Abstract
Background
Lafora's progressive myoclonic epilepsy (Lafora's disease) is an autosomal recessive neurodegenerative disorder characterised by the presence of polyglucosan intracellular inclusions called Lafora bodies. Mutations in two genes, EPM2A and NHLRC1, have been shown to cause the disease. A previous study showed mutations in the EPM2A gene in 14 Lafora's disease families and excluded the involvement of this gene in five other families who were biopsy proven to have the disease.
Objective
To relate the genetic findings to the clinical course of the disease.
Methods
As part of an ongoing mutational study of the Lafora's disease genes, five new families with the disease were recruited and the genetic analysis was extended to screen the entire coding region of the NHLRC1 gene. Genotype–phenotype correlations were carried out.
Results
Seven NHLRC1 mutations were identified, including five novel mutations (E91K, D195N, P218S, F216_D233del, and V359fs32), in eight families with Lafora's disease. On relating the genetic findings to the clinical course of the disease it was shown that patients with NHLRC1 mutations had a slower rate of disease progression (p<0.0001) and thus appeared to live longer than those with EPM2A mutations. A simple DNA based test is described to detect the missense mutation C26S (c.76T→A) in the NHLRC1 gene, which is prevalent among French Canadians.
Conclusions
Patients with NHLRC1 mutations have a slower rate of disease progression than those with EPM2A mutations.
Keywords: Lafora's disease, protein‐ubiquitin ligase, polyglucosan bodies, neurodegeneration
Lafora's progressive myoclonus epilepsy, or Lafora's disease (OMIM 254780), is a fatal autosomal recessive disorder with pathognomonic periodic acid Schiff positive (PAS+) staining intracellular inclusion bodies.1,2,3 Symptoms for Lafora's disease usually start in the teenage years in the form of grand mal seizures or myoclonus, followed by rapid and severe mental deterioration, often with psychotic features.1,2,3 Survival is short, often less than 10 years after the onset. Lafora's disease is caused by mutations in the EPM2A4,5,6,7 or the NHLRC1 genes,8 encoding the laforin dual specificity protein phosphatase6 and the malin ubiquitin E3 ligase,9,10 respectively. Malin has been shown to ubiquitinate and promote the degradation of laforin.9 There is also evidence for a third as yet unidentified locus for Lafora's disease.11
In an earlier 2002 study,7 we related mutations in EPM2A with the phenotypes of 22 patients (14 families) and identified two subsyndromes. The first subsyndrome is classic Lafora's disease with adolescent onset, stimulus sensitive grand mal, absences, and myoclonic seizures, followed by dementia and neurological deterioration. Adolescent onset classic Lafora's disease is associated mainly with mutations in exon 4 of EPM2A gene. The second subsyndrome is childhood onset Lafora's disease, with dyslexia and learning disorder followed by epilepsy and neurological deterioration. Childhood Lafora's disease is associated mainly with mutations in exon 1 of the EPM2A gene. Childhood onset Lafora's disease was recently confirmed by a separate group of investigators from Italy.12 In the same earlier study, we were unable to detect mutations in the entire coding region and the relevant intron boundaries of EPM2A in five other Lafora's disease families,7 suggesting a second gene for the disease. As part of our ongoing mutational studies of the Lafora's disease genes, we recruited five new families and extended our analysis to screen the entire coding regions of the NHLRC1 and EPM2A genes. In this paper, we relate our genetic findings to the clinical course of the disease and show that patients with NHLRC1 mutations are less severely affected and appear to live longer than those with EPM2A mutations.
