Abstract
We report on a multigenerational family with isolated Hirschsprung's disease (HSCR). Five patients were affected by either short segment or long segment HSCR. The family consists of two main branches: one with four patients (three siblings and one maternal uncle) and one with one patient. Analysis of the RET gene, the major gene involved in HSCR susceptibility, revealed neither linkage nor mutations. A genome wide linkage analysis was performed, revealing suggestive linkage to a region on 4q31–q32 with a maximum parametric multipoint LOD score of 2.7. Furthermore, non‐parametric linkage (NPL) analysis of the genome wide scan data revealed a NPL score of 2.54 (p = 0.003) for the same region on chromosome 4q (D4S413–D4S3351). The minimum linkage interval spans a region of 11.7 cM (12.2 Mb). No genes within this chromosomal interval have previously been implicated in HSCR. Considering the low penetrance of disease in this family, the 4q locus may be necessary but not sufficient to cause HSCR in the absence of modifying loci elsewhere in the genome. Our results suggest the existence of a new susceptibility locus for HSCR at 4q31.3‐q32.3.
Keywords: Hirschsprung's disease, RET , 4q31.3–q32.3
Hirschsprung's disease (HSCR; OMIM #143623) is a congenital disorder characterised by the absence of enteric neurones, which are derived from the neural crest, in the digestive tract. Delayed passage of meconium is the cardinal symptom in neonates with HSCR. If untreated, bowel hypomotility leads to severe constipation, often associated with obstruction, gross distension of the bowel, and vomiting. The prevalence of HSCR is approximately 1 in every 5000 liveborn infants. In the majority of cases (75–80%) the aganglionosis typically involves the rectum and the sigmoid (short segment HSCR; SS‐HSCR). In 20–25% of patients the aganglionosis extends proximally of the rectosigmoid, and the disease is called long segment HSCR (LS‐HSCR).1 HSCR mostly presents as an isolated congenital malformation (non‐syndromic HSCR), but can be found in association with other congenital abnormalities (syndromic HSCR).2
So far mutations in 10 genes (RET, GDNF, EDNRB, EDN3, ECE1, SOX10, ZFHX1B, NTN, PHOX2B, and KIAA1279) have been implicated in HSCR.3,4,5,6,7,8,9,10,11,12,13,14,15,16 The RET gene, located at 10q11.2, is the major susceptibility locus in HSCR; 15%–35% of the sporadic patients have inactivating mutations in the coding sequence of RET,17,18,19,20 and linkage analysis has shown that all autosomal dominant families but one are linked to RET.21 High penetrance mutations in the coding sequence of the RET gene, however, are found in only 50% of the RET linked families. RET linked multigenerational families without a coding sequence mutation may have a mutation in the non‐coding sequence of the RET gene, including alterations in intronic and promoter sequences, or may harbour (frequent) variants that change the function of the RET protein slightly.21 Furthermore, similar haplotypes are found in the 5′ region of the RET locus in HSCR patient populations from all over the world, indicating the segregation of identical ancestral variant(s).22,23,24,25,26 Evidence is accumulating that specific (common) non‐coding low penetrance variants just before the gene and within intron 1 of RET are associated with susceptibility to HSCR.22,25,27,28,29,30
Single mutations leading to either isolated or syndromic HSCR have been found in the aforementioned 10 genes, although evidence is building up that HSCR is a multigenic congenital malformation in the majority of HSCR patients. A “multiplicative model” has been suggested, which assumes that additional loci are involved apart from the RET locus and that their individual effects can be multiplied.31
Furthermore, four HSCR susceptibility loci (9q31, 3p21, 19q12, and 16q23) have been identified, harbouring unidentified genes.21,31,32 Linkage at 9q31 was reported in five families that also showed linkage with RET; however, no causative RET mutation could be identified. A sixth family that did not show linkage to RET was linked to the locus at 9q31.21 Furthermore, susceptibility loci at 3p21 and 19q12 have been identified in affected nuclear families, suggesting that these two loci probably function as RET dependent modifiers. Non‐random allele sharing was also found at 9q31 in nuclear families in which no RET mutation was identified, confirming the segregation of the 9q31 locus in multiplex families.31,21
In an inbred Mennonite population, RET not only interacts with EDNRB, but also with an unknown gene on chromosome 16q23. This locus is probably only of importance in this genetically isolated population,32 in which HSCR can be associated with symptoms also found in Shah‐Waardenburg syndrome.7 Conversely, no linkage with the other susceptibility loci at 3p21, 9q31 and 19q12 was found in this Mennonite kindred.
Clearly, HSCR is a heterogeneous congenital malformation. It is estimated that only 30% of cases can be attributed to mutations of the known genes.33 Thus, a considerable number of additional genes involved in enteric nervous system development could be identified in the future.28 Here, we describe a five generation family with non‐syndromic HSCR with suggestive evidence for a new susceptibility locus on chromosome 4.
