Abstract
Objective
To study the SHOX gene and the PAR1 region in individuals with short stature.
Methods
The study involved 56 cases of dyschondrosteosis and 84 cases of idiopathic short stature (ISS). The study was designed to determine the following: the prevalence of SHOX anomalies in ISS; the frequency of Madelung deformity in individuals with SHOX anomalies; and the value of a family history of short stature in deciding whether to test for the SHOX gene.
Results
54 SHOX anomalies were observed, including 42 (68%) in the dyschondrosteosis group and 12 (15%) in the ISS group. The high frequency of SHOX anomalies in the ISS group can be explained by the large proportion of boys in this group, reflecting the difficulty in diagnosing dyschondrosteosis in young boys. Clinical evidence of Madelung deformity in six parents of ISS individuals emphasised the importance of family evaluation. Among the 54 SHOX anomalies, 33 PAR1 deletions were identified encompassing the SHOX gene (62%), one partial intragenic deletion (2%), nine deletions located downstream of the SHOX gene (16%), and 11 point mutations (20%).
Conclusions
These data emphasise the value of using microsatellite markers located within and downstream of the SHOX gene.
Keywords: SHOX, PAR1, dyschondrosteosis, short stature
The SHOX gene is located on the pseudoautosomal regions (PAR1) of the X and Y chromosomes. This gene has been shown to account both for some cases of idiopathic short stature (ISS) and for the short stature observed in Turner's syndrome.1,2 The prevalence of SHOX anomalies in ISS has been estimated at 2.4% in a large series of ISS individuals.3 In 1998, SHOX was also shown to account for dyschondrosteosis. Dyschondrosteosis is a mesomelic dysplasia characterised by the association of moderate short stature (below −2 SD) caused by shortening of the forelegs, and Madelung deformity of the forearm.4,5 Dyschondrosteosis has an autosomal dominant mode of inheritance and a broad phenotypic variability, with females usually being more severely affected than males. With up to 70% molecular anomalies, including large scale deletions and point mutations, SHOX has proved to be the major dyschondrosteosis gene.
In view of the remarkably variable clinical severity of SHOX mutations in dyschondrosteosis families and recent advances in the molecular analyses of the PAR1 region, we have carried out an extensive analysis of the SHOX gene region in a series of 140 individuals with short stature who were being followed by paediatric endocrinologists. Particular attention was paid to the prevalence of SHOX anomalies in ISS individuals, to the frequency of Madelung deformity in individuals with SHOX anomalies, and to the value of the family history of short stature for deciding whether the SHOX gene should be tested.
Methods
Subjects
This study was part of the GeNeSIS International Observational Study (Genetics and Neuroendocrinology of Short Stature International Study), conducted by Eli Lilly. All patients were screened through the usual short stature diagnostic work‐up in paediatric endocrinology centres, and the main causes of short stature such as growth hormone deficiency, chronic renal failure, and malabsorption were ruled out. Criteria for inclusion in the study were short stature (height between −1.8 and −3.5 SD) and a normal endocrine screen.
In all, 140 individuals were included (86 females and 54 males, ranging in age from 2 to 17 years). They were split into two groups:
Dyschondrosteosis, based on the clinical examination (n = 56 (40%); 19 males and 37 females);
Idiopathic short stature with normal proportions and no clinical symptoms of dyschondrosteosis (n = 84 (60%); 35 males and 49 females).
Molecular analyses
Genomic DNA from probands and parents was extracted from 5 ml of EDTA blood, using an extraction kit (Flexigen; Qiagen Inc, Valencia, California, USA). Molecular studies included the segregation of four highly polymorphic microsatellite markers flanking the SHOX locus on Xp22.3, namely the CASHOX and GASHOX (DXYS10092) repeats located distal to SHOX, DXYS233 located 278 kb proximal to SHOX, and the intragenic marker CTSHOX (DXYS10093) (fig 1). Polymorphic loci were polymerase chain reaction (PCR) amplified with fluorescent primers and separated by electrophoresis on an ABI 3100 DNA fragment analyser. Alleles were scored manually, using the Genscan v3.1 and Genotyper v2.0 softwares (Applied Biosystems, Foster City, California, USA).
