Abstract
Background
The 3243A→G is a common pathogenic mitochondrial DNA (mtDNA) point mutation causing a variety of different phenotypes. Segregation of this mutation to different tissues during embryonic life and postnatally is still enigmatic.
Objective
To investigate the tissue distribution of this mutation.
Methods
In 65 individuals from nine families segregating the 3243A→G mutation, the mutation load (% mutated mtDNA) was determined in various tissues. Mutation load was measured in two to four cell types—blood leucocytes, buccal cells, skeletal muscle cells, and urine epithelial cells (UEC)—derived from all three embryogenic germ layers.
Results
There was a significant correlation among mutation loads in the four tissues (r = 0.80–0.89, p<0.0001). With blood serving as reference, the mutation load was increased by 16% in buccal mucosa, by 31% in UEC, and by 37% in muscle. There were significant differences between the mitotic tissues blood, buccal mucosa, and UEC (p<0.0001), but no difference between UEC and muscle. Using the present data as a cross sectional investigation, a negative correlation of age with the mutation load was found in blood, while the mutation load in muscle did not change with time; 75% of the children presented with higher mutation loads than their mothers in mitotic tissues but not in the post‐mitotic muscle.
Conclusions
There appears to be a uniform distribution of mutant mtDNA throughout the three germ layers in embryogenesis. The significant differences between mutation loads of the individual tissue types indicate tissue specific segregation of the 3243A→G mtDNA later in embryogenesis.
Keywords: 3243A→G, mtDNA, heteroplasmy, mutation distribution, tissue segregation
Mutations in mitochondrial DNA (mtDNA) are important causes of many different diseases, with a high variability in the phenotypic presentation.1,2 The symptoms are often neurological, presenting as encephalopathy, stroke‐like episodes, hearing loss, migraine, ataxia, myopathy, and ophthalmoplegia, but non‐neurological symptoms such as cardiomyopathy and diabetes mellitus are also common.3
The presentation may be mono‐symptomatic, but often multiple organs are involved, and the same mutation may cause very different phenotypes, varying from asymptomatic carriers to severely affected individuals within the same family. Pathogenic mtDNA point mutations usually co‐exist with wild‐type mtDNA in the same cell (mtDNA heteroplasmy). In contrast, healthy subjects generally carry identical mtDNA molecules in their cells (mtDNA homoplasmy). For some mtDNA mutations the degree of heteroplasmy—that is, the mutation load—varies considerably among tissues,4,5 while for others, such as 8993T→G/C, no such variation occurs.6 Additionally, there seems to be both a mutation specific as well as a tissue specific threshold for the mutation load before symptoms develop.7,8,9
Mitochondrial mutations are maternally inherited or occur sporadically. Mature oocytes contain up to 100 000 mitochondria. According to the generally accepted genetic bottleneck, the number of mtDNA molecules is reduced to during oogenesis before final replication. As a consequence of this bottleneck, a very limited number of mitochondrial genomes will found the next generation of mtDNA.10,11,12
During the last decade, the patterns of inheritance, segregation, and distribution of mtDNA heteroplasmy have been studied intensively in both man and mouse. Human studies are limited by the few families available, and most results come from heteroplasmic mice. It is, however, generally believed that random genetic drift is the main mechanism for the apparently unpredictable transmission of pathogenic mtDNA mutations from one generation to the next.13,14,15
Case reports on mutation positive human fetuses indicate that the mutation load is the same in different tissues, and thus that random segregation prevails in early development.16,17 A study of four tissues from one 3243A→G mtDNA positive family showed ranked hierarchy, indicating a non‐random tissue specific distribution of the mutated mtDNA.18
One of the most prevalent pathogenic mitochondrial mutations is the 3243A→G transition. Among the phenotypes this mutation may cause are: mitochondrial myopathy, encephalopathy, lactic acidosis and stroke‐like episodes (MELAS); chronic progressive external ophthalmoplegia (CPEO)19; myoclonus epilepsy with ragged red fibres (MERFF)20; and maternally inherited diabetes and deafness (MIDD).21
To shed more light on the tissue distribution of this mutation, we studied correlations between mutation loads in four different tissues in a cohort of 65 persons from nine 3243A→G mtDNA families; we studied the transmission of mutation load from mothers to their children; and we evaluated in a cross sectional design the age dependent development of the mutation load in the four tissues.