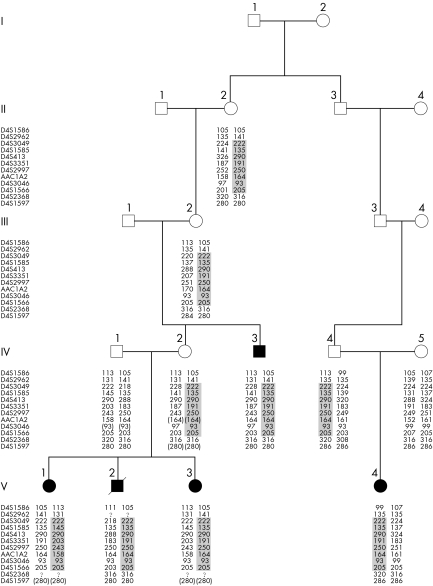
METHODS
Patients
The family described here is a five generation pedigree of native Dutch origin with five cases presenting HSCR. The segregation pattern is compatible with an autosomal dominant mode of inheritance with incomplete penetrance. One branch of the family consists of a sibship with three affected children and an affected uncle. Two sisters (V‐1 and V‐3) have SS‐HSCR. Their brother (V‐2) was diagnosed with total intestinal aganglionosis, both large and small intestines were aganglionic. Given the poor prognosis, a joint medical and parental decision was made for conservative care of HSCR; he died at the age of 1 month. Delayed passage of meconium led to the suspected diagnosis of HSCR in all three children. Suction and/or full thickness biopsies were consistent with this diagnosis. Their maternal uncle (IV‐3) had undergone surgery during childhood because of SS‐HSCR. The other branch of the family contains one affected female member with SS‐HSCR (V‐4). Her paternal grandfather (III‐3) is a cousin of the maternal grandmother (III‐2) of the three siblings detailed above (fig 1). Congenital malformations indicative of syndromic HSCR were lacking in all five patients as confirmed by two dysmorphologists (ASB and IvL). Karyotyping was normal in patients V‐1 and V‐2. Brainstem evoked response audiometry, which we performed because of his expected early death and to exclude hearing loss consistent with Shah‐Waardenburg syndrome, showed no abnormalities in patient V‐2. Chronic severe constipation was not reported in II‐2, III‐2, or IV‐4, although IV‐2 and her father (III‐1) experienced severe constipation in childhood. Informed consent was given by the parents of the children and by the adult patient (IV‐3). Genomic DNA was isolated from peripheral blood obtained from II‐2, III‐2, IV‐1, IV‐2, IV‐3, IV‐4, IV‐5, V‐1, V‐2, V‐3, and V‐4 using standard protocols.34
Figure 1 Pedigree structure and haplotypes. Patients are represented as filled symbols. The segregating 4q31‐q32 haplotype is depicted. The minimum critical region at chromosome 4 spans 11.7 cM between D4S3049 and D4S1566.
Analysis of markers encompassing the RET locus
The following markers D10S141 (−1 Mb to RET), RETint5, D10S1099 (1.4 Mb downstream to RET) and five single nucleotide polymorphisms (SNPs) (rs741763, rs2435362, rs2565206, rs2506004, and rs2435357) from the 5′RET region were analysed as reported by us and others.26,27,28
Mutational analysis of RET
Mutation analysis of the 21 exons of RET was performed in patients V‐1 and V‐4, as described previously.20
Genome wide linkage analysis
We performed a systematic genome scan using the ABI Prism MD‐10 set (Applied Biosystems) consisting of 382 markers (short tandem repeat polymorphisms; STRPs), with an average spacing of 10 cM. Additional markers for further characterisation of candidate regions were selected from the sex average Marshfield genetic map or were newly designed. Genomic DNA (20 ng) was used as a template in 7.5 μl PCR reactions, with 5 pmol of oligonucleotides, 0.3 U AmpliTaq Gold polymerase in Gold buffer (Applied Biosystems) and 2.5 mmol/l MgCl2. The thermal cycling consisted of an initial incubation at 95°C for 5 minutes, followed by 10 cycles of 95°C for 30 seconds, 55°C for 15 seconds, and 72°C for 30 seconds, then 25 cycles of 92°C for 30 seconds, 55°C for 15 seconds, and 72°C for 30 seconds. PCR products were pooled and loaded on an automated sequencer (ABI3100; Applied Biosystems). Data were analysed using GeneMapper software (version 2.0). Mega235 was used to process the genetic data into the appropriate format and perform data validation checks. Simulation analysis to estimate the probability of detecting genetic linkage given the pedigree structure (statistical power) was performed with the SLINK program.36 Because of uncertainties related to the correct genetic model in this pedigree, parametric and non‐parametric linkage analyses were performed using SimWalk2 software (version 2.9).37
For the parametric analysis, we specified an autosomal dominant mode of inheritance, a mutant allele frequency of 0.01% with a penetrance of 40% and equal marker allele frequencies. Pedigree location scores were calculated; these location scores are directly comparable to multipoint LOD scores. For the non‐parametric analysis, the maximum tree statistic (the largest number of affected members inheriting an allele from one founder allele) is reported. This statistic was designed for traits best modelled by dominant inheritance and was formerly known as STAT B. The NPL_ALL (STAT E) statistic, a measure of whether a few founder alleles are overly presented in affected memberss (suitable for an additive model) is reported as well. A large value of the statistic indicates a high degree of identity by descent allele sharing among the patients, and usually a result >2 can be considered significant. Empirical p values (10 000 simulations) were also obtained. This p value is the probability of obtaining a value for that statistic that is equal to or greater than the observed value, if the trait were not linked to the markers.
