Abstract
Background
Approximately half the cases of prelingual hearing loss are caused by genetic factors. Identification of genes causing deafness is a crucial first step in understanding the normal function of these genes in the auditory system. Recently, a mutant allele of Tmhs was reported to be associated with deafness and circling behaviour in the hurry‐scurry mouse. Tmhs encodes a predicted tetraspan protein of unknown function, which is expressed in inner ear hair cells. The human homologue of Tmhs is located on chromosome 6p.
Objective
To determine the cause of deafness in four consanguineous families segregating recessive deafness linked to markers on chromosome 6p21.1‐p22.3 defining a novel DFNB locus.
Results
A novel locus for non‐syndromic deafness DFNB67 was mapped in an interval of approximately 28.51 cM on human chromosome 6p21.1‐p22.3. DNA sequence analysis of TMHS revealed a homozygous frameshift mutation (246delC) and a missense mutation (Y127C) in affected individuals of two families segregating non‐syndromic deafness, one of which showed significant evidence of linkage to markers in the DFNB67 interval. The localisation of mTMHS in developing mouse inner ear hair cells was refined and found to be expressed briefly from E16.5 to P3.
Conclusions
These findings establish the importance of TMHS for normal sound transduction in humans.
There are approximately 100 genes that are associated with hearing loss in the mouse.1 In humans, more than 47 deafness loci have been mapped and 21 of the corresponding genes have been identified.2,3 Because of the similarities in the morphology of their auditory systems, deaf mice have provided a valuable resource for understanding the pathophysiology of human hereditary hearing disorders and the normal functions of these genes. Molecular genetic studies of deaf mice have been instrumental in identifying six orthologous deafness genes in humans, including MYO7A (USH1B), MYO15 (DFNB3), TMIE (DFNB6), PCDH15 (DFNB23/USH1F), WHRN (DFNB31), and SANS (USH1G).4,5,6,7,8,9,10,11,12,13,14,15
When a novel human deafness locus is mapped, the question arises as to whether or not there is a strain of deaf mouse that carries a mutated gene at a chromosomal map position suggesting conserved synteny with a human locus for deafness. Positional cloning in the mouse or phenotypic rescue using a BAC transgene13,16 can lead to gene identification more quickly than sequencing human genes in a large chromosomal interval of a deafness locus.17 Alternatively, identification of a gene responsible for deafness in a mouse may suggest a candidate human chromosomal location to screen for linkage of deafness segregating in large families that have a structure suitable for providing significant evidence of linkage.18 A combination of two of these strategies was used to identify mutations of TMHS (MIM_609427) as the gene on human chromosome 6p21.1‐p22.3 responsible for non‐syndromic deafness DFNB67, segregating in two consanguineous families.
Methods
Family enrolment
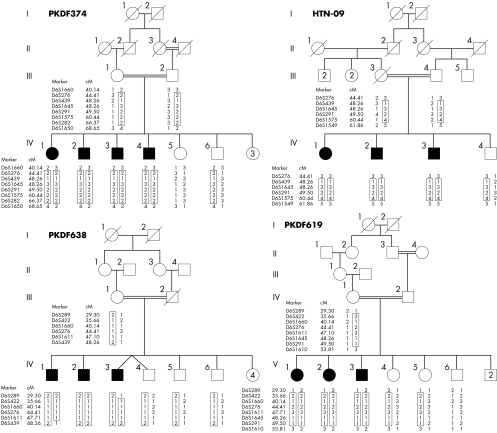
Approval for the study was obtained from the institutional review board at the National Centre of Excellence in Molecular Biology, Lahore, Pakistan (FWA00001758), the NIDCD/NINDS IRB at the National Institutes of Health, Bethesba, Maryland, USA (OH‐93‐N‐016) and the institutional review board at the All India Institute of Medical Sciences, Delhi, India (FWA00001997). Written informed consent was obtained from all the participants. Families PKDF374, PKDF619, and PKDF638 were ascertained from Sindh, Pakistan and family HTN‐09 lives in Chennai, India. The inheritance patterns of deafness segregating in families PKDF374, PKDF619, PKDF638, and HTN‐09 are consistent with an autosomal recessive trait (fig 1).