Methods
We studied patients whose clinical, electroencephalographic, and biopsy data proved the diagnosis of Lafora's disease. Each participating subject, or in the case of minors the responsible adult, signed an informed consent form as approved by the human subjects protection committee at the David Geffen School of Medicine at UCLA, the institute ethics committee for human genetics research at IIT Kanpur, and the review committees of the participating centres. Genomic DNA was extracted from blood samples using a QIAamp blood DNA purification kit (Qiagen Inc, Valencia, California, USA). The coding regions of the EPM2A and NHLRC1 genes were polymerase chain reaction (PCR) amplified using established primers,7,13 directly sequenced using the DTCS QuickStart sequencing kit (Beckman Coulter, Fullerton, California, USA) on a CEQ800 automated DNA sequencer (Beckman Coulter), and analysed using CEQuence investigator module (Beckman Coulter).13
A novel PCR based assay was developed to survey for the 76T→A mutation in genomic DNA. The 5′ segment of the NHLRC1 coding region was amplified using a forward (5′‐TGA CCA TGA CTG TGA CCG TGA‐3′) and a reverse mismatch primer (5′‐CAA ACT TCT CAA AGC ACA CCT AGC‐3′) (the mismatch was at the base underlined; see fig 3). A 236 base pair (bp) product was PCR amplified from ∼100 ng of genomic DNA using the following amplification conditions: 94°C for five minutes, followed by 35 cycles of 94°C for 45 seconds, 58°C for 45 seconds, and 72°C for 30 seconds, followed by a final 10 minutes' extension at 72°C. Aliquots were digested with AluI restriction enzyme (Fermentas, Burlington, Ontario, Canada), separated on 15% polyacrylamide gels, and stained with ethidium bromide.
Results
Mutational spectrum in Lafora's disease
Our analysis identified five novel mutations and two recurrent mutations in the NHLRC1 gene of probands belonging to eight Lafora's disease families (table 1). None of these individuals harboured mutations in the coding region of the EPM2A gene. Adding these data to those reported previously,8,13,14,15 there are 39 different mutations in the NHLRC1 gene known so far (fig 1A). Among a total of 26 Lafora's disease families in our present cohort, 23 families (84%) showed mutations in either the EPM2A or the NHLRC1 gene. Of these 26 families, 14 (54%) had mutations in the EPM2A gene and nine (34%) in the NHLRC1 gene. It is possible that the three families which did not reveal any mutation in the EPM2A or NHLRC1 gene harbour mutations in regulatory regions. Alternatively, they may harbour mutations in a third Lafora's disease gene.11 Finding an NHLRC1 mutation (D195N) in only one chromosome, such as in LDCH2, suggests that there are indeed unidentified mutations lying in non‐coding or regulatory regions of the NHLRC1 gene on another chromosome. Such heterozygous mutations in Lafora's disease have been reported previously.7,13,16 Mendelian inheritance of mutations was confirmed in families when the DNA samples of parents were available.
Table 1 Genotype–phenotype correlation: NHLRC1 mutations, clinical manifestations of Lafora disease, and age during appearance of signs and symptoms.
| Mutation* (nucleotide change followed by its predicted effect on protein) | Age (y) at seizure onset | Age (y) at onset of mild cognitive decline | Age (y) at onset of neurological deterioration | Reference | |||
|---|---|---|---|---|---|---|---|
| Ataxia±spasticity at latest evaluation† | Dementia, mutism, | Respiratory assistance and gastrostomy | |||||
| Family/individual | loss of independent | ||||||
| (ethnicity) | living activities | ||||||
| LDCH2‐1 (Chinese) | c.583G→A‡ | NI | NI | NI | NI | NI | Present study |
| D195N | |||||||
| LD28‐1 (French‐Canadian) | c.76T→A | 12 | 14 | 16 | 14/26 | 26 | Present study |