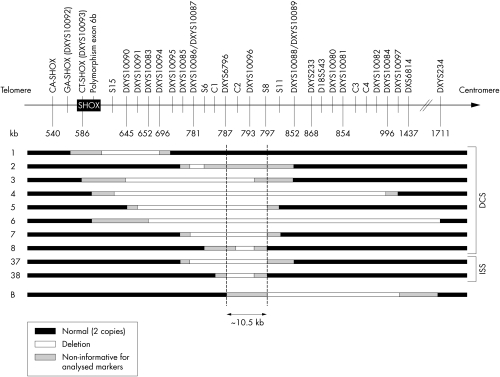
Figure 1 PAR1 deletions located downstream of the SHOX gene.
After having excluded a deletion, a direct sequencing of the SHOX gene was undertaken. Genomic DNA was PCR amplified and sequenced on an ABI 3100 automatic sequencer using the fluorescent dideoxy‐terminator method, and the six coding exons were amplified using previously described primers (exons 2–6b).6
In individuals carrying no SHOX anomaly, extensive genotyping of the PAR1 region—using 20 microsatellites and 49 SNPs, spanning 650 kb of PAR1—was carried out as previously described7 (table 1).
Table 1 Oligonucleotide sequences and polymerase chain reaction conditions of the PAR1‐analysed single nucleotide polymorphisms.
| Amplicon ID | dbSNP ID | Variation | Sense oligo 5′–3′ | Antisense oligo 5′–3′ | Annealing temp | Amplicon size | ||
|---|---|---|---|---|---|---|---|---|
| (°C) | range (pb) | |||||||
| C1 | rs5988280 | T/C | CAG CCC CAA AAT ACT TGC AAA TAC | TAC ACG ATC ACA GAA CCT AGG AAT | 57 | 721 | ||
| rs17148742 | G/A | |||||||
| rs5988281 | G/C | |||||||
| rs5946329 | T/C | |||||||
| rs7062093 | C/T | |||||||
| rs5988432 | C/T | |||||||
| rs784986 | G/A | |||||||
| rs5946331 | A/G | |||||||
| rs5946505 | C/G | |||||||
| rs5946506 | C/G | |||||||
| rs6579619 | A/G | |||||||
| C2 | rs6644384 | G/A | CAG AAG GAG GTT TAT CTC CAC CAC | GAATGCAAAATCACAGAAGTGACTG | 57 | 568 | ||
| rs6644385 | C/T | |||||||
| C3 | rs5946642 | G/A | TGA GAC GGA GTC TTG CTC TTG TC | CTGCAGAGAGCTCACGAGCACA | 57 | 453 | ||
| rs5988571 | G/A | |||||||
| rs5988328 | G/A | |||||||
| rs5946643 | A/G | |||||||
| rs5988329 | G/A | |||||||
| C4 | rs6644498 | T/C | TCC CGC ACC TGT GAT AAC TTC GGC | ACA TAC GTG CGC ATG TGT GTT TAT A | 57 | 641 | ||
| rs6579699 | T/A | |||||||
| rs6579700 | G/T | |||||||
| rs6579701 | A/G | |||||||
| rs6644499 | G/A | |||||||
| rs5946384 | C/T | |||||||
| rs5946385 | G/A |
S15, S6, S8, and S11 have been reported previously.7
SNP, single nucleotide polymorphism; temp, temperature.
Results
The first screen detected SHOX anomalies in 54% of the dyschondrosteosis group (30 of 56) and in 19% of the ISS group (16 of 84). These anomalies included deletions (35 of 46) and point mutations (11 of 46). The extent of the deletions was variable, ranging from complete SHOX gene deletions (32 of 35) to partial deletions (3 of 35, including two deletions of the extragenic marker DXYS233 and one deletion of the intragenic marker CTSHOX). The 11 point mutations were located throughout the SHOX gene, and one missense mutation located in exon 6a was found in both dyschondrosteosis and ISS individuals (I276T) (table 2).