Figure 1 Chromosome 6 markers that co‐segregate with deafness in families PKDF374, PKDF619, PKDF638, and HTN‐09. STR marker positions are in cM according to the Marshfield human genetic map (http://research.marshfieldclinic.org/genetics/). The linked haplotypes are boxed. Filled symbols denote profound sensorineural hearing loss. The DFNB67 interval is defined by meiotic breakpoints in family PKDF374. Multipoint linkage analyses provided Zmax (maximum LOD scores) of 3.2 for family PKDF374 (markers D6S439 and D6S1645), 1.80 for family HTN‐09 (markers D6S439 and D6S1645), 2.35 for family PKDF638 (markers D6S276 and D6S1611), and 2.80 for family PKDF619 (markers D6S1660 and D6S276).
Clinical evaluation
All participating members of these families were evaluated by a physician to rule out obvious extra‐auditory phenotypes associated with common syndromic forms of deafness. Air conduction pure tone audiometry tests were carried out under quiet ambient conditions at octave frequencies ranging from 250 to 8000 Hz. Vestibular function was evaluated by tandem gait and Romberg testing.
Linkage analysis
Genomic DNA was extracted from peripheral venous blood samples by a standard protocol.19 Samples were genotyped for markers flanking known DFNB loci, using marker information provided by the Hereditary Hearing Loss Homepage (as of August 2004 http://webhost.ua.ac.be/hhh/). For genome‐wide screens, we used the ABI Prism v2.5 Linkage Mapping Set (panels 1 to 27; Applied Biosystems, Foster City, California, USA) containing 388 fluorescently labelled microsatellite markers spaced at an average interval of 10 cM. Short tandem repeat polymorphisms (STRPs) were amplified by polymerase chain reaction (PCR), alleles were assigned using Genescan 3.7 and Genotyper 3.7 (Applied Biosystems), and LOD scores were calculated using LINKMAP as described.9,20,21
Candidate gene screening
Candidate genes were identified using the UCSC Genome Bioinformatics web browser (http://genome.ucsc.edu/) and selected for mutation screening on the basis of their potential role in the inner ear. Primers used for PCR amplification and subsequent sequencing of COL11A2 and TMHS exons were designed using the Primer3 Web site (http://frodo.wi.mit.edu/cgi‐bin/primer3/primer3_www.cgi). PCR primers are from sequence flanking each exon (supplementary tables 1 and 2; the supplementary tables can be seen on the journal website: http://www.jmedgenet.co/supplemental). Amplification, sequencing reactions, and mutation analysis were carried out as described.8
Immunocytochemistry
In order to characterise in greater detail the cellular localisation and developmental profile of TMHS with immunofluorescence confocal microscopy, we used a previously reported affinity purified rabbit polyclonal antiserum generously provided by Ken Johnson.22 Immunostaining was carried out as described.11,23 After fixation in 4% paraformaldehyde for two hours at room temperature, organs of Corti and vestibular end organs of mice were dissected in phosphate buffered saline (PBS). Samples were permeabilised in 0.5% Triton X‐100 for 30 minutes and then washed in PBS. Non‐specific binding sites were blocked using 5% normal goat serum (Life Technologies, Gaithersburg, Maryland, USA) and 2% bovine serum albumin (ICN, Aurora, Ohio, USA) in PBS. Samples were incubated for two hours in the anti‐mTMHS antisera at a concentration of approximately 5 μg/ml in blocking solution. After three rinses in PBS, samples were incubated in a 1:200 dilution of the FITC conjugated anti‐rabbit IgG for 30 minutes, washed again three times with PBS, mounted using the ProLong Antifade kit (Molecular Probes, Eugene, Oregon, USA), and viewed with a LSM510 Zeiss confocal microscope.23
Results
Phenotype
Affected individuals in families PKDF374, PKDF619, PKDF638, and HTN‐09 had congenital bilateral profound hearing loss. Although, we cannot rule out a mixed hearing loss, the expression pattern of TMHS in the inner ear suggests a sensorineural deficit. No vestibular dysfunction was detected using tandem gait or Romberg testing. Clinical evaluation revealed no ophthalmological, skin, or renal anomalies. Fundoscopic examination of IV:2 (22 years) and IV:4 (35 years) from family PKDF374 (fig 1) showed no signs of retinitis pigmentosa.