RESULTS
Linkage analysis to the RET locus and sequence analysis of the RET gene
The family we investigated is a five generation pedigree of native Dutch origin with five cases presenting HSCR (fig 1). Because the majority of the multigenerational families with HSCR show linkage to the RET gene,21 we investigated the RET locus by haplotype and mutational analysis. We observed that not all five patients (IV‐3, V‐1, V‐2, V‐3, and V‐4) shared the same haplotype at the RET locus (table 1), excluding linkage with the RET locus in both branches of this family. However, IV‐2, IV‐3, V‐1, V‐2, and V‐3 from branch 1 did share the same haplotype encompassing the RET locus, which must be inherited from III‐1. We also performed sequence analysis of the entire coding region of the RET gene. Direct sequencing revealed no mutations in patients V‐1 (branch 1) and V‐4 (branch 2).
Table 1 RET haplotypes.
| II‐2 | III‐2 | IV‐1 | IV‐2 | IV‐3 | IV‐4 | IV‐5 | V‐1 | V‐2 | V‐3 | V‐4 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D10S141 | 3 1 | 3 2 | 2 2 | 4 3 | 4 2 | 3 4 | 2 2 | 2 4 | 2 4 | 2 4 | 3 2 | |||||||||||
| rs741763 | C G | C G | G G | C C | C G | C C | G G | G C | G C | G C | C G | |||||||||||
| rs2435362 | C C | C C | A A | C C | C C | C C | C A | A C | A C | A C | C C | |||||||||||
| rs2435357 | C C | C C | T T | C C | C C | C C | C T | T C | T C | T C | C C | |||||||||||
| rs2506004 | C C | C C | A A | C C | C C | C C | C A | A C | A C | A C | C C | |||||||||||
| rs2565206 | G T | G T | G G | G G | G T | G G | T G | G G | G G | G G | G T | |||||||||||
| Retint5 | 3 2 | 3 2 | 1 3 | 2 3 | 2 2 | 3 1 | 2 1 | 1 2 | 1 2 | 3 2 | 3 2 | |||||||||||
| D10S1099 | 5 2 | 5 5 | 5 4 | 5 5 | 5 5 | 2 3 | 5 1 | 5 5 | 5 5 | 4 5 | 2 5 |
Affected family members are in bold.
Genome search
Simulation analysis (using SLINK) yielded an average LOD score of 1.54 and a maximum of 2.15. We performed a genome wide search using 382 STRPs. Results from the parametric linkage analysis excluded most of the genome (data not shown). Only two genomic regions displayed LOD scores >1, a region on chromosome 17 (D17S798, mLOD score = 1.15) and on chromosome 4 (mLOD = 1.84). We tested additional markers and performed haplotype analysis. The region on chromosome 17 was rapidly excluded because one patient (IV‐3) was not sharing the haplotype observed in patients from branch 1.
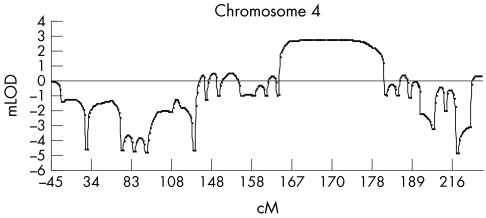
The highest multipoint LOD score (mLOD = 1.84) was obtained for chromosome 4 between markers D4S424 and D4S413. This region fully segregated with the disease phenotype (fig 1). When we saturated the chromosome 4 region with additional markers, a maximum mLOD of 2.7 (fig 2) was reached between markers D4S1585 and D4S3351, which is consistent with suggestive linkage according to the Lander‐Kruglyak guidelines for significance thresholds.38
Figure 2 Multipoint LOD score analysis with several markers on chromosome 4. The x axis represents the chromosomal position of the markers. The y axis indicates the multipoint LOD score, showing a peak LOD score of 2.7.