| C26S | |||||||
| LD28‐2 (French‐Canadian) | c.76T→A | 12 | 16 | 16 | 20/31 | 31 | Present study |
| C26S | |||||||
| LD28‐3 (French‐Canadian) | c.76T→A | 12 | 16 | 16 | 20/32 | 32 | Present study |
| C26S | |||||||
| LD6‐1 (French Canadian) | c.76T→A | Visual hallucination; | 16–20 | 16/20 | 28 | 30 | Chan et al8 |
| C26S | 11.5–12 | ||||||
| LD6‐2 (French Canadian) | c.76T→A | 12 | 16–20 | 16 | 28 | 30 | Chan et al8 |
| C26S | |||||||
| LD27‐1 (French Canadian) | c.76T→A | 13 | 14 | 14/20 | 19 | 29 | Chan et al8 |
| C26S | |||||||
| LD27‐2 (French Canadian) | c.76T→A | 15 | 15 | 15 | NI | NI | Chan et al8 |
| C26S | |||||||
| LD39‐1 (Italian) | c.260T→C | 13 | 14 | 27 | 30 | 36 | Present study |
| L87P | |||||||
| LD41‐1 (Italian) | c.271G→A | Febrile seizures at 4 y; | 27 | 27 | 30 | 37 | Present study |
| E91K | 15 (no visual seizures) | ||||||
| LD10‐1 (American) | c.271G→A | 9 (male); hyperactive, impulsive, hyperkinetic | 9 | 10.5 | NI | NI | Present study |
| E91K | |||||||
| LD36‐1 (Italian) | c.652C→T | 12 (no visual seizures) | 16 | 16–18 | 28 | 32 | Present study |
| P218S | |||||||
| LD38‐1 (Italian) | c.645_699del54 | 14 | – | 21 | NI | 33 | Present study |
| F216_D233del | |||||||
| LD11‐1 (Spanish) | c.1076insT | 7 (male), hyperactive, hyperkinetic, hypotonic | 14 | 4, 20 | 28 | 35 | Present study |
| V359fs32 | |||||||
*The nucleotide and amino acid positions were assigned based on the GenBank reference sequence for the NHLRC1 gene, NM_198586 (RING, RING finger domain; NHL, NHL repeats).
†During period of preserved daily living activities.
‡The mutant allele was identified in heterozygous condition.
NI, no information available; y, years.
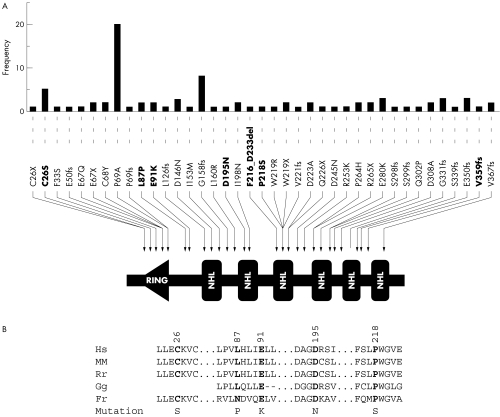
Figure 1 (A) Representation of 39 known mutations in the NHLRC1 gene. This schematic diagram shows the domain organisation of the malin protein, the positions of various mutations found in Lafora's disease families, and their frequency (the number of independent families with a given mutation). Mutations were tabulated from PubMed indexed papers in English reporting the mutations.8,13,14,15 Mutations identified in the present study are shown in bold font. The amino acid positions were assigned based on the GenBank reference sequence for the NHLRC1 gene, NM_198586 (RING, RING finger domain; NHL, NHL repeats). (B) Evolutionary conservation of amino acid residues altered by the five missense mutations identified in the present study. A comparison of amino acids and the flanking sequence of malin orthologues from human (Hs), mouse (Mm), rat (Rn), chicken (Gg), and puffer fish (Fr) is depicted. Positions of the altered residue and the amino acid change are also shown. Amino acid sequence were derived from GenBank deposits NP_940988 (Hs), NP_780549 (Mm), NP_954706 (Rn), and XP_426034 (Gg).
Three of the five novel mutations reported here are missense mutations, resulting in the following amino acid substitutions: E91K, D195N, and P218S. To test whether these mutations might be normal polymorphism, we sequenced the coding region NHLRC1 in 100 control individuals and did not detect any of these variants. Furthermore, these variations were not present in any expressed sequence tag (EST) sequences representing the NHLRC1 gene. Missense mutation E91K was identified in two independent and ethnically different families, and thus appears to be a recurrent mutational event (table 1). Likewise, mutations C26S and L87P were recurring mutations reported previously by Chan and colleagues.8 The residues affected by the five distinct missense mutations identified in the present study are highly conserved across orthologues, reflecting the evolutionary constraints placed on these residues (fig 1B). We therefore consider them as deleterious mutations.