Mapping of DFNB67
Over 600 families segregating profound congenital deafness were ascertained in Pakistan and were suitable for genetic linkage analyses. After excluding linkage to known DFNB loci,3 a genome‐wide linkage analysis was initially undertaken using DNA samples from four affected and four unaffected members of family PKDF374. Initial evidence of linkage was on chromosome 6p21.1‐p22.3. Additional short tandem repeat (STR) markers were genotyped for all the participating family members, and haplotype analysis revealed a region of homozygosity of approximately 29 centi‐Morgans (cM) delimited by markers D6S1660 (40.14 cM) and D6S1650 (68.65 cM) (fig 1). Multipoint linkage analysis provided a Zmax (maximum LOD score) of 3.2 for the markers D6S439 (48.26 cM) and D6S1645 (48.26 cM). We then discovered three additional families (PKDF619, PKDF638, and HTN‐09) segregating deafness consistent with linkage to markers in the 6p21.1‐p22.3 interval (fig 1). The Human Genome Organization (HUGO) (http://www.gene.ucl.ac.uk/hugo) nomenclature committee24 assigned DFNB67 as the designation for this locus for non‐syndromic deafness.
Genetic and physical map
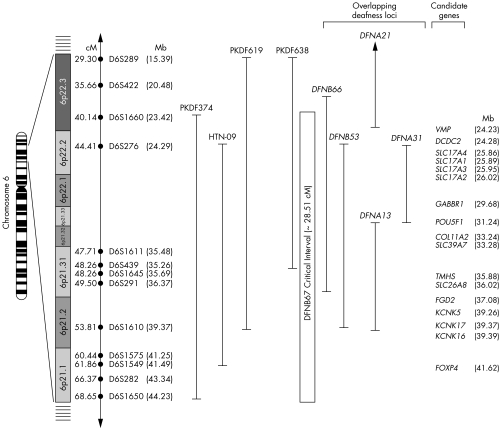
A genetic and physical map of the DFNB67 interval is shown in fig 2. This interval has more than 70 annotated genes and approximately 100 predicted genes (UCSC Genome Bioinformatics: http://genome.ucsc.edu). COL11A2 was a candidate in these families, as allelic variants of this gene are associated with recessive deafness DFNB5325 and nonocular Stickler syndrome.3 However, DNA sequence analysis of the 66 exons of COL11A2 from two affected individuals from each of the four DFNB67 families did not reveal any disease associated variants.
Figure 2 Chromosome 6p21.1‐p22.3 showing the linkage interval of DFNB67. Short tandem repeat (STR) markers are represented by filled circles. The sex‐averaged recombination distances in cM and in Mb are indicated along with STR markers. The DFNB67 interval is based on the meiotic breakpoints in family PKDF374. Linkage regions of five overlapping autosomal deafness loci (DFNB53, DFNB66, DFNA13, DFNA21, and DFNA31) are also illustrated. Cytogenetic locations of several candidate genes are indicated. The Mb positions of the markers and the genes are according to the May 2004 NCBI build 35 of the human genome browser assembly (http://genome.cse.ucsc.edu/cgi‐bin/hgGateway).
