Abstract
Background
Oculodentodigital syndrome (ODD) is a pleiotropic congenital disorder characterised by abnormalities of the face, eyes, dentition, and limbs. ODD, which is inherited as an autosomal dominant trait, results from missense mutations in the gap junction protein connexin 43.
Objective
To analyse a family with a history of ODD which is inherited in an autosomal recessive manner
Results
ODD in this family resulted from the homozygous mutation R33X in the first transmembrane domain of connexin 43.
Conclusions
The findings provide clear genetic evidence that ODD can be inherited in an autosomal recessive manner and that a dominant negative mechanism underlies autosomal dominant ODD.
Keywords: oculodentodigital syndrome, GJA1 , connexin 43
Oculodentodigital syndrome (ODD; OMIM 164200) is a congenital disorder characterised by abnormalities of the face, eyes, dentition, and limbs. Affected individuals have a long, narrow nose with hypoplastic alae and a prominent nasal bridge, short palpebral fissures, and bilateral microcornea, often with iris anomalies.1,2 The characteristic digital abnormality is bilateral complete syndactyly of the fourth and fifth fingers (type III syndactyly); the third finger may also be involved.3 In addition, microdontia and enamel hypoplasia—which tend to affect both the primary and secondary dentitions—are often observed.3,4
ODD is inherited in an autosomal dominant fashion with high penetrance but variable expression.5 A potential case of autosomal recessive inheritance of ODD has also been documented.6 Recently, Paznekas and co‐workers reported that ODD arises as the result of heterozygous mutations in GJA1, the gene encoding the gap junction protein connexin 43.7 These findings have been confirmed and extended by a series of studies which demonstrate clearly that the pleiotropic features observed in ODD result from mutation of GJA1.8,9,10,11,12 All but two of over 30 mutations that have been reported to date result in missense changes in highly conserved amino acid residues in the N‐terminal two thirds of connexin 43, the exceptions being the mutations F52dup and C260fsX307.7,11 The paucity of deletions or mutations resulting in the introduction of a termination codon into the protein suggests that the underlying mechanism for ODD is either a dominant negative effect or a gain of function rather than haploinsufficiency. In the current investigation, we provide clear genetic evidence that ODD can be inherited in an autosomal recessive manner and that a dominant negative mechanism underlies autosomal dominant ODD.
Methods
Genetic analysis
GJA1 contains two exons, the coding sequence being encompassed in its entirety by the second one. The coding sequence of GJA1 was amplified in two overlapping segments using the primers 5′‐AAT ACG TGA AAC CGT TGG TAG‐3′ and 5′‐CTC TTT CCC TTA ACC CGA TC‐3′, which amplified a product of 855 base pairs (bp), and 5′‐TCT TTG AGG TGG CCT TCT TG‐3′ and 5′‐TAA GGC TGT TGA GTA CCA CC‐3′, which amplified a product of 773 bp. After amplification, the products were excised from a 1% agarose gel and sequenced directly by the dideoxy chain termination method using dye primer chemistry. Primers were designed to avoid amplification of the processed pseudogene GJA1P1 on human chromosome 5q21‐q22.
Results
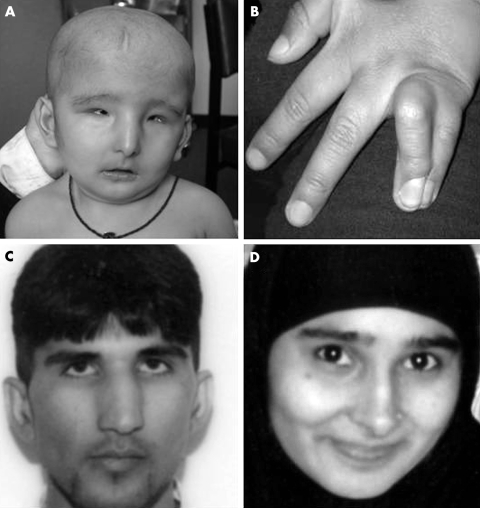
The proband is the first child of a consanguineous couple of Pakistani origin. At birth, she was noted to be hypotonic with large fontanelles and widely separated sutures, small deep set eyes, a pinched nasal appearance with hypoplastic alae nasi, and micrognathia (fig 1A). She had complete skin syndactyly of the left ring and little fingers and the right middle, ring, and little fingers (fig 1B). Ophthalmological assessment showed bilateral microphthalmia, persistent pupillary membrane, and cataract (fig 1A). Tooth eruption was delayed and her deciduous teeth, which were severely hypoplastic, had to be removed in her third year because of chronic abscesses. She has sparse, fine hair which did not appear until her second year. She has gross motor and speech delay and has been investigated for failure to thrive. Her karyotype was 46,XX. Renal and cardiac ultrasound scans were normal. Computed tomography (CT) of the brain showed grossly abnormal grey and white matter differentiation, particularly in the frontal and occipital regions and cerebellum.