Methods
Subjects
The cohort consisted of nine 3243A→G mtDNA mutation positive white Danish families from which 65 members agreed to participate. The probands in six of the families presented with MIDD and were recruited from an endocrinological clinic, while three probands presented with epilepsy and exercise intolerance (EIT) and were recruited through a neurological clinic. The median age was 41 years (7 to 74). Twenty six were male and 39 female. The median age of the mothers was 60 years (38 to 66) and the median age of their children 34 years (9 to 43). At the time of investigation, 42 subjects (64%) had developed one or more symptoms associated with the 3243A→G mtDNA mutation.
In each subject, two to four of the following tissues were sampled for determination of mutation load: blood, skeletal muscle, buccal mucosa, and urine epithelial cells (UEC). Most samples were collected on the same day. The time interval between the sampling of different tissues was not more than one year.
Skeletal muscle biopsy was not done in subjects below 18 years of age.
The project was approved by the scientific‐ethics committees of the hospitals in Ringkoebing, Ribe, and South Jutland Counties (M‐2305‐01), the Hospitals of Copenhagen (KF 01‐075/00), and the Danish Data Inspection Board (2001‐41‐1648). Signed informed consent was obtained from all subjects, and subjects younger than 18 years (n = 9) were only tested after signed informed consent from their parents. All families received genetic counselling.
Tissue sampling and isolation of DNA
DNA from blood was isolated using a routine salting out procedure. For skeletal muscle, a needle biopsy was done in the left lateral vastus muscle. The biopsy was immediately frozen in isopentane, cooled by liquid nitrogen, and placed at −80°C until analysis. Buccal mucosa cells were obtained by brushing the cheek in the oral cavity with a cytobrush for 30 seconds. Samples were stored at 5°C until DNA isolation. Twenty millilitres of urine were sampled, and the UEC was spun down and resuspended in Tris buffer. DNA extractions were carried out using the QIAamp® DNA Mini Kit (Qiagen GmbH, Hilden, Germany), using the manufacturer's protocol.
Molecular analyses
An allele specific polymerase chain reaction (PCR) assay was used for the detection of the 3243A→G mutation by a direct PCR reaction. Two sets of primers were used in one reaction tube: 3014L (forward): GTG CAG CCG CTA TTA AAG GT; and 3262HM17 (reverse, with two mismatches): TTT TAT GCG ATT ACCGCG CC. The PCR product was 249 base pairs (bp). A control product was co‐amplified using the primers: 5289–5309 (forward) and 5903–5884 (reverse). The PCR product was 614 bp.
Determination of heteroplasmy
The ratio between wild‐type (3243A) and mutant mtDNA (3243G) was determined using solid phase minisequencing.22 PCR products spanning the position in question were generated by the 5′‐biotinylated primer (3014–3033) and the primer (3376–3357). PCR products were immobilised on a streptavidin coated solid support (96‐well plate) and denatured by sodium hydroxide. To quantify the ratios of 3243A and 3243G in the sample, a sequencing primer (3263–3244) was designed to anneal adjacently (downstream) to nt 3243. The nucleotide at the site of the mutation was analysed by primer extension reaction, in which a [3H] labelled dNTP corresponding to either the 3243A (dTTP) or 3243G (dCTP) nucleotide was added to two parallel reactions. After washing, the elongated primers were eluted by sodium hydroxide and the amounts of incorporated [3H]dNMP were determined using a liquid scintillation counter. The ratio of T to C incorporated into each sequencing primer was determined and compared with a standard curve constructed using known proportions of cloned mt3243A and mt3243G. The sensitivity of solid phase minisequencing is 1%. Controls negative for the 3243A→G mutation had an average test result for mutation load of 0.37% (range 0 to 1.01%).
Mutation load below 1.5% was considered equal to zero.