We also performed non‐parametric analysis for our genome wide scan data. We found non‐parametric linkage (NPL) scores >1 only for this region on chromosome 4q. The maximum NPL was found for marker D4S413 (158.0 Mb, NCBI build 35.1) located between markers D4S1585 and D4S3351 (NPL = 2.55, p = 0.003), under both dominant and additive mode of inheritance (table 2).
Table 2 Results from the non‐parametric analysis on chromosome 4q after fine mapping.
| Marker | Genetic position (cM*) | Physical position (Mb)† | Max tree‡ | NPL_ All§ | Empirical p value | ||||
|---|---|---|---|---|---|---|---|---|---|
| D4S1575 | 132.1 | 135.1 | 0.35 | 0.38 | 0.418 | ||||
| D4S1644 | 143.3 | 142.1 | 0.80 | 0.83 | 0.148 | ||||
| D4S424 | 144.6 | 142.6 | 0.82 | 0.84 | 0.143 | ||||
| D4S1625 | 146.0 | 143.9 | 0.84 | 0.85 | 0.141 | ||||
| D4S1586 | 147.1 | 147.1 | 0.87 | 0.86 | 0.136 | ||||
| EDNRA | – | 148.8 | 0.86 | 0.86 | 0.137 | ||||
| D4S2962 | 153.0 | 150.7 | 0.87 | 0.86 | 0.137 | ||||
| D4S3049 | 155.2 | 155.1 | 1.60 | 1.60 | 0.025 | ||||
| D4S1585 | 158.0 | 157.9 | 2.42 | 2.42 | 0.004 | ||||
| D4S413 | 158.0 | 158.7 | 2.54 | 2.55 | 0.003 | ||||
| D4S3351 | 158.7 | 159.9 | 2.54 | 2.54 | 0.003 | ||||
| D4S2997 | 158.7 | 160.0 | 2.52 | 2.51 | 0.003 | ||||
| AAC1A2 | 158.0 | 160.0 | 2.51 | 2.51 | 0.003 | ||||
| D4S3046 | 162.5 | 163.7 | 1.55 | 1.57 | 0.030 | ||||
| D4S1566 | 166.9 | 167.3 | 0.99 | 0.99 | 0.102 | ||||
| D4S2368 | 167.6 | 169.1 | 0.83 | 0.84 | 0.144 | ||||
| D4S1597 | 169.4 | 170.2 | 0.74 | 0.79 | 0.163 | ||||
| D4S2431 | 176.2 | 175.2 | 0.51 | 0.63 | 0.234 | ||||
| D4S415 | 181.4 | 179.1 | 0.10 | 0.11 | 0.776 |
*According to the Marshfield sex average genetic map; †according to the NCBI physical map, build 35.1; ‡the allele sharing statistic for traits best modelled as dominant inheritance; §the statistic for traits following an additive inheritance. Significant p values are shown in bold type.
Recombination events can be identified in individual IV‐4 and show that marker D4S2962 at the centromeric site, and D4S2368 at the telomeric site limits the critical region. The maximum critical region between D4S2962 and D4S2368 spans approximately 16.4 cM (19.7 Mb according to NCBI physical map, build 35.1), the minimum shared region extend from marker D4S3049 until D4S1566 (11.7 cM or 12.2 Mb).
5′ RET common risk haplotype analysis
To test whether non‐coding low penetrance variants just before or within intron 1 of RET were associated with HSCR susceptibility in a part of this family, we typed five SNPs all being part of an ancestral haplotype.27 Spouse IV‐1 (who had married into the family) was homozygous for the GATAG haplotype that is part of the core risk SNP haplotype detected in European, European American, and Asian American patients with sporadic HSCR.27,28,39 The three affected siblings (V‐1, V‐2, and V‐3) were thus carriers of the core risk haplotype. However, the other two patients (IV‐3 and V‐4) did not carry the 5′ RET common risk haplotype (table 1).
DISCUSSION
We identified a five generation family with five patients affected with HSCR. The patients were connected to a common ancestor within 3–4 generations. Two branches with HSCR patients within the family were identified. The inheritance pattern in the family is compatible with an autosomal dominant mode of inheritance with reduced penetrance of a single mutated gene.
Because RET is the major gene involved in HSCR susceptibility in multigenerational families,21 linkage to the RET locus and mutations in the coding sequence of RET were first excluded in both branches. However, under the assumption that HSCR is caused by two separate genes in the two branches, an alternative hypothesis would be that the individuals of the main branch with four affected individuals (the core two generation family) carry a hitherto unidentified RET mutation (introduced into the family by III‐1). As an oligogenic model with the contribution of two or more loci has been previously demonstrated for HSCR (9q31 and RET), we hypothesised that even if there is true linkage to RET in branch 1, this does not mutually exclude the existence of an additional locus in this family. Subsequently, we performed a genome wide scan to map additional disease gene(s) in this family. Model free or non‐parametric linkage analysis methods are more robust than parametric or model dependent analysis when the mode of inheritance or the genetic model is uncertain, such as this pedigree. Consequently, we performed both parametric and non‐parametric linkage analysis. Both methods highlighted the same region on chromosome 4q, and no other known HSCR susceptibility loci such as those at chromosomes 3, 9, and 1921,31 showed positive LOD scores. When adjacent markers for chromosome 4 were tested, the evidence of linkage became stronger and we could observe a common haplotype, extending at least 11.7 cM, which was inherited by all the affected individuals from their common ancestor. Our results indicate the existence of a novel HSCR susceptibility locus on chromosome 4q.