Identification of DFNB67 gene
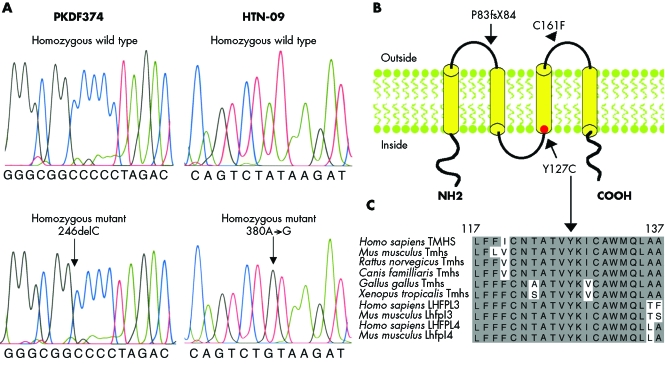
TMHS was also a candidate gene for DFNB67 (fig 2) as a missense mutation of the mouse orthologue, Tmhs, was reported to cause deafness and vestibular dysfunction in the hurry‐scurry (hscy) mouse, and TMHS is expressed in the inner ear hair cell stereocilia of mice.22 Like Tmhs, human TMHS has four exons (NM_182548), which encode a 2162 base pair mRNA. The deduced translation of this cDNA yields a protein of 219 amino acids that has four predicted transmembrane helices. We screened the protein coding sequence, adjacent intronic sequence, and the 5′ and 3′ UTRs of TMHS in two affected individuals from each of the DFNB67 linked families. Affected individuals of family PKDF374 had a homozygous deletion of a single nucleotide at position 246 (246delC) in the first exon of TMHS (fig 3A), which co‐segregated with the hearing loss. This mutant allele is predicted to cause a frameshift and a subsequent truncation of the deduced protein at amino acid position 84 (P83fsX84; fig 3B). A homozygous missense mutation (380A→G) was found in all the affected individuals of family HTN‐09 (fig 3A). This allele results in an amino acid substitution of a conserved tyrosine residue at position 127 (Y127C; fig 3, panels B and C). These two mutations (246delC and 380A→G) were not found in 200 chromosomes from ethnically and geographically matched normally hearing individuals from Sindh province of Pakistan and Chennai, India.
Figure 3 TMHS mutations. (A) Wild type and mutant alleles from unaffected and affected members of families PKDF374 and HTN‐09, respectively. (B) Schematic representation of the predicted TMHS structure (modified from Longo‐Guess et al22) showing the stop codon P83fsX84 (which is caused by the 246delC frameshift mutation) in the sequence encoding the first extracellular loop. The missense mutation Y127C is located in the predicted third transmembrane domain. The location of C161F reported for the hscy mouse22 is indicated by an arrowhead. (C) Alignment of TMHS amino acids from various species shows that Y127 (arrow) is conserved (shaded background, similar amino acids; light background, non‐conserved amino acids).
TMHS is expressed transiently in inner ear hair cell stereociliary bundles
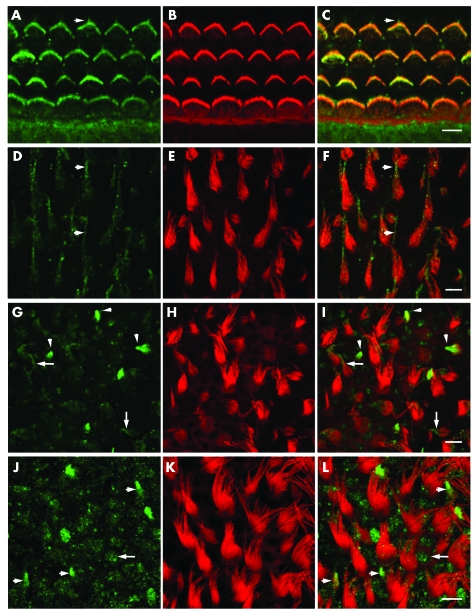
For the developmental profile of the expression pattern of TMHS, we immunostained the inner ears from P0, P3, and P10 mice. TMHS immunoreactivity was detected in the organ of Corti and vestibular hair cells of C57BL/6 mouse as well as in the other cell types such as supporting cells of cochlear and vestibular sensory epithelia, and internal and external sulcus cells of the organ of Corti (fig 4 and data not shown). At high resolution, TMHS immunoreactivity was detected in the kinocilium and along the length of P0 cochlear hair cell stereocilia and appeared to be more concentrated toward the tips (fig 4, panels A to C). A similar pattern was observed in hair cells of the saccular (fig 4, panels D to F) and utricular maculae (fig 4, panels G to I). The amount of TMHS immunoreactivity in the stereocilia appears to vary among different hair bundles. Hair cells with immature stereociliary bundles have a strong TMHS signal but as the hair cells mature there is a gradual reduction in immunoreactivity (fig 4, panels D and G), consistent with the report by Longo‐Guess and co‐workers.22 TMHS immunoreactivity in hair cell stereocilia and kinocilia appears at approximately E16.5 and rapidly disappears by P3, after which we could not detect a signal in stereocilia or in kinocilia except in the vestibular sensory epithelium associated with immature hair cells and in the non‐sensory cells (fig 4, panels J to L).