Besides these missense mutations, we identified two other forms of mutation: a 54 bp microdeletion resulting in an 18 amino acid deletion (F216_D233del), and a single base pair insertion resulting in a frame shift mutation (V359fs32). The deletion mutation affects the third NHL repeat whereas the frameshift mutation removes the carboxyl terminal segment of malin (fig 1A).
Our studies presented here and elsewhere7 reveal an interesting distribution of Lafora's disease mutations in different geographical populations. Four of five Italian families of our cohort had mutations in the NHLRC1, whereas six of eight Spanish families had mutations in the EPM2A gene.
Genotype–phenotype correlations
We reviewed in detail the clinical manifestations of 13 Lafora's disease patients belonging to nine families, and related their disease phenotypes to NHLRC1 mutations. For this genotype–phenotype correlation, we added four Lafora's disease patients belonging to two families (LD6 and LD27), whose mutations had been published previously8 (see table 1) and which we reconfirmed for the present study. These four patients from two separate Lafora's disease families were added to the 10 patients belonging to nine families listed in table 1. Clinical details were not available for patient LDCH2; hence, we excluded this patient for genotype–phenotype correlations. This left us with 13 patients belonging to nine families with NHLRC1 mutations, which we summarised in table 1. We compared the clinical course of these 13 patients with NHLRC1 mutations with the clinical course of 22 patients with EMP2A mutations. We did not find any significant difference in age at onset between the two patient groups (12 years for NHLRC1 (n = 13) and 11.5 years for EPM2A (n = 22)). However, patients bearing the NHLRC1 gene defects had a slower rate of disease progression and thus appeared to live longer. For example, patients with NHLRC1 mutations were provided with respiratory assistance at a mean of around 20 years after disease onset (n = 11; table 1) compared with 6.5 years for the EPM2A patients (n = 12) (p<0.0001; Student's t test).7 Cognitive decline, ataxia, and spasticity appeared two to four years after epilepsy onset in both EPM2A and NHLRC1 mutations. However, independent living activities were lost later in NHLRC1 mutations than in EPM2A mutations (about 26 to 32 years in NHLRC1 mutations, at a time when patients with the EMP2A mutations had usually already died).
The genotype and phenotype details discussed in the present study have been submitted to the Lafora progressive myoclonus epilepsy mutation and polymorphism database17 (http://projects.tcag.ca/lafora/).
A simple diagnostic test to detect the recurrent missense mutation, C26S
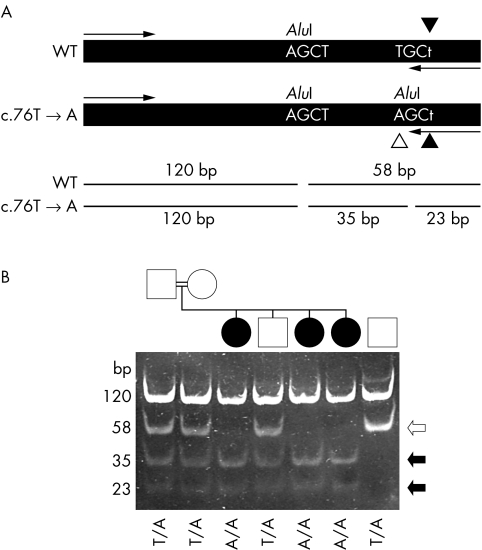
The missense mutation C26S (76T→A)—identified in the French Canadian families LD6, LD17, and 28—was previously shown to be prevalent in the French Canadian isolate, perhaps because of a founder effect.7,16 We therefore devised a novel DNA based diagnostic test for prenatal and carrier screening of this mutation for use in the French Canadian population (fig 2). A 236 bp long PCR fragment of the NHLRC1 gene coding sequence was amplified by a forward and reverse mismatch primer (fig 2A). PCR fragments amplified from the 76T→A mutant allele created an additional site for the restriction enzyme AluI, thus providing a rapid method to screen for this mutation. Restriction digests of control DNA yielded two fragments of 120 and 58 bp in molecular size. In samples containing the 76T→A mutation, the 58 bp product was cleaved in two, producing 35 and 23 bp fragments. In fig 2B, lanes 3, 5, and 6 show that the three affected individuals were homozygous for the 76T→A mutation. The parents (lanes 1 and 2) and an affected brother (lane 4) were heterozygous. These results are consistent with an autosomal recessive mode of inheritance for this mutation, and were independently confirmed by direct sequencing of the PCR product.