Clearly, the chromosome 4q locus has incomplete penetrance. Does this 4q locus solely lead to HSCR or alternatively, is the phenotype only expressed in the presence of other susceptibility loci? Modifier loci either can increase susceptibility and severity of the phenotype or can act protectively to confer resistance to the disease in the face of a predisposing mutation.40 Can variants within the RET gene explain the difference in penetrance observed in both branches? We investigated whether all patients shared a haplotype similar to the common risk haplotype defined by SNPs located in the 5′ region of the RET locus reported in Dutch HSCR patients.26 We identified non‐risk haplotypes in patients IV‐3 and V‐4. However, the spouse IV‐1 and the three affected children (V‐1, V‐2, and V‐3) were homozygous and heterozygous, respectively, for the GATAG haplotype, which contains the core risk haplotype detected in European, European American, and Asian American patients with sporadic HSCR.27,28 The third and fourth SNPs (RET3+, rs2435357 and IVS1+9494, rs2506004) are particularly interesting as disease associated RET variants, because of the homology and evolutionary conservation between rodents and primates and the differences in allele/genotype frequencies among patients and controls.27 Recent data show that RET3+ might lie within, and might compromise the activity of an enhancer‐like sequence in the RET intron 1.28 These findings make us hypothesise that the 5′ RET risk haplotype in combination with the identified chromosome 4 locus might play a role in the penetrance and severity observed in the sibship with three affected children.
We looked for candidate genes in the minimum 12.2 Mb linked region at 4q31.3–32.3 (between markers D4S3049 and D4S1566) in the human genome sequence. This region contains at least 57 genes, in accordance with the National Center for Biotechnology Information (build 35.1) of the human genome and the Ensemble Genome Browser. The maximum 20 Mb linked region between D4S2962 and D4S1597 encompasses 93 genes, including several interesting functional candidate genes that are proposed to be involved in neural crest development or neuronal development.
Unfortunately, the maximum genetic interval contains far too many candidates to begin functional evaluation of each gene individually; the best positional and functional candidate we could identify is Mab21L2 (named after male abnormal 21 in Caenorhabtidis elegans). Mab21L2 is expressed in the central nervous system and neural crest in midgestation embryogenesis in mice.41 Furthermore, Mab21L2 is linked to the transforming growth factor‐β signalling pathway to which ZFHX1, the gene involved in Mowat‐Wilson syndrome, a syndromic form of HSCR, also belongs.13,42ZFHX1B functions as a transcriptional repressor.42 For these reasons we sequenced the complete coding region of the Mab21L2 gene for mutations; however, we did not identify a sequence variant in Mab21L2. Besides Mab21L2, many other candidate genes are located in the region. Several proteins encoding neuropeptide Y receptors (NPY1R, NPY2R, and NPY5R) are located in the minimum 12.2 Mb linked region. In mammals, NPY, the ligand, is mainly found in cells derived from the neural crest, and is widely distributed in the central and peripheral nervous system.43 Furthermore, the gene for the secreted frizzled related protein 2 (SFRP2) is located in this region.44 Wnt‐frizzled signalling is involved in neural crest formation.45
ELECTRONIC DATABASE INFORMATION
Online Mendelian Inheritance in Man (OMIM): http://www.ncbi.nlm.nih.gov/Omim (for MIM 142623)
The 4q locus may be necessary but not sufficient to cause HSCR in the absence of modifying loci elsewhere in the genome. Linkage analysis in additional multigenerational HSCR families and association studies in sporadic patients with high density marker sets covering the entire interval will be necessary to confirm this suggestive linkage to chromosome 4q. Eventually, identification of the causative gene defect will specify the susceptibility to HSCR conferred by this novel locus at 4q31.3‐q32.3.
ACKNOWLEDGEMENTS
We are grateful to the family who made this study possible. This work was supported in part by The Termeulen foundation (grant to ASB) and the Nederlandse organisatie voor Wetenschappelijk Onderzoek (grants 901‐04‐210 and 901‐04‐225 to RMWH).