Statistics
The program “Intercooled Stata version 8.1 for Windows” was applied for statistical calculations. Spearman rank coefficients were calculated for correlations between mutation load in the different cell types and for the correlations among tissue mutation loads and age of subject at time of testing. A one way analysis of variance (ANOVA) was applied for comparing degrees of heteroplasmy among tissues using multiple comparison estimation (Bonferroni's correction). The Mann–Whitney two sample rank‐sum test was used to compare differences among groups. Scatterplots with regression lines and 95% confidence intervals are presented. Values are mean (SD) unless stated otherwise. A significance level of 0.05 or less (two way testing) was considered statistically significant.
Results
Heteroplasmy in the tissues
We determined the mutation load in blood, buccal mucosa, UEC, and skeletal muscle from 65 subjects from the nine families segregating 3243A→G mutation. All the data are listed in table 1.
Table 1 Mutation load of skeletal muscle, urine epithelial cells (UEC), buccal mucosa, and blood leucocytes (A), with differences (Δ) between mutation loads in tissues (B).
| n | Median % | Min % | Max % | |
|---|---|---|---|---|
| A Tissue | ||||
| Muscle | 43 | 56 | 0 | 92 |
| UEC | 55 | 50 | 0 | 96 |
| Buccal mucosa | 54 | 35 | 0 | 79 |
| Blood | 57 | 19 | 0 | 78 |
| B Differences between tissues | ||||
| ΔMuscle − blood | 31 | 33 | −2 | 72 |
| ΔUEC − blood | 50 | 26 | −2 | 76 |
| ΔBuccal mucosa − blood | 48 | 9 | −5 | 43 |
| ΔMuscle − UEC | 35 | 2 | −27 | 51 |
Max, maximum; min, minimum; UEC, urine epithelial cells.
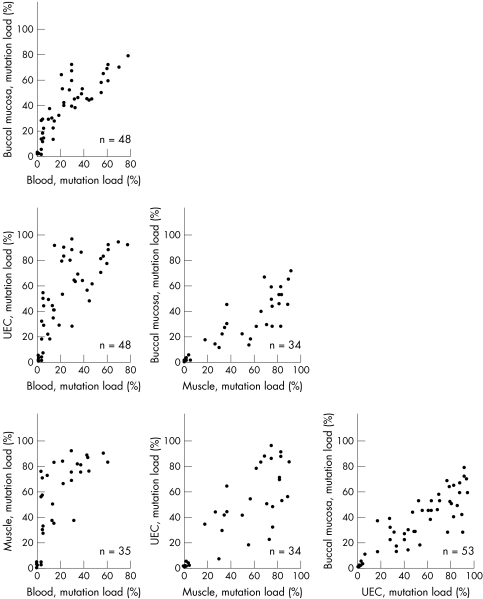
Although mutation load differed considerably among tissues, there were strong pairwise correlations for mutation load among all four tissues (r = 0.80–0.89, p<0.00001) (fig 1).
Figure 1 Correlation between heteroplasmy in blood, buccal mucosa, urine epithelial cells (UEC), and skeletal muscle. Numbers on the x and y axes designate percentage of heteroplasmy; n = number of subjects.
There were significant differences (p<0.0001) among mutation loads of the three mitotic tissues. With blood serving as a reference, the median value of the mutation load was increased by 16% in buccal mucosa, by 31% in UEC, and by 37% in skeletal muscle. The median mutation load in UEC was the same as in the post‐mitotic skeletal muscle, and was higher (p<0.0001) than in blood leucocytes and buccal mucosa.
In all individuals in whom the mutation was detectable in blood, it was also detected in other tissues. In some individuals in whom the mutation was undetectable in blood it was clearly present, but at low concentration, in other tissues.
The four tissues were not tested in all subjects. This was either because muscle biopsies were not taken in persons below 18 years of age, or because subjects refused to have further tests done, or because of the unavailability of some subjects.