Table 2 SHOX mutations.
| Patient | Exon | Nucleotide | Mutations |
|---|---|---|---|
| group | change | ||
| ISS | 2 | 178 A→C | T60P |
| DCS | 2 | 105 C→A | Y34X |
| DCS | 2 | 274 G→T | E92X |
| DCS | 3 | 380 Del A | E127G and frameshift causing a stop 2 codons downstream |
| DCS | 3 | 463 G→C | G155R |
| ISS | 4 | 492 G→A | W164X |
| DCS | 4 | 502 C→T | R168W |
| ISS | 6a | 645 C→T | Q215X |
| DCS | 6a | 805 Del A | S269A and frameshift causing a deletion of terminal stop |
| ISS | 6a | 827 T→C | I276T |
| DCS | 6a | 827 T→C | I276T |
The absence of SHOX anomalies in 26 of the 56 dyschondrosteosis cases and 68 of the 84 ISS individuals prompted us to analyse the PAR1 region further, using 20 microsatellites and 49 single nucleotide polymorphisms (SNPs), especially as two deletions encompassing only the extragenic marker DXYS233 were observed. Solely by extending the study in the unexplained familial dyschondrosteosis cases and in the 25 ISS individuals uninformative for DXYS233, we were able to confirm the two deletions encompassing only DXYS233 and to detect seven additional deletions located downstream to the SHOX gene (five in the dyschondrosteosis group and two in the ISS group) (fig1). Six of these deletions have been reported previously 7 (fig 1: families 2, 4–7, and 37) and four were novel deletions. In family 1, none of the intragenic markers was informative and the analysis of the PAR1 region allows us to confirm the presence of a PAR1 deletion which may encompass the SHOX gene.
The deletions were variable in size and location. Additional SNP genotyping defined a minimal common deletion of 12 kb in our series, and comparison with the previously reported series7 (fig 1: family B) allowed us to reduce the commonly deleted region to 10.5 kb outside the SHOX gene. Taken together, anomalies of the SHOX gene/region were detected in 64% of the dyschondrosteosis cases (36 of 56) and in 21% of the ISS cases (18 of 84). Our second screen allowed us to increase our detection rate of 8.3% in dyschondrosteosis.
Finally, we attempted to correlate mutant phenotypes with clinical features in our series (table 3). In the dyschondrosteosis cases carrying SHOX anomalies (36 patients), there was a large majority of females (29 of 36 (80.5%); mean age, 11 years; mean (SD) height, −2.4 (0.8) SD). Among the affected males (seven of 36 (19.4%)), mean age was 11.5 years and mean height −2 (0.3) SD. Seven cases were sporadic (five de novo deletions, two de novo mutations) and 29 were familial. The anomalies were inherited from the mother in 15 of 29 cases, and clinical features of the mother included a mean height of −2 (1.2) SD and a Madelung deformity in 14 of 15 cases. When inherited from the father (14 of 29 cases), the mean height of the father was −2 (1.1) SD and a Madelung deformity was present in nine of 14 cases.
Table 3 Patients and parents in the dyschondrosteosis and idiopathic short stature groups with SHOX anomalies.