Abbreviations
HSCR - Hirschsprung's disease
LS‐HSCR - long segment Hirschsprung's disease
NPL - non‐parametric linkage
SNP - single nucleotide polymorphism
SS‐HSCR - short segment Hirschsprung's disease
STRP - short tandem repeat polymorphism
Footnotes
Competing interests: there are no competing interests
References
- 1.Puri P. Hirschsprung's disease: Clinical Generalities. In: Holschneider A, ed. Hirschsprung's disease. 2nd ed. Amsterdam: Harwood Academic Publishers, 2000129–135.
- 2.Amiel J, Lyonnet S. Hirschsprung disease, associated syndromes, and genetics: a review. J Med Genet 200138729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romeo G, Ronchetto P, Luo Y, Barone V, Seri M, Ceccherini I, Pasini B, Bocciardi R, Lerone M, Kaariainen H, Martucciello G. Point mutations affecting the tyrosine kinase domain of the RET proto‐oncogene in Hirschsprung's disease. Nature 1994367377–378. [DOI] [PubMed] [Google Scholar]
- 4.Edery P, Lyonnet S, Mulligan L M, Pelet A, Dow E, Abel L, Holder S, Nihoul‐Fekete C, Ponder B A, Munnich A. Mutations of the RET proto‐oncogene in Hirschsprung's disease. Nature 1994367378–380. [DOI] [PubMed] [Google Scholar]
- 5.Angrist M, Bolk S, Halushka M, Lapchak P A, Chakravarti A. Germline mutations in glial cell line‐derived neurotrophic factor (GDNF) and RET in a Hirschsprung disease patient. Nat Genet 199614341–344. [DOI] [PubMed] [Google Scholar]
- 6.Salomon R, Attie T, Pelet A, Bidaud C, Eng C, Amiel J, Sarnacki S, Goulet O, Ricour C, Nihoul‐Fekete C, Munnich A, Lyonnet S. Germline mutations of the RET ligand GDNF are not sufficient to cause Hirschsprung disease. Nat Genet 199614345–347. [DOI] [PubMed] [Google Scholar]
- 7.Puffenberger E G, Hosoda K, Washington S S, Nakao K, deWit D, Yanagisawa M, Chakravart A. A missense mutation of the endothelin‐B receptor gene in multigenic Hirschsprung's disease. Cell 1994791257–1266. [DOI] [PubMed] [Google Scholar]
- 8.Hofstra R M, Osinga J, Tan‐Sindhunata G, Wu Y, Kamsteeg E J, Stulp R P, van Ravenswaaij‐Arts C, Majoor‐Krakauer D, Angrist M, Chakravarti A, Meijers C, Buys C H. A homozygous mutation in the endothelin‐3 gene associated with a combined Waardenburg type 2 and Hirschsprung phenotype (Shah‐Waardenburg syndrome). Nat Genet 199612445–447. [DOI] [PubMed] [Google Scholar]
- 9.Edery P, Attie T, Amiel J, Pelet A, Eng C, Hofstra R M, Martelli H, Bidaud C, Munnich A, Lyonnet S. Mutation of the endothelin‐3 gene in the Waardenburg‐Hirschsprung disease (Shah‐Waardenburg syndrome). Nat Genet 199612442–444. [DOI] [PubMed] [Google Scholar]
- 10.Hofstra R M, Valdenaire O, Arch E, Osinga J, Kroes H, Loffler B M, Hamosh A, Meijers C, Buys C H. A loss‐of‐function mutation in the endothelin‐converting enzyme 1 (ECE‐1) associated with Hirschsprung disease, cardiac defects, and autonomic dysfunction. Am J Hum Genet 199964304–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pingault V, Bondurand N, Kuhlbrodt K, Goerich D E, Prehu M O, Puliti A, Herbarth B, Hermans‐Borgmeyer I, Legius E, Matthijs G, Amiel J, Lyonnet S, Ceccherini I, Romeo G, Smith J C, Read A P, Wegner M, Goossens M. SOX10 mutations in patients with Waardenburg‐Hirschsprung disease. Nat Genet 199818171–173. [DOI] [PubMed] [Google Scholar]
- 12.Cacheux V, Dastot‐Le Moal F, Kaariainen H, Bondurand N, Rintala R, Boissier B, Wilson M, Mowat D, Goossens M. Loss‐of‐function mutations in SIP1 Smad interacting protein 1 result in a syndromic Hirschsprung disease. Hum Mol Genet 2001101503–1510. [DOI] [PubMed] [Google Scholar]
- 13.Wakamatsu N, Yamada Y, Yamada K, Ono T, Nomura N, Taniguchi H, Kitoh H, Mutoh N, Yamanaka T, Mushiake K, Kato K, Sonta S, Nagaya M. Mutations in SIP1, encoding Smad interacting protein‐1, cause a form of Hirschsprung disease. Nat Genet 200127369–370. [DOI] [PubMed] [Google Scholar]
- 14.Doray B, Salomon R, Amiel J, Pelet A, Touraine R, Billaud M, Attie T, Bachy B, Munnich A, Lyonnet S. Mutation of the RET ligand, neurturin, supports multigenic inheritance in Hirschsprung disease. Hum Mol Genet 199871449–1452. [DOI] [PubMed] [Google Scholar]