Figure 1 Photographs of the family in which the oculodentodigital syndrome (ODD) phenotype is inherited as an autosomal recessive trait. (A) The facial appearance of the proband at eight months showing bilateral microphthalmia, a pinched nasal appearance with hypoplastic alae nasi and micrognathia. (B) The left hand of the proband demonstrating the 4–5 syndactyly. The father (C) and the mother (D) appear to be unaffected clinically. All photographs are reproduced with the written permission of the parents.
The proband's parents went on to have a second daughter. She has a similar facial appearance to her sister; however, she has bilateral syndactyly of her ring and little fingers only. At one year she still has no teeth. She is not delayed in her gross motor milestones and has not had any investigations. Neither parent has similar facial features or syndactyly (fig 1). In addition, dental and ophthalmological examinations were normal and, although brain CT was not done, neither parent reported any neurological symptoms.
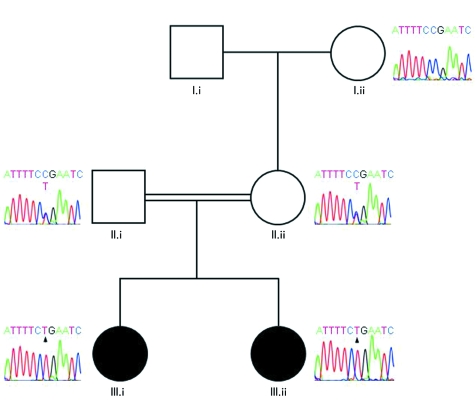
Molecular analysis of DNA samples from the proband and her sister revealed the homozygous nonsense mutation R33X in the first transmembrane domain of connexin 43 (fig 2). Both of the child's parents were heterozygous for this mutation, confirming autosomal recessive inheritance of ODD in this family (fig 2). This mutation was not detected in a panel of 50 control alleles. Our results therefore document the first case of autosomal recessive inheritance of ODD that has been confirmed by molecular analysis and the first nonsense mutation reported to underlie this condition.
Figure 2 Pedigree of the family with partial sequence chromatograms. The maternal grandmother (I.ii) is homozygous for the wild‐type allele, while the unaffected parents (II.i and II.ii) are heterozygous for the R33X mutation. Both of the affected children (III.i and III.ii) are homozygous for the mutated allele (arrowed). DNA samples from remaining family members were not available for analysis.
Discussion
Connexin 43 belongs to a family of proteins which are composed of four transmembrane domains linked by two extracellular and one intracellular loop and intracytoplasmic amino and carboxy termini. Six connexins form a connexon which spans the cell membrane allowing connexons from neighbouring cells to dock to form a complete gap junction. Gap junctions allow the exchange of secondary messengers, ions, and other small molecules up to the size of 1 kDa between adjacent cells, thereby allowing cell–cell communication. Gap junction communication plays a crucial role not only in normal tissue physiology but in the regulation of information flow required during embryonic morphogenesis.13
Mutations in connexin 43 have been shown to cause ODD, which is usually inherited in an autosomal dominant manner. Intriguingly, the vast majority of the mutations that have been reported to date lead to missense changes in connexin 43. This observation, together with the paucity of deletions or mutations resulting in the introduction of a termination codon into the protein, suggests that either a dominant negative or a gain of function mechanism underlies ODD. In the current study, we have provided clear genetic evidence that ODD can be inherited as an autosomal recessive trait. The only previous report of recessive inheritance of an ODD‐like phenotype, which has been confirmed by molecular analysis, was in an individual with a phenotype that bridged the Hallermann‐Streiff/ODD spectrum.10 The mutation that leads to classical ODD in the family analysed in the current study is a nonsense mutation that falls towards the middle of the first transmembrane domain of connexin 43. As GJA1 is a two exon gene with the entire coding sequence contained within exon 2, nonsense mediated mRNA degradation is unlikely to operate in this case.14 Therefore, the consequence of the R33X mutation reported in the current study is that the protein product of the mutant allele will consist solely of the N‐terminal extracellular domain and a portion of first transmembrane domain. Given that the integrity of the four transmembrane domains is known to be essential for correct transport of connexins into the plasma membrane15 and that the extracellular loops play a central role in the specific docking of two hemichannel connexons to form a functional gap junction,16 the mutant protein is likely to be functionless. Evidence from animal models and other autosomal recessive connexinopathies strongly suggests that the deletion of one copy of a connexin gene does not appear to be of major functional significance.17,18,19,20 In contrast, the loss of both copies will be of consequence in cells and tissues where the particular connexin is crucial or where other connexin family members cannot compensate.