Mutation load and age
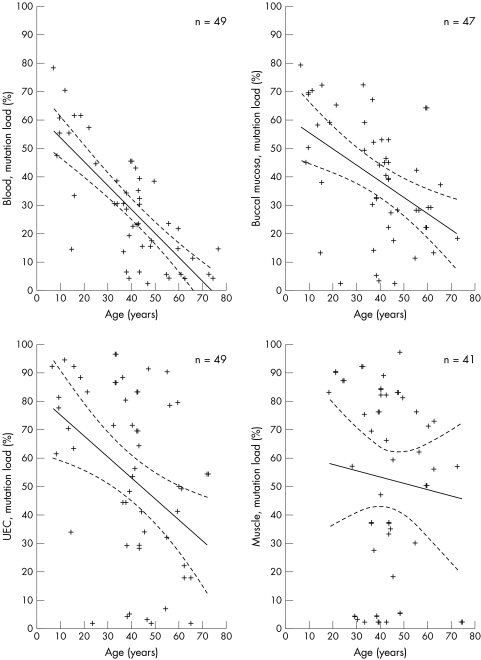
When correlating age of the subjects to the mutation load in the four tissues, a negative correlation was identified for blood (r = −0.73, p<0.00001) whereas correlations for the remaining two mitotic tissues were not obvious (buccal mucosa: r = −0.50, p<0.00004; UEC: r = −0.44, p<0.0002; fig 2). There was no correlation between age and level of heteroplasmy in muscle. The mutation load in muscle did not change with time. This was a cross sectional study and we did not determine the mutation load in a person more than once.
Figure 2 Correlation between the age of subjects and the mutation load (heteroplasmy) in blood, buccal mucosa, urine epithelial cells (UEC), and skeletal muscle. Each cross represents a subject; n = number of subjects. Solid line designates Spearman's correlation coefficient line. Dotted line designates the 95% confidence intervals.
Offspring analysis
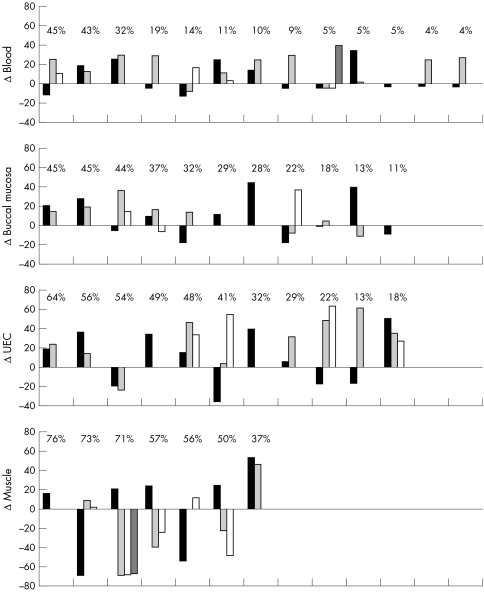
Levels of heteroplasmy in the mothers were compared with the levels found in their children (fig 3). Within families, there was no consistent pattern in mother to child mutation transmission. In mitotic tissue, 69% of the children had higher mutation loads than their mother. In blood the average mutation load was higher (mean 11%; range −13 to +39) in 19 of 30 children, in buccal mucosa it was higher (mean 10%; range −19 to +44) in 14 of 22, and in UEC 19 of 24 had a higher mutation load (mean 22%; range −36 to 64). In skeletal muscle the average mutation load in eight of 17 children was higher than that in their mothers (mean −17%; range −69 to +53).
Figure 3 Differences in heteroplasmy of the 3243A→G mutation between the mothers and their offspring. Differences in mutation load among mothers and children were calculated by subtracting the tissue mutation loads in children from the load in corresponding tissues of the mothers. Each column represents a child, and siblings are arranged in appearance of birth. The level of heteroplasmy of their mother is shown above the columns. The data are ordered according to the mutation load in the mothers. UEC, urine epithelial cells.
Discussion
We present a large and comprehensive study of mutation load in four different tissues originating from the three germ layers, in a cohort of persons with the 3243A→G mutation of the mtDNA, where both symptomatic and asymptomatic individuals were studied. The symptoms ranged from one person with MELAS to impaired glucose intolerance or hearing impairment as the only symptoms (table 2).