| Case | Sex | Height | MD | Father | Mother | SHOX anomalies | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| No | (SD) | Sample | Height | MD | Sample | Height | MD | |||
| DCS group | ||||||||||
| 1 | F | −2.8 | + | + | −1.2 | − | + | −2.9 | + | Complete deletion inherited from mother |
| 2 | F | −2.8 | + | + | −0.8 | + | + | −0.3 | − | Del PAR1 inherited from father |
| 3 | F | −2.2 | + | + | −2.0 | − | + | −0.5 | + | Del PAR1 inherited from mother |
| 4 | F | −2.0 | + | − | −0.8 | − | + | −2.3 | + | Del PAR1 inherited from mother |
| 5 | F | −2.4 | + | − | −0.7 | − | + | −2.3 | + | Del PAR1 inherited from mother |
| 6 | M | −2.0 | + | − | / | − | + | −2.3 | + | Del PAR1 inherited from mother |
| 7 | F | −1.8 | + | + | / | − | + | −2.3 | + | Del PAR1 inherited from mother |
| 8 | F | −1.8 | + | + | −1.2 | + | + | +1.8 | − | Del PAR1 inherited from father |
| 9 | F | −3.5 | + | + | −1.3 | + | + | −1.3 | − | Ex6a:DelA805 codon269 inherited from father |
| 10 | F | −3.0 | + | + | −2.0 | + | + | −0.5 | − | Ex2:E92X inherited from father |
| 11 | F | −2.7 | + | + | −0.8 | − | + | −2.5 | − | Neomutation Ex3:delA380 codon127 |
| 12 | M | −1.8 | + | + | −0.5 | − | + | −2.0 | − | Ex3:G‐3.3SDSR inherited from father |
| 13 | F | −3.2 | + | + | −0.8 | − | + | −0.5 | − | Neomutation Ex4:R‐1.2SDSW |
| 14 | F | −1.8 | + | − | / | / | + | −1.8 | + | Ex2:Y34X inherited from mother |
| 15 | F | −3.5 | + | + | −1.0 | + | + | −2.9 | − | Ex6a:I276T inherited from father |
| 16 | F | −2.5 | + | + | −0.5 | + | + | −0.5 | / | Complete deletion inherited from father |
| 17 | F | −2.5 | + | + | −0.8 | − | + | +1.3 | − | Complete deletion inherited from father |
| 18 | F | −2.0 | + | + | −1.7 | − | − | −3.3 | − | Complete neodeletion |
| 19 | M | −2.0 | + | + | −0.7 | − | + | −1.3 | − | Complete neodeletion |
| 20 | F | −2.5 | + | + | / | − | + | −2.2 | − | Complete deletion inherited from mother |
| 21 | M | −2.0 | + | + | −1.5 | + | + | +0.3 | − | Complete deletion inherited from father |
| 22 | M | −1.8 | + | + | −3.3 | − | + | −1.2 | − | Complete deletion inherited from father |
| 23 | F | −1.8 | + | + | −3.5 | + | + | −1.2 | − | Complete deletion inherited from father |
| 24 | F | −2.0 | + | + | −0.8 | − | + | −2.4 | + | Complete deletion inherited from mother |
| 25 | F | −1.8 | + | + | −0.3 | − | + | −0.5 | − | Complete neodeletion |
| 26 | F | −1.8 | + | + | +1.3 | − | + | −0.2 | − | Complete neodeletion |
| 27 | F | −3.5 | + | + | +0.5 | − | + | −1.8 | + | Complete deletion inherited from mother |
| 28 | M | −2.0 | + | + | −2.0 | + | + | −2.3 | − | Complete deletion inherited from father |
| 29 | F | −2.0 | + | + | −0.8 | − | + | −2.3 | + | Complete deletion inherited from mother |
| 30 | F | −2.1 | + | − | −2.0 | − | + | −1.8 | + | Complete deletion inherited from mother |
| 31 | F | −3.2 | + | + | −0.7 | − | + | −1.8 | + | Complete deletion inherited from mother |
| 32 | F | −2.6 | + | + | −1.8 | − | + | −2.3 | − | Complete deletion inherited from father |
| 33 | F | −2.0 | + | + | −2.0 | + | + | −1.8 | − | Complete deletion inherited from father |
| 34 | F | −2.2 | + | + | / | − | + | / | − | Complete neodeletion |
| 35 | F | −1.8 | + | − | / | − | + | −1.3 | + | Complete deletion inherited from mother |
| 36 | M | −2.5 | + | − | −2.6 | − | + | −2.3 | + | Complete deletion inherited from mother |
| ISS group | ||||||||||
| 37 | F | −1.8 | − | + | +0.8 | − | + | −0.5 | + | Del PAR1 inherited from mother |