Figure 4 Immunolocalisation of TMHS in the organ of Corti and vestibular sensory epithelia of a P0 C57BL/6 mouse. (A to C) Localisation of TMHS in cochlear hair cell stereocilia. (A) Anti‐TMHS antibody staining (green channel). Arrowhead indicates the staining of kinocilia. (B) Rhodamine‐phalloidin staining of filamentous actin in stereocilia of one row of inner hair cells and three rows of outer hair cells (red channel). (C) Merged image for TMHS and F‐actin (green and red, respectively). (D to F) TMHS was detected predominantly in immature stereociliary bundles of saccular hair cells and its expression level decreases as bundles mature, as previously reported.22 The arrowheads in panel F indicate kinocilia labelling of mature stereociliary bundles and residual staining of the upper portion of stereociliary bundles. (G to I) In P0 utricular hair cells the pattern of stereociliary staining with anti‐TMHS antibody was similar to that of saccular hair cell stereocilia. The arrowheads point to intensely stained immature hair cell stereociliary bundles and an arrow points to a mature hair cell with staining in the upper portion of the stereociliary bundle. (J to L) In P10 saccule only immature hair cell stereociliary bundles (arrowheads) and non‐sensory cells, such as supporting cells (arrow), are stained with anti‐TMHS antibodies, while no staining was observed in stereociliary bundles of mature hair cells. Scale bar in (C), (F), (I), and (L) is 5 μm.
Discussion
In the hscy mouse, a mutation of Tmhs causes deafness and vestibular dysfunction manifested by circling behaviour.22 The identification of recessive mutations of TMHS indicates an essential role for TMHS in the human auditory system. There are at least two possibilities to explain the discrepancy between the loss of vestibular function in Tmhs mutant mice22 but not in humans. TMHS may be required for normal development of mouse vestibular hair cells but is not required for development of human vestibular hair cells. Alternatively, unlike mice, humans may be better able to compensate partially for loss of vestibular dysfunction by somatosensory and visual input. Subjects in our study were examined in the field only by tandem gait and Romberg testing, which probably excludes severe bilateral vestibular failure but may have missed a more subtle compensated vestibular disorder. More sensitive and informative vestibular testing using posturography was not available to us in India and Pakistan.
The protein encoded by TMHS is a member of a superfamily of tetraspan proteins, which includes the claudin tight junction proteins, gap junction proteins, peripheral myelin, and epithelial membrane proteins as well as calcium channel‐like proteins. Several genes of this superfamily have been reported to be necessary for hearing in either humans or mice, or both.1,3 The predicted structure of TMHS consists of four transmembrane helices with two extracellular loops.22 The missense mutation (C161F) previously reported in hscy mice may disrupt a disulphide bond present in the second extracellular loop and destabilise the secondary structure.22
The frameshift mutation that we found in family PKDF374 (246delC; P83fsX84) is predicted to introduce a stop codon in the first extracellular loop (fig 3B). In vivo, the mutant mRNA would either be translated into a truncated TMHS protein with only one transmembrane domain or be degraded by nonsense mediated decay (NMD).26 The Y127C missense mutation (380A→G) found in family HTN‐09 causes a substitution of the second residue of the third transmembrane domain of TMHS (fig 3B). Tyrosine is a relatively non‐polar hydrophobic amino acid with an aromatic side chain, whereas cysteine is a weakly polar hydrophilic amino acid with a thiol side chain. The large difference between the two amino acids may cause disruption of the third transmembrane domain and may lead to mislocalisation of the encoded protein.