Our results provide the first evidence from human genetic analysis that the missense mutations underlying autosomal dominant ODD act in a dominant negative fashion. Our results therefore confirm previous in vitro studies in which the localisation of the ODD mutations G21R and G138R was examined in NRK and HeLa cells. Although GFP tagged mutant proteins were transported to the plasma membrane, they did not form functional gap junction channels. Moreover, both mutants were found to exert dominant negative effects when expressed in connexin 43 positive cells, thereby inhibiting the function of endogenous connexin 43.21 Further support for a dominant negative effect is provided by the study of mice with the ENU induced mutation G60S in the first extracellular loop of connexin 43. Unlike mice heterozygous for a Gja1‐null allele,17Gja1+/G60S mice phenocopy patients affected by ODD.22 In vitro and in vivo studies using the Gja1+/G60S mice indicated that, although gap junctions formed and localised to the to the cell surface in plaque‐like structures, they were considerably less numerous and were non‐functional. Importantly, western analysis indicated that total connexin 43 levels in connexin 43‐expressing tissues of Gja1+/G60S mice were reduced substantially below 50%. These results, together with the data presented in the current paper, confirm that human ODD mutations are not due merely to loss of connexin 43 function; rather, they act in a dominant negative fashion.
Acknowledgements
We thank the family for participating in the study. This work was supported by grants from the Wellcome Trust (058423, 069243).
Abbreviations
ODD - oculodentodigital syndrome
Footnotes
Conflicts of interest: none declared
References
- 1.Sugar H S, Thompson J P, Davis J D. The oculodento‐digital dysplasia syndrome. Am J Ophthalmol 1966611448–1451. [DOI] [PubMed] [Google Scholar]
- 2.Dudgeon J, Chisholm I A. Oculodentodigital dysplasia. Trans Ophthalmol Soc UK 197494203–210. [Google Scholar]
- 3.Reisner S H, Kott E, Bornstein B, Salinger H, Kaplan I, Gorlin R J. Oculodentodigital dysplasia. Am J Dis Child 1969118600–607. [DOI] [PubMed] [Google Scholar]
- 4.Fara M, Horak I, Hrivnakova J, Kapras J, Nova M, Stloukalova M. Oculodentodigital dysplasia. Acta Chir Plast 197719110–122. [PubMed] [Google Scholar]
- 5.Judisch G F, Martin‐Casals A, Hanson J W, Olin W H. Oculodentodigital dysplasia: four new reports and a literature review. Arch Ophthalmol 197997878–884. [DOI] [PubMed] [Google Scholar]
- 6.Frasson M, Calixto N, Cronemberger S, de Aguiar R A L P, Leao L L, de Aguiar M J B. Oculodentodigital dysplasia: study of ophthalmological and clinical manifestations in three boys with probably autosomal recessive inheritance. Ophthalmic Genet 200425227–236. [DOI] [PubMed] [Google Scholar]
- 7.Paznekas W A, Boyadjiev S A, Shapiro R E, Daniels O, Wollnik B, Keegan C E, Innis J W, Dinulos M B, Christian C, Hannibal M C, Jabs E W. Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am J Hum Genet 200372408–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richardson R J, Donnai D, Meire F, Dixon M J. Expression of Gja1 correlates with the phenotype observed in oculodentodigital syndrome/type III syndactyly. J Med Genet 20044160–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kjaer K W, Hansen L, Eiberg H, Leicht P, Opitz J M, Tommerup N. Novel Connexin 43 (GJA1) mutation causes oculo‐dento‐digital dysplasia with curly hair. Am J Med Genet A 2004127152–157. [DOI] [PubMed] [Google Scholar]