Table 2 Clinical symptoms and mutation load in the four tissues.
| Clinical symptoms | n | Mutation load (%) | ||||
|---|---|---|---|---|---|---|
| Age | Blood | Buccal | UEC | Muscle | ||
| (years) | mucosa | |||||
| MELAS, epilepsy, HI, ataxia | 2 | 18 to 25 | 30 to 44 | 71 | 72 | 87 to 92 |
| MIDD, encephalopathy, EIT | 5 | 21 to 42 | 30 to 57 | 44 to 65 | 48 to 96 | 37 to 90 |
| MIDD | 8 | 37 to 74 | 10 to 39 | 29 to 67 | 18 to 90 | 69 |
| MIDD, EIT | 10 | 35 to 65 | 9 to 38 | 28 to 53 | 18 to 91 | 62 to 84 |
| IGT, HI, EIT | 4 | 60 to 74 | 4 to 57 | 18 to 22 | 41 to 83 | 50 to 90 |
| IGT | 5 | 37 to 61 | 4 to 19 | 11 to 32 | 7 to 34 | 18 to 56 |
| HI | 2 | 10 to 48 | 17 to 70 | 70 | 94 | 71 to 82 |
| CU | 23 | 7 to 66 | 0 to 61 | 0 to 79 | 0 to 92 | 0 to 83 |
| Other* | 6 | 13 to 62 | 0 to 55 | 0 to 58 | 0 to 70 | 0 to 76 |
*Gastropathy, nephropathy, ataxia, cardiomyopathy, tinnitus.
CU, clinically unaffected; EIT, exercise intolerance; HI, hearing impairment; IGT, impaired glucose tolerance; MIDD, maternally inherited deafness and diabetes mellitus; n, number of subjects.
Although mutation load differed considerably among tissues, we found a strong pairwise correlations for mutation load among all four tissues.
Tissue‐specific levels of pathogenic mtDNA mutations are well known. The mutation load of mtDNA deletions are often confined to muscle, and levels of tRNA point mutations are generally higher in muscle than in blood. Several small studies have shown a correlation of the levels of 3243A→G mtDNA in different types of tissues. Chinnery et al analysed skeletal muscle, hair follicles, buccal mucosa, and blood in five individuals from a single MIDD family and found a non‐random tissue distribution of the 3243A→G mtDNA.18 In members of MELAS pedigrees, a high level was found in UEC.5,23 In the larger study of MELAS pedigrees (61 persons) by Shanske et al,24 the mutation load was studied in four tissues (blood, skin fibroblasts, buccal mucosa, and UEC) but not in skeletal muscle. The highest level was found in UEC, being three times higher than the level in blood. Our results fully agree with these findings. Furthermore we have extended the analysis to include skeletal muscle, and have showed that the mutation load in UEC is as high as in muscle.
The tissues studied in this investigation are derived from all three germ layers: the endoderm (UEC), the mesoderm (muscle and blood), and the ectoderm (buccal mucosa). It is generally believed that higher levels of mutation load are found in postmitotic tissues such as muscle, whereas lower levels are found in highly mitotic tissues such as leucocytes. The significant tissue correlations and the differences in tissue mutation loads support the view that distribution of the 3243A→G mutation is not a random process,18 but is highly tissue specific. The high mutation level found in UEC and buccal mucosa cells suggests that in these cells with a relatively short life span, no selection against the cells harbouring the mutation occurs and this may reflect the low energy demand of these tissues. However, our results may also be explained by a negative selection in the process of mitotic activity, because, as reported earlier,25 a negative correlation between age and the mutation load of mitotic tissues was identified (fig 2). The age associated decrease in mutation load was strongest for blood and less for buccal mucosa and UEC. In the post‐mitotic skeletal muscle, the mitotic elimination of cells with high levels of mutant mtDNA is not an option. The lack of correlation between age and muscle mutation load in the present study supports the hypothesis that mutant load remains constant during life in skeletal muscle. Follow up studies are needed to determine whether there is an age associated decrease for other mitotic tissues. The correlation between age and tissue heteroplasmy levels was based on a cross sectional survey and could therefore be biased. However, the conclusion was based on findings in a very large cohort of 3243A→G mutation positive persons with a wide range of age, including nine under 18 years of age.
The tight coupling between mutation load in each individual further supports the view that the distribution of the 3243A→G mutation is uniform during early embryogenesis.23,26
The data on levels of the pathogenic mtDNA mutation in human embryos are very limited. A uniform distribution of mutation load was reported in two 3243A→G mutation positive, 24 to 25 week old human fetuses16,17 and together with findings from heteroplasmic bovine models27 this has led to the hypothesis that no major segregation takes place in tissues in the prenatal state.