The disappearance of TMHS from hair cell stereociliary bundles at P3 occurs just before the gradual loss of cadherin 23 from stereocilia, which appears to be complete at P16.27,28 A role for TMHS in organising a transient cytoskeleton–membrane interaction in sensory hair cells would be consistent with the stereociliary pathology found in hscy mice22 and the developmental expression profile of TMHS (fig 4).22
We found no disease associated mutations in TMHS or COL11A2 in affected members of families PKDF619 and PKDF638. The linkage interval defined by these two families (PKDF619, PKDF638) is approximately 19cM delimited by markers D6S289 (29.30cM) and D6S439 (48.26). It is possible that these individuals harbour mutations of cis acting regulatory elements of either COL11A2 or TMHS. The deafness linked haplotype of affected individuals in family PKDF638 excludes the protein coding region of TMHS (fig 2).
There are three additional loci for hearing loss defined by dominant mutant alleles (DFNA13, DFNA21, and DFNA31) at chromosome 6p21.2‐p22.3 and two recessive deafness loci (DFNB53 and DFNB66) in this interval (fig 2). In the single family used to map DFNB66, TMHS was screened for mutations and none was found.29 It seems plausible that a mutation of a second gene in this interval is associated with hearing loss in families PKDF619 and PKDF638. Other candidates in the 6p21.2‐p22.3 interval include five solute carrier family members (SLC17A1, SLC17A2, SLC17A3, SLC17A4, and SLC39A7), POU5F1, a POU domain containing transcription factor and γ‐aminobutyric acid B receptor 1 (GABBR1). Genes encoding SLCs and POU domain transcription factors are important for normal hearing.3,30 Another possibility for not finding mutations in TMHS in two of the four families (fig 1) is that the LOD scores for deafness segregating in PKDF619 and PKDF638 (Zmax 2.80 and 2.35, respectively) do not rise to the level of statistical significance for linkage, and thus there may be spurious association of deafness segregating in these two families with STRs on chromosome 6p. The actual deafness‐causing mutations in families PKDF619 and PKDF638 may be somewhere else in the genome.
In summary, we have mapped a new non‐syndromic recessive deafness locus DFNB67 on chromosome 6p21.1‐p22.3. In two consanguineous families, we have identified two likely pathogenic mutations of TMHS that co‐segregate with deafness.
The supplementary tables are accessible on the journal website: http://www.jmedgenet.co/supplemental
Acknowledgements
We are grateful to the members of the four families for their participation in this study and to K Johnson for generously providing anti‐TMHS antisera. We also thank Dennis Drayna, Andrew Griffith, Karen Friderici, Shin‐ichiro Kitajiri, Rob Morell, Julie Schultz, Sabiha Nazli, and Saeeda Kalsoom for their suggestions in relation to the manuscript. The study was supported by the Higher Education Commission, Islamabad, Pakistan; Ministry of Science and Technology, Islamabad, Pakistan; the International Centre for Genetic Engineering and Biotechnology, Trieste, Italy under project CRP/PAK02‐01 (contract No 02/013); and intramural funds from the National Institute on Deafness and Other Communication Disorders (1 ZO1 DC000035‐07 and 1 ZO1 DC000039‐07).