Figure 2 Analysis of the c.76T→A mutation. (A) Schematic diagram illustrating the approach taken to detect the mutation. A 236 bp fragment was amplified from genomic DNA is shown along with the primer annealing sites (arrows). The PCR using a mismatched reverse primer (c.79A→T change, indicated by filled arrowheads) was designed so that it introduced an AluI restriction enzyme digestion site into the PCR product only if the mutant (c.76T→A) gene sequence was present (identified by a white arrowhead). The restriction site was not introduced if the wild type sequence was present. An additional AluI site present in the amplified section from both normal and mutant alleles acted as a positive control for restriction enzyme digestion. Digestion of the amplified fragment with AluI at position 120 would produce 120 and 58 bp fragments. In the presence of the c.76T→A mutation, the 58 bp fragment would be cleaved into 35 and 23 bp fragments. (B) Analysis of samples from the French Canadian family LD28 (lanes 1 to 6) and a wild type control subject (lane 7). Samples in all lanes were digested with AluI. The genotype (c.76T→A mutation) of each individual is identified at the bottom of the gel picture. The sizes of each fragment are identified on the right side. Black and white arrows indicate fragments generated from the mutant and wild type alleles, respectively.
Discussion
We describe here mutations in the NHLRC1 gene in Lafora's disease families primarily from European populations. Given that only three mutations detected in the present study have been reported previously, it is likely that the majority of the Lafora's disease mutations arise as a single event and that only a very small proportion of mutant alleles can be predicted in certain populations. The C26S missense mutation, which is yet to be detected in other populations, was previously identified in four independent families which were exclusively of French Canadian descent, and which originated from the same region of east Quebec.8,18 The affected individuals of the four families reported by Chan and colleagues8,18 share the same haplotype around the NHLRC1 locus, suggesting a founder effect for the mutation.3 Our identification of a yet another French Canadian Lafora's disease family with the same mutation suggests that this mutation could be the cause for the prevalence of Lafora's disease in this ethnic community. Thus there is a strong rationale to screen for this mutation in any patient, from the French Canadian isolate, who is suspected of having Lafora's disease. The diagnostic test described in our report using genomic DNA offers a rapid and accurate method.
The missense mutation C26S is the third most frequent mutation observed for NHLRC1 and targets the RING domain (fig 1A). Biochemical studies have shown that mutation C26S indeed affects the ubiquitin ligase activity of malin.9 The functional implications of the four other missense mutations identified in the present study are unknown. It is of interest to note that missense mutations D195N and P218S target the second and third NHL domains of malin, respectively (fig 1A), and mutations affecting the NHL domain are proven to affect the ubiquitin ligase activity of malin.9 As the assay systems for ubiquitin ligase activities have already been established, it would be of interest now to check the effects of these mutations on malin's ubiquitin ligase activity.