Heteroplasmic healthy mouse models have previously demonstrated such tissue specific segregation and indicated that it is both mtDNA genotype dependent as well as under nuclear genomic control.28 In theory, this may also be the case for certain human pathogenic mtDNA mutations as the 8993T→G and T→C seem equally distributed, in contrast to the above findings concerning 3243A→G.
Because of the unpredictable phenotypic presentations and transmission patterns from mother to child, it has proved difficult to offer genetic counselling to women with point mutations of the mtDNA.29 The bottleneck has been found to be the major component of the variable transmission of mutations, as shown both in a patient with Kearns‐Sayre syndrome and in heteroplasmic mouse models.11,30 Chinnery et al15 studied the transmission of six of the most common pathogenic point mutations: 8344A→G, 8993T→C, 3460G→A, 3243A→G, 8993T→G, and 11778G→A. In a meta‐analysis they calculated the differences in mutation load between mothers and their children. They found that for the transmission of 3243A→G, 8993T→G, and 11778G→A the mean value was significantly greater than zero, suggesting a preferential transmission of mutant genomes. This study was based on the mutation load in blood, and our results are in agreement as far as mutation load in blood is concerned. The mean difference in our study, based on 30 transmissions, was 11% compared with 9.6% in the study by Chinnery et al based on 80 transmissions. We found the same pattern in buccal mucosa and UEC. However, our study in muscle, based on 17 transmissions, showed a mean value of −17%, suggesting the preferential transmission of mutant genome. As several studies including our own have shown a marked decrease in the mutation load in blood with age it is questionable whether blood should be used in the study of transmission. Our study has shown that the mutation load in muscle is the most stable and therefore the best choice. However, although our results in muscle show a mean difference of −17%, the data were widely scattered. The women gave birth both to severely disabled children with high mutation loads and to healthy children with very low mutation levels (<2%). This variance in mother to child transmission—that is, the result of the bottleneck—of the 3243A→G mtDNA was clearly demonstrated in our cohort for all four tissues (fig 3).
Our results concerning the mutation loads in muscle indicate that this mutation load may be predicted by determining the load in the fetus or the embryo. Thus our findings support the suggestion that preimplantation genetic diagnosis could be of help in some mitochondrial diseases.29,30,31
Genetic investigation of mitochondrial disease often uses blood leucocyte mtDNA as the reference for testing. Nevertheless, this tissue is not adequate for studying pathogenic mtDNA point mutations. In our study, the level of heteroplasmy in buccal mucosa and UEC was consistently higher than the level in blood, and also in asymptomatic persons. Thus tissues other than blood should be used diagnostically for the detection of the 3243A→G mutation, as previously proposed.5,23,24 However, although UEC, buccal cells, or hair epithelial cells are very attractive diagnostic alternatives to a muscle biopsy, these tissues cannot be used for histochemical analyses, which are necessary in most patients with unknown mutations. Furthermore, sporadic mutations—such as the single large scale mtDNA deletions—may be confined to muscle and therefore not present in either buccal cells or UEC.
Genetic testing of buccal cells and UEC may prove an important supplement to the diagnosis and counselling of patients with mitochondrial diseases, but cannot replace skeletal muscle as the most informative tissue for the diagnosis of mitochondrial disease.
Acknowledgements
We wish to thank Lars Korsholm, Department of Statistics, University of Southern Denmark (SDU) for excellent statistical assistance and fruitful discussions. The excellent technical assistance by Dorthe Munkløv is highly appreciated. The study was financially supported by grants from the Novo Nordic Foundation, Copenhagen Hospital Community Foundation, The Danish National Research Council (No 22‐03‐0474), and Fabrikant Vilhelm Pedersen og Hustrus legat on recommendation from The Novo Nordisk Foundation.