- 15.Amiel J, Laudier B, Attie‐Bitach T, Trang H, de Pontual L, Gener B, Trochet D, Etchevers H, Ray P, Simonneau M, Vekemans M, Munnich A, Gaultier C, Lyonnet S. Polyalanine expansion and frameshift mutations of the paired‐like homeobox gene PHOX2B in congenital central hypoventilation syndrome. Nat Genet 200333459–461. [DOI] [PubMed] [Google Scholar]
- 16.Brooks A S, Bertoli‐Avella A M, Burzynski G M, Breedveld G J, Osinga J, Boven L G, Hurst J A, Mancini G M, Lequin M H, de Coo R F, Matera I, de Graaff E, Meijers C, Willems P J, Tibboel D, Oostra B A, Hofstra R M. Homozygous nonsense mutations in KIAA1279 are associated with malformations of the central and enteric nervous systems. Am J Hum Genet 200577120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Angrist M, Bolk S, Thiel B, Puffenberger E G, Hofstra R M, Buys C H, Cass D T, Chakravarti A. Mutation analysis of the RET receptor tyrosine kinase in Hirschsprung disease. Hum Mol Genet 19954821–830. [DOI] [PubMed] [Google Scholar]
- 18.Attie T, Pelet A, Edery P, Eng C, Mulligan L M, Amiel J, Boutrand L, Beldjord C, Nihoul‐Fekete C, Munnich A.et al Diversity of RET proto‐oncogene mutations in familial and sporadic Hirschsprung disease. Hum Mol Genet 199541381–1386. [DOI] [PubMed] [Google Scholar]
- 19.Seri M, Yin L, Barone V, Bolino A, Celli I, Bocciardi R, Pasini B, Ceccherini I, Lerone M, Kristoffersson U, Larsson L T, Casasa J M, Cass D T, Abramowicz M J, Vanderwinden J M, Kravcenkiene I, Baric I, Silengo M, Martucciello G, Romeo G. Frequency of RET mutations in long‐ and short‐segment Hirschsprung disease. Hum Mutat 19979243–249. [DOI] [PubMed] [Google Scholar]
- 20.Hofstra R M, Wu Y, Stulp R P, Elfferich P, Osinga J, Maas S M, Siderius L, Brooks A S, vd Ende J J, Heydendael V M, Severijnen R S, Bax K M, Meijers C, Buys C H. RET and GDNF gene scanning in Hirschsprung patients using two dual denaturing gel systems. Hum Mutat 200015418–429. [DOI] [PubMed] [Google Scholar]
- 21.Bolk S, Pelet A, Hofstra R M, Angrist M, Salomon R, Croaker D, Buys C H, Lyonnet S, Chakravarti A. A human model for multigenic inheritance: phenotypic expression in Hirschsprung disease requires both the RET gene and a new 9q31 locus. Proc Natl Acad Sci U S A 200097268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borrego S, Wright F A, Fernandez R M, Williams N, Lopez‐Alonso M, Davuluri R, Antinolo G, Eng C. A founding locus within the RET proto‐oncogene may account for a large proportion of apparently sporadic Hirschsprung disease and a subset of cases of sporadic medullary thyroid carcinoma. Am J Hum Genet 20037288–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fitze G, Schierz M, Kuhlisch E, Schreiber M, Ziegler A, Roesner D, Schackert H K. Novel intronic polymorphisms in the RET proto‐oncogene and their association with Hirschsprung disease. Hum Mutat 200322177. [DOI] [PubMed] [Google Scholar]
- 24.Sancandi M, Griseri P, Pesce B, Patrone G, Puppo F, Lerone M, Martucciello G, Romeo G, Ravazzolo R, Devoto M, Ceccherini I. Single nucleotide polymorphic alleles in the 5′ region of the RET proto‐oncogene define a risk haplotype in Hirschsprung's disease. J Med Genet 200340714–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griseri P, Bachetti T, Puppo F, Lantieri F, Ravazzolo R, Devoto M, Ceccherini I. A common haplotype at the 5′ end of the RET proto‐oncogene, overrepresented in Hirschsprung patients, is associated with reduced gene expression. Hum Mutat 200525189–195. [DOI] [PubMed] [Google Scholar]
- 26.Burzynski G M, Nolte I M, Osinga J, Ceccherini I, Twigt B, Maas S, Brooks A, Verheij J, Plaza Menacho I, Buys C H, Hofstra R M. Localizing a putative mutation as the major contributor to the development of sporadic Hirschsprung disease to the RET genomic sequence between the promoter region and exon 2. Eur J Hum Genet 200412604–612. [DOI] [PubMed] [Google Scholar]