| 38 | M | −1.8 | − | − | / | − | + | −0.5 | − | Del PAR1 inherited from mother |
| 39 | F | −3.2 | − | + | −2.6 | − | + | −3.0 | − | Ex6a:I276T inherited from father |
| 40 | M | −1.8 | − | + | −1.2 | − | + | −2.0 | − | Ex6a:Q215X inherited from father |
| 41 | F | −1.8 | − | + | / | − | + | −0.2 | − | Neomutation: Ex4 W164X |
| 42 | M | −3.0 | − | + | −2.8 | − | + | −0.9 | + | Ex2:T60P inherited from mother |
| 43 | F | −3.5 | − | + | −3.2 | − | + | −2.2 | − | Complete deletion inherited from father |
| 44 | M | −1.8 | − | + | −4.2 | − | + | / | / | Complete deletion inherited from father |
| 45 | F | −1.8 | − | − | −1.2 | − | + | +0.7 | − | Complete deletion inherited from father |
| 46 | F | −2.0 | − | − | −2.3 | − | + | +0.9 | − | Complete deletion inherited from father |
| 47 | F | −3.0 | − | + | +0.3 | − | + | −2.3 | − | Complete neodeletion |
| 48 | M | −1.8 | − | + | +0.5 | − | + | 0.0 | − | Complete neodeletion |
| 49 | F | −2.0 | − | + | −2.0 | − | + | −1.8 | − | Complete deletion inherited from father |
| 50 | F | −2.5 | − | + | +0.5 | − | + | −0.5 | − | Complete neodeletion |
| 51 | M | −2.0 | − | + | −3.7 | + | + | −0.7 | − | Complete deletion inherited from father |
| 52 | M | −2.0 | − | + | −1.3 | + | + | / | − | Complete deletion inherited from father |
| 53 | M | −1.8 | − | + | +1.8 | − | + | −1.2 | + | Complete deletion inherited from mother |
| 54 | F | −1.8 | − | + | −1.7 | − | + | −4.2 | + | Complete deletion inherited from mother |
DCS, dyschondrosteosis; ISS, idiopathic short stature; MD, Madelung deformity.
Among the 18 ISS individuals with SHOX anomalies, there were 10 females (55.6%, mean age 9.5 years, mean height –2.4 (0.8) SD) and eight males (44.4%, mean age 10 years, mean height –2.0 (0.6) SD). Four cases were sporadic (three de novo deletions, one de novo mutation) and 14 were familial. The anomalies were inherited from the mother in five cases (mean height –1.8 (1.8) SD) and a Madelung deformity was present in four. When inherited from the father (nine of 14, mean height –2.4 (1.6) SD), a Madelung deformity was present in two. Based on the family evaluation, at least six individuals considered to have ISS belonged to a dyschondrosteosis family. When we included the ISS individuals with at least one parent with Madelung deformity in the dyschondrosteosis group, the frequency of SHOX anomalies in the that group became 68% (36+6/56+6), and in the final analysis there were 42 families or patients with Madelung deformity among the 54 families or patients with SHOX anomalies (77.8%).
We identified 43 familial cases and among these, 30 parents presented with a short stature (69.8%) and 26 parents with a Madelung deformity (60.5%). It is important to emphasise that four fathers and one mother had neither short stature nor Madelung deformity.
With respect to the 11 sporadic cases, we identified eight de novo deletions which all occurred on the paternal allele (data not shown). Finally, the last group, composed of 20 individuals with Madelung deformity but no SHOX anomalies, included eight females (mean age 9 years, mean height –1.9 (0.2) SD) and 12 males (mean age 11 years, mean height –2.6 (0.8) SD). Fifteen cases were familial with a family history of isolated short stature in eight and short stature plus Madelung deformity in seven. In this group, linkage analysis clearly excluded PAR1 in one large family (data not shown).