Footnotes
Conflicts of interest: none declared
The supplementary tables are accessible on the journal website: http://www.jmedgenet.co/supplemental
References
- 1.Anagnostopoulos A V. A compendium of mouse knockouts with inner ear defects. Trends Genet 200218499. [DOI] [PubMed] [Google Scholar]
- 2.Finsterer J, Fellinger J. Nuclear and mitochondrial genes mutated in nonsyndromic impaired hearing. Int J Pediatr Otorhinolaryngol 200569621–627. [DOI] [PubMed] [Google Scholar]
- 3.Friedman T B, Griffith A J. Human nonsyndromic sensorineural deafness. Annu Rev Genomics Hum Genet 20034341–402. [DOI] [PubMed] [Google Scholar]
- 4.Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston M D.et al Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 199537460–61. [DOI] [PubMed] [Google Scholar]
- 5.Friedman T B, Liang Y, Weber J L, Hinnant J T, Barber T D, Winata S, Arhya I N, Asher J H. A gene for congenital, recessive deafness DFNB3 maps to the pericentromeric region of chromosome 17. Nat Genet 1995986–91. [DOI] [PubMed] [Google Scholar]
- 6.Wang A, Liang Y, Fridell R A, Probst F J, Wilcox E R, Touchman J W, Morton C C, Morell R J, Noben‐Trauth K, Camper S A, Friedman T B. Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science 19982801447–1451. [DOI] [PubMed] [Google Scholar]
- 7.Fukushima K, Ramesh A, Srisailapathy C R, Ni L, Wayne S, O'Neill M E, Van Camp G, Coucke P, Jain P, Wilcox E R.et al An autosomal recessive nonsyndromic form of sensorineural hearing loss maps to 3p‐DFNB6. Genome Res 19955305–308. [DOI] [PubMed] [Google Scholar]
- 8.Naz S, Giguere C M, Kohrman D C, Mitchem K L, Riazuddin S, Morell R J, Ramesh A, Srisailpathy S, Deshmukh D, Riazuddin S, Griffith A J, Friedman T B, Smith R J, Wilcox E R. Mutations in a novel gene, TMIE, are associated with hearing loss linked to the DFNB6 locus. Am J Hum Genet 200271632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmed Z M, Riazuddin S, Bernstein S L, Ahmed Z, Khan S, Griffith A J, Morell R J, Friedman T B, Riazuddin S, Wilcox E R. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet 20016925–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alagramam K N, Yuan H, Kuehn M H, Murcia C L, Wayne S, Srisailpathy C R, Lowry R B, Knaus R, Van Laer L, Bernier F P, Schwartz S, Lee C, Morton C C, Mullins R F, Ramesh A, Van Camp G, Hageman G S, Woychik R P, Smith R J. Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum Mol Genet 2001101709–1718. [DOI] [PubMed] [Google Scholar]
- 11.Ahmed Z M, Riazuddin S, Ahmad J, Bernstein S L, Guo Y, Sabar M F, Sieving P, Riazuddin S, Griffith A J, Friedman T B, Belyantseva I A, Wilcox E R. PCDH15 is expressed in the neurosensory epithelium of the eye and ear and mutant alleles are responsible for both USH1F and DFNB23. Hum Mol Genet 2003123215–3223. [DOI] [PubMed] [Google Scholar]
- 12.Mustapha M, Chouery E, Chardenoux S, Naboulsi M, Paronnaud J, Lemainque A, Megarbane A, Loiselet J, Weil D, Lathrop M, Petit C. DFNB31, a recessive form of sensorineural hearing loss, maps to chromosome 9q32–34. Eur J Hum Genet 200210210–212. [DOI] [PubMed] [Google Scholar]
- 13.Mburu P, Mustapha M, Varela A, Weil D, El‐Amraoui A, Holme R H, Rump A, Hardisty R E, Blanchard S, Coimbra R S, Perfettini I, Parkinson N, Mallon A M, Glenister P, Rogers M J, Paige A J, Moir L, Clay J, Rosenthal A, Liu X Z, Blanco G, Steel K P, Petit C, Brown S D. Defects in whirlin, a PDZ domain molecule involved in stereocilia elongation, cause deafness in the whirler mouse and families with DFNB31. Nat Genet 200334421–428. [DOI] [PubMed] [Google Scholar]
- 14.Weil D, El‐Amraoui A, Masmoudi S, Mustapha M, Kikkawa Y, Laine S, Delmaghani S, Adato A, Nadifi S, Zina Z B, Hamel C, Gal A, Ayadi H, Yonekawa H, Petit C. Usher syndrome type I G (USH1G) is caused by mutations in the gene encoding SANS, a protein that associates with the USH1C protein, harmonin. Hum Mol Genet 200312463–471. [DOI] [PubMed] [Google Scholar]