The identification of disease causing mutations in a large panel of Lafora's disease families provided us with a good basis for studying phenotype–genotype relations within and between families for both EPM2A and NHLRC1 mutations. As of now, we are unable to assign any specific phenotypic variation that relates to specific mutations in the NHLRC1 gene. However, the present study clearly indicates that disease progression is longer in patients with NHLRC1 mutations as against those with EPM2A mutations. While our studies were being prepared for publication, similar observations were reported by Gomez‐Abad and colleagues,14 who noted that patients with NHLRC1 mutations had a slightly milder clinical course. The recent demonstration that laforin is a substrate for malin E3 ubiquitin ligase implies that the malin could probably act upstream of laforin in the cellular cascade.9,10 Considering the difference in the rate of disease progression in patients with NHLRC1 gene defects (in the present study and that of Gomez‐Abad et al14), it is tempting to speculate that at least some functions of laforin are regulated by multiple factors and that malin could be only one of them. Thus, while loss of laforin may result in rapid progression of the disease, the effect of malin defects would be restricted to a few and not all functions of laforin, leading to a slower clinical course. This suggestion also supports a role for the protein product of a third Lafora's disease gene, as well as other factors that could regulate the function of laforin.3 Further studies exploring the biochemical pathways that link the regulators of laforin to glycogen metabolism might therefore unravel the molecular mechanisms that lead to polyglucosan accumulation and cell death in Lafora's disease.
Acknowledgements
We gratefully acknowledge our Lafora's disease patients and their parents, families, and collaborating physicians for their participation in this study. This study was supported in part by research grants from the Department of Science and Technology, Government of India (SG), RIKEN Brain Science Institute (KY), NIH Grant 5P01‐NS21908 (AVDE), and by contributions from the Quebec (Mrs Odette Malenfant) and Sweden (Mrs Vera Faludi) Lafora Disease Associations. SS was supported by a research fellowship from the Council of Scientific and Industrial Research, Government of India.
Footnotes
Conflicts of interest: none declared
References
- 1.Van Heycop Ten Ham M W. Lafora disease, a form of progressive myoclonus epilepsy. In: Vinken PJ, Bruyn GW, editors. The epilepsies. Handbook of clinical neurology , vol 15. Amsterdam: North‐Holland, 1975382–422.
- 2.Delgado‐Escueta A V, Ganesh S, Yamakawa K. Advances in the genetics of progressive myoclonus epilepsy. Am J Med Genet 2001106129–138. [DOI] [PubMed] [Google Scholar]
- 3.Ganesh S, Puri R, Singh S, Mittal S, Dubey D. Recent advances in the molecular basis of Lafora's progressive myoclonus epilepsy. J Hum Genet 2006511–8. [DOI] [PubMed] [Google Scholar]
- 4.Minassian B A, Lee J R, Herbrick J A, Huizenga J, Soder S, Mungall A J, Dunham I, Gardner R, Fong C Y, Carpenter S, Jardim L, Satishchandra P, Andermann E, Snead O C, Lopes‐Cendes I, Tsui L C, Delgado‐Escueta A V, Rouleau G A, Scherer S W. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat Genet 199820171–174. [DOI] [PubMed] [Google Scholar]
- 5.Serratosa J M, Gomez‐Garre P, Gallardo M E, Anta B, de Bernabe D B, Lindhout D, Augustijn P B, Tassinari C A, Malafosse R M, Topcu M, Grid D, Dravet C, Berkovic S F, de Cordoba S R. A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2). Hum Mol Genet 19998345–352. [DOI] [PubMed] [Google Scholar]