- 10.Pizzuti A, Flex E, Mingarelli R, Salpietro C, Zelante L, Dallapiccola B. A homozygous GJA1 gene mutation causes a Hallermann‐Streiff/ODDD spectrum phenotype. Hum Mutat 200423286. [DOI] [PubMed] [Google Scholar]
- 11.van Steensel M A, Spruijt L, van der Burgt I, Bladergroen R S, Vermeer M, Steijlen P M, van Geel M. A 2‐bp deletion in the GJA1 gene is associated with oculo‐dento‐digital dysplasia with palmoplantar keratoderma. Am J Med Genet A 2005132171–174. [DOI] [PubMed] [Google Scholar]
- 12.Vitiello C, D'Adamo P, Gentile F, Vingolo E M, Gasparini P, Banfi S. A novel GJA1 mutation causes oculodentodigital dysplasia without syndactyly. Am J Med Genet A 200513358–60. [DOI] [PubMed] [Google Scholar]
- 13.Levin M. Isolation and community: A review of the role of gap‐junctional communication in embryonic patterning. J Membrane Biol 2001185177–192. [DOI] [PubMed] [Google Scholar]
- 14.Frischmeyer P A, Dietz H C. Nonsense‐mediated mRNA decay in health and disease. Hum Mol Genet 199981893–1900. [DOI] [PubMed] [Google Scholar]
- 15.Leube R E. The topogenic fate of the polytopic transmembrane proteins, synaptophysin and connexin, is determined by their membrane‐spanning domains. J Cell Sci 1995108883–894. [DOI] [PubMed] [Google Scholar]
- 16.Foote C I, Zhou L, Zhu X, Nicholson B J. The pattern of disulfide linkages in the extracellular loop regions of connexin 32 suggests a model for the docking interface of gap junctions. J Cell Biol 19981401187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reaume A G, de Sousa P A, Kulkarni S, Langille B L, Zhu D, Davies T C, Juneja S C, Kidder G M, Rossant J. Cardiac malformation in neonatal mice lacking connexin43. Science 19952671831–1834. [DOI] [PubMed] [Google Scholar]
- 18.Krutovskikh V, Yamasaki H. Connexin gene mutations in human genetic diseases. Mutat Res 2000462197–207. [DOI] [PubMed] [Google Scholar]
- 19.Kelsell D P, Dunlop J, Hodgins M B. Human diseases: clues to cracking the connexin code. Trends Cell Biol 2001112–6. [DOI] [PubMed] [Google Scholar]
- 20.Rabionet R, Lopez‐Bigas N, Arbones M L, Estivill X. Connexin mutations in hearing loss, dermatological and neurological disorders. Trends Mol Med 20028205–212. [DOI] [PubMed] [Google Scholar]
- 21.Roscoe W, Veitch G I L, Gong X ‐ Q, Pellegrino E, Bai D, MacLachlan E, Shao Q, Kidder G M, Laird D W. Oculodentodigital dysplasia‐causing connexin 43 mutants are non‐functional and exhibit dominant effects on wild‐type connexin 43. J Biol Chem 200528011458–11466. [DOI] [PubMed] [Google Scholar]
- 22.Flenniken A M, Osborne L R, Anderson N, Ciliberti N, Fleming C, Gittens J E, Gong X Q, Kelsey L B, Lounsbury C, Moreno L, Nieman B J, Peterson K, Qu D, Roscoe W, Shao Q, Tong D, Veitch G I, Voronina I, Vukobradovic I, Wood G A, Zhu Y, Zirngibl R A, Aubin J E, Bai D, Bruneau B G, Grynpas M, Henderson J E, Henkelman R M, McKerlie C, Sled J G, Stanford W L, Laird D W, Kidder G M, Adamson S L, Rossant J. A Gja1 missense mutation in a mouse model of oculodentodigital dysplasia. Development 20051324375–4386. [DOI] [PubMed] [Google Scholar]