Abbreviations
MELAS - mitochondrial myopathy, encephalopathy, lactic acidosis and stroke‐like episodes
MIDD - maternally inherited diabetes and deafness
MtDNA - mitochondrial DNA
UEC - urine epithelial cells
Footnotes
Conflicts of interest: none declared
References
- 1.Chinnery P F, Johnson M A, Wardell T M, Singh‐Kler R, Hayes C, Brown D T, Taylor R W, Bindoff L A, Turnbull D M. The epidemiology of pathogenic mitochondrial DNA mutations. Ann Neurol 200048188–193. [PubMed] [Google Scholar]
- 2.Schaefer A M, Taylor R W, Turnbull D M, Chinnery P F. The epidemiology of mitochondrial disorders – past, present and future. Biochim Biophys Acta 20041659115–120. [DOI] [PubMed] [Google Scholar]
- 3.Taylor R W, Turnbull D M. Mitochondrial DNA mutations in human disease. Nat Rev Genet 20056389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lestienne P, Ponsot G. Kearns‐Sayre syndrome with muscle mitochondrial DNA deletion. Lancet . 1988;i885. [DOI] [PubMed]
- 5.Dubeau F, De Stefano N, Zifkin B G, Arnold D L, Shoubridge E A. Oxidative phosphorylation defect in the brains of carriers of the tRNAleu(UUR) A3243G mutation in a MELAS pedigree. Ann Neurol 200047179–185. [PubMed] [Google Scholar]
- 6.White S L, Collins V R, Wolfe R, Cleary M A, Shanske S, DiMauro S, Dahl H H, Thorburn D R. Genetic counseling and prenatal diagnosis for the mitochondrial DNA mutations at nucleotide 8993. Am J Hum Genet 199965474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mariotti C, Savarese N, Suomalainen A, Rimoldi M, Comi G, Prelle A, Antozzi C, Servidei S, Jarre L, DiDonato S.et al Genotype to phenotype correlations in mitochondrial encephalomyopathies associated with the A3243G mutation of mitochondrial DNA. J Neurol 1995242304–312. [DOI] [PubMed] [Google Scholar]
- 8.van den Ouweland J M, Maechler P, Wollheim C B, Attardi G, Maassen J A. Functional and morphological abnormalities of mitochondria harbouring the tRNALeu(UUR) mutation in mitochondrial DNA derived from patients with maternally inherited diabetes and deafness (MIDD) and progressive kidney disease. Diabetologia 199942485–492. [DOI] [PubMed] [Google Scholar]
- 9.Jeppesen T D, Schwartz M, Olsen D B, Vissing J. Oxidative capacity correlates with muscle mutation load in mitochondrial myopathy. Ann Neurol 20035486–92. [DOI] [PubMed] [Google Scholar]
- 10.Jansen R P. Germline passage of mitochondria: quantitative considerations and possible embryological sequelae. Hum Reprod 200015(suppl 2)112–128. [DOI] [PubMed] [Google Scholar]
- 11.Marchington D R, Macaulay V, Hartshorne G M, Barlow D, Poulton J. Evidence from human oocytes for a genetic bottleneck in an mtDNA disease. Am J Hum Genet 199863769–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poulton J, Marchington D R. Segregation of mitochondrial DNA (mtDNA) in human oocytes and in animal models of mtDNA disease: clinical implications. Reproduction 2002123751–755. [DOI] [PubMed] [Google Scholar]
- 13.Blok R B, Gook D A, Thorburn D R, Dahl H H. Skewed segregation of the mtDNA nt 8993 (T→G) mutation in human oocytes. Am J Hum Genet 1997601495–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marchington D R, Hartshorne G M, Barlow D, Poulton J. Homopolymeric tract heteroplasmy in mtDNA from tissues and single oocytes: support for a genetic bottleneck. Am J Hum Genet 199760408–416. [PMC free article] [PubMed] [Google Scholar]
- 15.Chinnery P F, Thorburn D R, Samuels D C, White S L, Dahl H M, Turnbull D M, Lightowlers R N, Howell N. The inheritance of mitochondrial DNA heteroplasmy: random drift, selection or both? Trends Genet 200016500–505. [DOI] [PubMed] [Google Scholar]