- 27.Burzynski G M, Nolte I M, Bronda A, Bos K K, Osinga J, Plaza Menacho I, Twigt B, Maas S, Brooks A S, Verheij J B, Buys C H, Hofstra R M. Identifying candidate Hirschsprung disease‐associated RET variants. Am J Hum Genet 200576850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emison E S, McCallion A S, Kashuk C S, Bush R T, Grice E, Lin S, Portnoy M E, Cutler D J, Green E D, Chakravarti A. A common sex‐dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature 2005434857–863. [DOI] [PubMed] [Google Scholar]
- 29.Pelet A, de Pontual L, Clement‐Ziza M, Salomon R, Mugnier C, Matsuda F, Lathrop M, Munnich A, Feingold J, Lyonnet S, Abel L, Amiel J. Homozygosity for a frequent and weakly penetrant predisposing allele at the RET locus in sporadic Hirschsprung disease. J Med Genet 200542e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernandez R M, Boru G, Pecina A, Jones K, Lopez‐Alonso M, Antinolo G, Borrego S, Eng C. Ancestral RET haplotype associated with Hirschsprung's disease shows linkage disequilibrium breakpoint at ‐1249. J Med Genet 200542322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gabriel S B, Salomon R, Pelet A, Angrist M, Amiel J, Fornage M, Attie‐Bitach T, Olson J M, Hofstra R, Buys C, Steffann J, Munnich A, Lyonnet S, Chakravarti A. Segregation at three loci explains familial and population risk in Hirschsprung disease. Nat Genet 20023189–93. [DOI] [PubMed] [Google Scholar]
- 32.Carrasquillo M M, McCallion A S, Puffenberger E G, Kashuk C S, Nouri N, Chakravarti A. Genome‐wide association study and mouse model identify interaction between RET and EDNRB pathways in Hirschsprung disease. Nat Genet 200232237–244. [DOI] [PubMed] [Google Scholar]
- 33.Chakravarti A. Hirschsprung disease. In: Scriver E, ed. The metabolic and molecular bases of inherited disease. 8 ed. New York: McGraw‐Hill, 20016231–6255.
- 34.Miller S A, Dykes D D, Polesky H F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988161215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill W P, Weeks D E. Mega2: data‐handling for facilitating genetic linkage and association analyses. Bioinformatics 2005212556–2557. [DOI] [PubMed] [Google Scholar]
- 36.Weeks D E, Ott J. Risk calculations under heterogeneity. Am J Hum Genet 198945819–821. [PMC free article] [PubMed] [Google Scholar]
- 37.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker‐sharing statistics. Am J Hum Genet 1996581323–1337. [PMC free article] [PubMed] [Google Scholar]
- 38.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 199511241–247. [DOI] [PubMed] [Google Scholar]
- 39.Garcia‐Barcelo M, Ganster R W, Lui V C, Leon T Y, So M T, Lau A M, Fu M, Sham M H, Knight J, Zannini M S, Sham P C, Tam P K. TTF‐1 and RET promoter SNPs: regulation of RET transcription in Hirschsprung's disease. Hum Mol Genet 200514191–204. [DOI] [PubMed] [Google Scholar]
- 40.Nadeau J H. Modifier genes and protective alleles in humans and mice. Curr Opin Genet Dev 200313290–295. [DOI] [PubMed] [Google Scholar]
- 41.Mariani M, Baldessari D, Francisconi S, Viggiano L, Rocchi M, Zappavigna V, Malgaretti N, Consalez G G. Two murine and human homologs of mab‐21, a cell fate determination gene involved in Caenorhabditis elegans neural development. Hum Mol Genet 199982397–2406. [DOI] [PubMed] [Google Scholar]
- 42.Verschueren K, Remacle J E, Collart C, Kraft H, Baker B S, Tylzanowski P, Nelles L, Wuytens G, Su M T, Bodmer R, Smith J C, Huylebroeck D. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5′‐CACCT sequences in candidate target genes. J Biol Chem 199927420489–20498. [DOI] [PubMed] [Google Scholar]
- 43.Thorsell A, Heilig M. Diverse functions of neuropeptide Y revealed using genetically modified animals. Neuropeptides 200236182–193. [DOI] [PubMed] [Google Scholar]
- 44.Chang J T, Esumi N, Moore K, Li Y, Zhang S, Chew C, Goodman B, Rattner A, Moody S, Stetten G, Campochiaro P A, Zack D J. Cloning and characterization of a secreted frizzled‐related protein that is expressed by the retinal pigment epithelium. Hum Mol Genet 19998575–583. [DOI] [PubMed] [Google Scholar]
- 45.Wu J, Saint‐Jeannet J P, Klein P S. Wnt‐frizzled signaling in neural crest formation. Trends Neurosci 20032640–45. [DOI] [PubMed] [Google Scholar]