- 15.Mustapha M, Chouery E, Torchard‐Pagnez D, Nouaille S, Khrais A, Sayegh F N, Megarbane A, Loiselet J, Lathrop M, Petit C, Weil D. A novel locus for Usher syndrome type I, USH1G, maps to chromosome 17q24‐25. Hum Genet 2002110348–350. [DOI] [PubMed] [Google Scholar]
- 16.Probst F J, Fridell R A, Raphael Y, Saunders T L, Wang A, Liang Y, Morell R J, Touchman J W, Lyons R H, Noben‐Trauth K, Friedman T B, Camper S A. Correction of deafness in shaker‐2 mice by an unconventional myosin in a BAC transgene. Science 19982801444–1447. [DOI] [PubMed] [Google Scholar]
- 17.Johnson K R. Mouse models of human hearing disorders. Curr Genom 2001255–69. [Google Scholar]
- 18.Steel K P, Kros C J. A genetic approach to understanding auditory function. Nat Genet 200127143–149. [DOI] [PubMed] [Google Scholar]
- 19.Grimberg J, Nawoschik S, Belluscio L, McKee R, Turck A, Eisenberg A. A simple and efficient non‐organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res 19891783–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmad J, Khan S N, Khan S Y, Ramzan K, Riazuddin S, Ahmed Z M, Wilcox E R, Friedman T B, Riazuddin S. DFNB48, a new nonsyndromic recessive deafness locus, maps to chromosome 15q23‐q25.1. Hum Genet 2005116407–412. [DOI] [PubMed] [Google Scholar]
- 21.Ramzan K, Shaikh S R, Ahmad J, Khan S N, Riazuddin S, Ahmed Z M, Friedman T B, Wilcox E R, Riazuddin S. A new locus for nonsyndromic deafness DFNB49 maps to chromosome 5q12.3‐q14.1. Hum Genet 200511617–22. [DOI] [PubMed] [Google Scholar]
- 22.Longo‐Guess C M, Gagnon L H, Cook S A, Wu J, Zheng O Y, Johnson K R. A missense mutation in the previously undescribed gene Tmhs underlies deafness in hurry‐scurry (hscy) mice. Proc Natl Acad Sci USA 20051027894–7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belyantseva I A, Boger E T, Naz S, Frolenkov G I, Sellers J R, Ahmed Z M, Griffith A J, Friedman T B. Myosin‐XVa is required for tip localization of whirlin and differential elongation of hair‐cell stereocilia. Nat Cell Biol 20057148–156. [DOI] [PubMed] [Google Scholar]
- 24.Povey S, Lovering R, Bruford E, Wright M, Lush M, Wain H. The HUGO Gene Nomenclature Committee (HGNC). Hum Genet 2001109678–680. [DOI] [PubMed] [Google Scholar]
- 25.Chen W, Kahrizi K, Meyer N C, Riazulhosseini Y, Van Camp G, Najamabadi H, Smith R J H. Mutation of COL11A2 causes autosomal recessive non‐syndromic hearing loss at DFNB53 locus. J Med Genet 200542e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maquat L E. Nonsense‐mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 2004589–99. [DOI] [PubMed] [Google Scholar]
- 27.Lagziel A, Ahmed Z M, Schultz J M, Morell R J, Belyantseva I A, Friedman T B. Spatiotemporal pattern and isoforms of cadherin 23 in wild type and waltzer mice during inner ear hair cell development. Dev Biol 2005280295–306. [DOI] [PubMed] [Google Scholar]
- 28.Michel V, Goodyear R J, Weil D, Marcotti W, Perfettini I, Wolfrum U, Kros C J, Richardson G P, Petit C. Cadherin 23 is a component of the transient lateral links in the developing hair bundles of cochlear sensory cells. Dev Biol 2005280281–294. [DOI] [PubMed] [Google Scholar]
- 29.Tlili A, Mannikko M, Charfedine I, Lahmar I, Benzina Z, Ben Amor M, Driss N, Ala‐Kokko L, Drira M, Masmoudi S, Ayadi H. A novel autosomal recessive non‐syndromic deafness locus, DFNB66, maps to chromosome 6p21.2‐22.3 in a large Tunisian consanguineous family. Hum Hered 200560123–128. [DOI] [PubMed] [Google Scholar]
- 30.Petit C, Levilliers J, Hardelin J P. Molecular genetics of hearing loss. Annu Rev Genet 200135589–646. [DOI] [PubMed] [Google Scholar]