- 16.Matthews P M, Hopkin J, Brown R M, Stephenson J B, Hilton‐Jones D, Brown G K. Comparison of the relative levels of the 3243 (A>G) mtDNA mutation in heteroplasmic adult and fetal tissues. J Med Genet 19943141–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardaioli E, Fabrizi G M, Grieco G S, Dotti M T, Federico A. Heteroplasmy of the A3243G transition of mitochondrial tRNALeu(UUR) in a MELAS case and in a 25‐week‐old miscarried fetus. J Neurol 2000247885–887. [DOI] [PubMed] [Google Scholar]
- 18.Chinnery P F, Zwijnenburg P J, Walker M, Howell N, Taylor R W, Lightowlers R N, Bindoff L, Turnbull D M. Nonrandom tissue distribution of mutant mtDNA. Am J Med Genet 199985498–501. [PubMed] [Google Scholar]
- 19.Goto Y, Nonaka I, Horai S. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 1990348651–653. [DOI] [PubMed] [Google Scholar]
- 20.Crimmins D, Morris J G, Walker G L, Sue C M, Byrne E, Stevens S, Jean‐Francis B, Yiannikas C, Pamphlett R. Mitochondrial encephalomyopathy: variable clinical expression within a single kindred. J Neurol Neurosurg Psychiatry 199356900–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van den Ouweland J M, Lemkes H H, Ruitenbeek W, Sandkuijl L A, de Vijlder M F, Struyvenberg P A, van de Kamp J J, Maassen J A. Mutation in mitochondrial tRNALeu(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat Genet 19921368–371. [DOI] [PubMed] [Google Scholar]
- 22.Suomalainen A, Syvanen A C. Quantitative analysis of human DNA sequences by PCR and solid‐phase minisequencing. Mol Biotechnol 200015123–131. [DOI] [PubMed] [Google Scholar]
- 23.McDonnell M T, Schaefer A M, Blakely E L, McFarland R, Chinnery P F, Turnbull D M, Taylor R W. Noninvasive diagnosis of the 3243A>G mitochondrial DNA mutation using urinary epithelial cells. Eur J Hum Genet 200412778–781. [DOI] [PubMed] [Google Scholar]
- 24.Shanske S, Pancrudo J, Kaufmann P, Engelstad K, Jhung S, Lu J, Naini A, DiMauro S, De Vivo D C. Varying loads of the mitochondrial DNA A3243G mutation in different tissues: implications for diagnosis. Am J Med Genet A 2004130134–137. [DOI] [PubMed] [Google Scholar]
- 25.t Hart L M, Jansen J J, Lemkes H H, Stolk R P, Feskens E J, Jansen J J, van der Does F E, Grobbee D E, Kromhout D, van den Ouweland J M.et al Heteroplasmy levels of a mitochondrial gene mutation associated with diabetes mellitus decrease in leucocyte DNA upon aging. Hum Mutat 19967193–197. [DOI] [PubMed] [Google Scholar]
- 26.Brown D T, Samuels D C, Michael E M, Turnbull D M, Chinnery P F. Random genetic drift determines the level of mutant mtDNA in human primary oocytes. Am J Hum Genet 200168533–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinborn R, Schinogl P, Zakhartchenko V, Achmann R, Schernthaner W, Stojkovic M, Wolf E, Muller M, Brem G. Mitochondrial DNA heteroplasmy in cloned cattle produced by fetal and adult cell cloning. Nat Genet 200025255–257. [DOI] [PubMed] [Google Scholar]
- 28.Battersby B J, Loredo‐Osti J C, Shoubridge E A. Nuclear genetic control of mitochondrial DNA segregation. Nat Genet 200333183–186. [DOI] [PubMed] [Google Scholar]
- 29.Thorburn D R, Dahl H H. Mitochondrial disorders: genetics, counseling, prenatal diagnosis and reproductive options. Am J Med Genet 2001106102–114. [DOI] [PubMed] [Google Scholar]
- 30.Dean N L, Battersby B J, Ao A, Gosden R G, Tan S L, Shoubridge E A, Molnar M J. Prospect of preimplantation genetic diagnosis for heritable mitochondrial DNA diseases. Mol Hum Reprod 20039631–638. [DOI] [PubMed] [Google Scholar]
- 31.Steffann J, Frydman N, Gigarel N, Burlet P, Ray P F, Fanchin R, Feyereisen E, Kerbrat V, Tachdjian G, Bonnefont J P, Frydman R, Munnich A. Analysis of mtDNA variant segregation during early human embryonic development: a tool for successful NARP preimplantation diagnosis. J Med Genet 200643244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]