- 6.Ganesh S, Agarwala K L, Ueda K, Akagi T, Shoda K, Usui T, Hashikawa T, Osada H, Delgado‐Escueta A V, Yamakawa K. Laforin, defective in the progressive myoclonus epilepsy of lafora type, is a dual‐specificity phosphatase associated with polyribosomes. Hum Mol Genet 200092251–2261. [DOI] [PubMed] [Google Scholar]
- 7.Ganesh S, Delgado‐Escueta A V, Suzuki T, Francheschetti S, Riggio C, Avanzini G, Rabinowicz A, Bohlega S, Bailey J, Alonso M E, Rasmussen A, Thomson A E, Ochoa A, Prado A J, Medina M T, Yamakawa K. Genotype‐phenotype correlations for EPM2A mutations in Lafora's progressive myoclonus epilepsy: exon 1 mutations associate with an early‐onset cognitive deficit subphenotype. Hum Mol Genet 2002111263–1271. [DOI] [PubMed] [Google Scholar]
- 8.Chan E M, Young E J, Ianzano L, Munteanu I, Zhao X, Christopoulos C C, Avanzini G, Elia M, Ackerley C A, Jovic N J, Bohlega S, Andermann E, Rouleau G A, Delgado‐Escueta A V, Minassian B A, Scherer S W. Mutations in NHLRC1 cause progressive myoclonus epilepsy. Nat Genet 200335125–127. [DOI] [PubMed] [Google Scholar]
- 9.Gentry M S, Worby C A, Dixon J E. Insights into Lafora disease: malin is an E3 ubiquitin ligase that ubiquitinates and promotes the degradation of laforin. Proc Natl Acad Sci USA 20051028501–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lohi H, Ianzano L, Zhao X C, Chan E M, Turnbull J, Scherer S W, Ackerley C A, Minassian B A. Novel glycogen synthase kinase 3 and ubiquitination pathways in progressive myoclonus epilepsy. Hum Mol Genet 2005142727–2736. [DOI] [PubMed] [Google Scholar]
- 11.Chan E M, Omer S, Ahmed M, Bridges L R, Bennett C, Scherer S W, Minassian B A. Progressive myoclonus epilepsy with polyglucosans (Lafora disease): evidence for a third locus. Neurology 200463565–567. [DOI] [PubMed] [Google Scholar]
- 12.Annesi G, Sofia V, Gambardella A, Candiano I C, Spadafora P, Annesi F, Cutuli N, De Marco E V, Civitelli D, Carrideo S, Tarantino P, Barone R, Zappia M, Quattrone A. A novel exon 1 mutation in a patient with atypical lafora progressive myoclonus epilepsy seen as childhood‐onset cognitive deficit. Epilepsia 200445294–295. [DOI] [PubMed] [Google Scholar]
- 13.Singh S, Suzuki T, Uchiyama A, Kumada S, Moriyama N, Hirose S, Takahashi Y, Inoue Y, Kimura K, Sawaishi S, Yamakawa K, Ganesh S. Mutations in the NHLRC1 gene are the common cause for Lafora disease in the Japanese population. J Hum Genet 200550347–352. [DOI] [PubMed] [Google Scholar]
- 14.Gomez‐Abad C, Gomez‐Garre P, Gutierrez‐Delicado E, Saygi S, Michelucci R, Tassinari C A, Rodriguez de Cordoba S, Serratosa J M. Lafora disease due to EPM2B mutations: a clinical and genetic study. Neurology 200564982–986. [DOI] [PubMed] [Google Scholar]
- 15.Baykan B, Striano P, Gianotti S, Bebek N, Gennaro E, Gurses C, Zara F. Late‐onset and Slow‐progressing Lafora Disease in Four Siblings with EPM2B Mutation. Epilepsia 2005461695–1697. [DOI] [PubMed] [Google Scholar]
- 16.Gomez‐Garre P, Sanz Y, Rodriguez De Cordoba S R, Serratosa J M. Mutational spectrum of the EPM2A gene in progressive myoclonus epilepsy of Lafora: high degree of allelic heterogeneity and prevalence of deletions. Eur J Hum Genet 20008946–954. [DOI] [PubMed] [Google Scholar]
- 17.Ianzano L, Zhang J, Chan E M, Zhao X C, Lohi H, Scherer S W, Minassian B A. Lafora progressive Myoclonus Epilepsy mutation database‐EPM2A and NHLRC1 (EMP2B) genes. Hum Mutat 200526397. [DOI] [PubMed] [Google Scholar]
- 18.Chan E M, Bulman D E, Paterson A D, Turnbull J, Andermann E, Andermann F, Rouleau G A, Delgado‐Escueta A V, Scherer S W, Minassian B A. Genetic mapping of a new Lafora progressive myoclonus epilepsy locus (EPM2B) on 6p22. J Med Genet 200340671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]