Discussion
Studying a large series of 140 individuals with short stature, we found a high incidence of SHOX anomalies (38.6%). Anomalies of the SHOX gene/region were present in 68% of dyschondrosteosis cases, which is similar to previous studies8,9,10,11 and in 15% of ISS individuals. The high frequency of SHOX anomalies in the ISS group is probably the result of a bias. Indeed, the larger proportion of males in the ISS group (44.4%) compared with the dyschondrosteosis group (19.4%) illustrates the frequent absence of clinical manifestations in dyschondrosteosis males, explaining why young boys with SHOX anomalies could have been regarded as ISS. The high frequency of SHOX anomalies observed in the ISS group reflects the difficulty in diagnosing dyschondrosteosis in young individuals at prepubertal stage and the possible absence of any clinically detectable Madelung deformity in males.
These findings show the importance of the parental examination and the value of a family history of short stature when deciding SHOX investigation. The observation of four fathers and one mother with no clinical evidence of dyschondrosteosis suggest also that clinical examination on its own is not sufficient to diagnose a dyschondrosteosis and emphasises the importance of including a forearm x ray in the family evaluation.
Finally, the high rate of SHOX anomalies in the ISS group may also partly reflect our extensive molecular screening. Indeed in previous studies, screening was carried out by single strand conformation polymorphism, fluorescent in situ hybridisation,3 or by the analysis of two microsatellite markers (CA‐SHOX repeat and DXYS233).12 In our study we included a complete microsatellite analysis of the PAR1 region as well as the direct sequencing of the SHOX gene. From a genetics point of view, the occurrence of de novo deletions on the paternal allele may be explained by the high rate of recombination events between sex chromosomes during male meiosis, which is restricted to PAR regions.13
Among a total of 54 SHOX gene/region anomalies, we observed a variety of distinct anomalies including a significant percentage of deletions located downstream the SHOX gene. A comparison of these deletions shows a common deletion of 10.5 kb encompassing the DXYS10096. We propose, therefore, routine molecular screening using six microsatellites: CASHOX, GASHOX (DXYS10092), CTSHOX (DXYS10093), DXS6796, DXYS10096, and DXYS233. In the cases where DXS6796 and DXYS10096 are uninformative, the use of additional SNPs (S6, C1, C2, S8, S2, S3, S4) and the direct sequencing of the SHOX gene should be of particular value.
Conclusions
SHOX anomalies and PAR1 deletions account for at least 68% of cases of classical dyschondrosteosis and for a significant fraction of ISS in young individuals. The identification of PAR1 deletions located downstream of the SHOX gene prompts to include additional microsatellite marker analysis in the routine molecular analysis of the SHOX/PAR1 region.
Electronic database information
UCSC Genome Bioinformatics, http://genome.ucsc.edu/
National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/
Online Mendelian Inheritance in Man (MIM), http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db = OMIM
The GDB Human Genome Database, http://www.gdb.org
Acknowledgements
This work was supported by a research grant from Lilly France. All the French patients were included in the SHOX module of the GeNeSIS study.
Abbreviations
GeNeSIS - Genetics and Neuroendocrinology of Short Stature International Study
ISS - idiopathic short stature
SNP - single nucleotide polymorphism
Appendix
MEMBERS OF THE FRENCH SHOX GeNeSIS MODULE
P Barat, H Bellon, P Berlier, A M Bertrand, C Blond‐Metz, H Bony‐Trifunovic, M Bost, R Brauner, S Cabrol, J C Carel, J B Cotton, M David, F Despert, T Edouard, L Faivre, P Goumy, P Jeannoël, M Jesuran‐Perelroizen, B Lebon‐Labich, C Lecointre, J Leger, A Lienhart, J M Limal, L Meyer, C Naud‐Saudreau, E Pichot, C Raynaud‐Ravni, S Soskin, M Tauber, C Thalassinos, J Weill, C Wright.
