupts a potential exonic splicing enhancer and causes exon skipping, but missense mutations in ASA are not associated with ESEs. Int J Mol Med 200414683–689. [PubMed] [Google Scholar]
- 19.van Bokhoven H, Hamel B C, Bamshad M, Sangiorgi E, Gurrieri F, Duijf P H, Vanmolkot K R, van Beusekom E, van Beersum S E, Celli J, Merkx G F, Tenconi R, Fryns J P, Verloes A, Newbury‐Ecob R A, Raas‐Rotschild A, Majewski F, Beemer F A, Janecke A, Chitayat D, Crisponi G, Kayserili H, Yates J R, Neri G, Brunner H G. p63 Gene mutations in eec syndrome, limb‐mammary syndrome, and isolated split hand‐split foot malformation suggest a genotype‐phenotype correlation. Am J Hum Genet 200169481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki Y, Jezewski P A, Machida J, Watanabe Y, Shi M, Cooper M E, Viet le T, Nguyen T D, Hai H, Natsume N, Shimozato K, Marazita M L, Murray J C. In a Vietnamese population, MSX1 variants contribute to cleft lip and palate. Genet Med 20046117–125. [DOI] [PubMed] [Google Scholar]
- 21.Pezzetti F, Martinelli M, Scapoli L, Carinci F, Palmieri A, Marchesini J, Carinci P, Caramelli E, Rullo R, Gombos F, Tognon M. Maternal MTHFR variant forms increase the risk in offspring of isolated nonsyndromic cleft lip with or without cleft palate. Hum Mutat 200424104–105. [DOI] [PubMed] [Google Scholar]
- 22.Gaspar D A, Matioli S R, de Cassia Pavanello R, Araujo B C, Alonso N, Wyszynski D, Passos‐Bueno M R. Maternal MTHFR interacts with the offspring's BCL3 genotypes, but not with TGFA, in increasing risk to nonsyndromic cleft lip with or without cleft palate. Eur J Hum Genet 200412521–526. [DOI] [PubMed] [Google Scholar]
- 23.Jugessur A, Wilcox A J, Lie R T, Murray J C, Taylor J A, Ulvik A, Drevon C A, Vindenes H A, Abyholm F E. Exploring the effects of methylenetetrahydrofolate reductase gene variants C677T and A1298C on the risk of orofacial clefts in 261 Norwegian case‐parent triads. Am J Epidemiol 20031571083–1091. [DOI] [PubMed] [Google Scholar]
- 24.Barrow L L, van Bokhoven H, Daack‐Hirsch S, Andersen T, van Beersum S E, Gorlin R, Murray J C. Analysis of the p63 gene in classical EEC syndrome, related syndromes, and non‐syndromic orofacial clefts. J Med Genet 200239559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]