References
- 1.Rao E, Weiss B, Fukami M, Rump A, Niesler B, Mertz A, Muroya K, Binder G, Kirsch S, Winkelmann M, Nordsiek G, Heinrich U, Breuning M H, Ranke M B, Rosenthal A, Ogata T, Rappold G A. Pseudoautosomal deletions encompassing a novel homeobox gene cause growth failure in idiopathic short stature and Turner syndrome. Nat Genet 19971654–63. [DOI] [PubMed] [Google Scholar]
- 2.Ellison J W, Wardak Z, Young M F, Gehron Robey P, Laig‐Webster M, Chiong W. PHOG, a candidate gene for involvement in the short stature of Turner syndrome. Hum Mol Genet 199761341–1347. [DOI] [PubMed] [Google Scholar]
- 3.Rappold G A, Fukami M, Niesler B, Schiller S, Zumkeller W, Bettendorf M, Heinrich U, Vlachopapadoupoulou E, Reinehr T, Onigata K, Ogata T. Deletions of the homeobox gene SHOX (short stature homeobox) are an important cause of growth failure in individuals with short stature. J Clin Endocrinol Metab 2002871402–1406. [DOI] [PubMed] [Google Scholar]
- 4.Belin V, Cusin V, Viot G, Girlich D, Toutain A, Moncla A, Vekemans M, Le Merrer M, Munnich A, Cormier‐Daire V. SHOX mutations in dyschondrosteosis (Leri‐Weill syndrome). Nat Genet 19981967–69. [DOI] [PubMed] [Google Scholar]
- 5.Shears D J, Vassal H J, Goodman F R, Palmer R W, Reardon W, Superti‐Furga A, Scambler P J, Winter R M. Mutation and deletion of the pseudoautosomal gene SHOX cause Leri‐Weill dyschondrosteosis. Nat Genet 19981970–73. [DOI] [PubMed] [Google Scholar]
- 6.Huber C, Cusin V, Le Merrer M, Mathieu M, Sulmont V, Dagoneau N, Munnich A, Cormier‐Daire V. SHOX point mutations in dyschondrosteosis. J Med Genet 200138323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benito‐Sanz S, Thomas N S, Huber C, Del Blanco D G, Aza‐Carmona M, Crolla J A, Maloney V, Argente J, Campos‐Barros A, Cormier‐Daire V, Heath K E. A Novel Class of Pseudoautosomal Region 1 Deletions Downstream of SHOX Is Associated with Leri‐Weill Dyschondrosteosis. Am J Hum Genet 20051577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schiller S, Spranger S, Schechinger B, Fukami M, Merker S, Drop S L, Troger J, Knoblauch H, Kunze J, Seidel J, Rappold G A. Phenotypic variation and genetic heterogeneity in Leri‐Weill syndrome. Eur J Hum Genet 2000854–62. [DOI] [PubMed] [Google Scholar]
- 9.Falcinelli C, Iughetti L, Percesepe A, Calabrese G, Chiarelli F, Cisternino M, De Sanctis L, Pucarelli I, Radetti G, Wasniewska M, Weber G, Stuppia L, Bernasconi S, Forabosco A SHOX point mutations and deletions in Leri‐Weill dyschondrosteosis J Med Genet. 2002;39:E33. doi: 10.1136/jmg.39.6.e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flanagan S F, Munns C F, Hayes M, Williams B, Berry M, Vickers D, Rao E, Rappold G A, Batch J A, Hyland V J, Glass I A. Prevalence of mutations in the short stature homeobox containing gene (SHOX) in Madelung deformity of childhood. J Med Genet 200239758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Binder G, Renz A, Martinez A, Keselman A, Hesse V, Riedl S W, Hausler G, Fricke‐Otto S, Frisch H, Heinrich J J, Ranke M B. SHOX haploinsufficiency and Leri‐Weill dyschondrosteosis: prevalence and growth failure in relation to mutation, sex, and degree of wrist deformity. J Clin Endocrinol Metab 2004894403–4408. [DOI] [PubMed] [Google Scholar]
- 12.Binder G, Ranke M B, Martin D D. Auxology is a valuable instrument for the clinical diagnosis of SHOX haploinsufficiency in school‐age individuals with unexplained short stature. J Clin Endocrinol Metab 2003884891–4896. [DOI] [PubMed] [Google Scholar]
- 13.Lien S, Szyda J, Schechinger B, Rappold G, Arnheim N. Evidence for heterogeneity in recombination in the human pseudoautosomal region: high resolution analysis by sperm typing and radiation‐hybrid mapping. Am J Hum Genet 200066557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]