Abstract
Background
A polymorphism in exon 4 (C77G) of CD45 that alters CD45 splicing has been associated with autoimmune and infectious diseases in humans.
Objective
To investigate the effect of C77G in hepatitis C virus (HCV) infected individuals and study the phenotype and function of peripheral blood mononuclear cells (PBMC) from healthy and hepatitis C infected C77G carriers.
Results
C77G individuals showed an increased proportion of primed CD45RA and effector memory CD8 T cells and more rapid activation of the lymphocyte specific protein tyrosine kinase (Lck) following CD3 stimulation. Transgenic mice with CD45 expression mimicking that in human C77G variants had more activated/memory T cells, more rapid proliferative responses, and activation of Lck.
Conclusions
Changes in CD45 isoform expression can alter immune function in human C77G variants and CD45 transgenic mice. The C77G allele may influence the outcome of HCV infection.
Keywords: CD45, C77G variant, hepatitis C, immune response
The leucocyte common (CD45) antigen is a transmembrane tyrosine phosphatase expressed on all nucleated haemopoietic cells.1,2 CD45 is essential for efficient T and B cell antigen receptor signal transduction, and Src family kinases are its primary targets. Multiple CD45 isoforms are generated by alternative splicing of exons 4 (A), 5 (B), and 6 (C) of the extracellular domain of the molecule. In man, naive T lymphocytes express high molecular weight (MW) isoforms containing the A exon (“CD45RA” cells) but following activation the low MW 180 kDa isoform is expressed (“CD45RO”cells) and maintained on the majority of primed (memory) cells.3 Although CD45 alternative splicing is highly conserved among vertebrates4 the function of different isoforms and the regulation of CD45 activity remain unclear.
Mutations in CD45 leading to failure of CD45 expression are a cause of severe combined immunodeficiency (SCID) in humans.5,6 More recently single nucleotide polymorphisms (SNPs) in CD45 that alter splicing have been described and are associated with infectious and autoimmune diseases.7,8,9,10,11,12 The exon 4 C77G transversion is a point mutation in a splice silencer region of the exon13,14 and individuals with C77G cannot splice out exon 4 normally so that their activated/memory lymphocytes co‐express CD45RA and CD45RO instead of CD45RO only. Although individuals heterozygous for the C77G allele are relatively rare (allele frequency from 0% to 3.5% in Europe and North America),8,9,15 the C77G variant is associated with multiple sclerosis in German7 and Italian16 studies but not in Swedish and North American cohorts.15,17 An increased frequency of C77G has been found in HIV infection,8 autoimmune hepatitis,18 systemic sclerosis,19 and Langerhans cell histiocytosis.11 No homozygotes have been reported.
Several other rare CD45 polymorphisms have been reported in Italy,20 and a new exon 4 (A54G) allele in Ugandan Africans (allele frequency 1.3%) is present with possibly increased frequency amongst HIV seropositive individuals.10 In contrast, the exon 6 A138G polymorphism is present at a high frequency (22.8%) in the Far East9 and promotes splicing towards low MW CD45 isoforms, resulting in an increased proportion of activated CD45RO cells and an increase in interferon γ (IFNγ) producing T cells. The A138G allele shows a protective effect in Graves' disease and hepatitis B virus infection.12
These studies indicate that CD45 is an immunomodulatory gene and that altered CD45 isoform expression can have profound effects on immune function, autoimmunity, and viral infections. Here we investigated the effect of C77G in HCV infected individuals and studied the phenotype and function of peripheral blood mononuclear cells (PBMC) from healthy and hepatitis C virus (HCV) infected C77G carriers. We also constructed a CD45RABC/+ transgenic (Tg) mouse model for C77G CD45 aberrant expression. Our results suggest that changes in CD45 isoform expression can alter immune function in human C77G carriers and CD45 Tg mice.
Methods
Materials
DNA or serum samples from 775 white patients with hepatitis C infection were obtained from the Trent Hepatitis C Cohort Study21 (369 patients) and from the HENCORE cohort (406 patients).22,23 In all, 616 patients were HCV RNA positive (persistently infected) and the remaining 159 were HCV RNA negative (resolvers). Among the 775 patients, 62% were male and 38% female. Fibrosis (mild <2 v severe >3) and necroinflammation (mild <5 v severe >5) were scored using Ishak's modification of the histology activity index. Information about interferon response (responders v non‐responders) was available on subsets of patients from the HENCORE cohort. None of the patients in the HENCORE cohort was co‐infected with HIV. There were no known HIV infected patients in the Trent cohort, although not all of them had been tested for HIV infection. As control we used 627 genomic DNA samples from healthy white British individuals (obtained from local blood donor banks (446 samples) or healthy donor volunteers (181 samples)), some of which have been described previously.8,9
Functional and phenotypic analysis was undertaken on cryopreserved PBMC from healthy C77G and control C77C individuals obtained from blood donor banks. C77G and C77C control hepatitis C PBMC samples were obtained from the Trent cohort. Age matched C77G and controls samples used in proliferative responses to mitogens and tetanus toxoid were between 20 and 58 years of age. The relevant local research ethics committees approved the collection of DNA samples and blood samples for functional experiments.
C77G genotyping
The C77G mutation was detected using alkaline mediated differential interaction (AMDI), which is based on the alkaline mediated detection of amplification refractory mutation system (ARMS)–polymerase chain reaction (PCR) products of two separate reaction mixes.9 The HENCORE samples were genotyped using the Sequenom Mass‐Array® MALDI‐TOF primer extension assay,24 (http://www.sequenom.de/) under standard conditions. Primers for C77G were P1: ACG TTG GAT GCT TTC AAG TGA CCC CTT ACC, P2: ACG TTG GAT GGT TGT GGT CTC TGA GAA GTC and extension P: CCA CTG CAT TCT CAC C. Sequenom and AMDI gave identical C77G frequencies.
CD45RABC/+ transgenic mice
We have previously described mice expressing a CD45RABC transgene on a CD45−/− background.25 We generated CD45RABC/+ mice by breeding single isoform CD45RABC Tg mice on a CD45−/− background with C57Bl/6 mice. The presence of the CD45RABC transgene in the F1 crosses was detected by PCR on tail genomic DNA, using forward 5′‐GAG CTC AGA ATC AAA AGA GGA‐3′ and reverse 5′‐TAA TTC ACA GTA ATG TTC CCA AAC ATG GC ‐3′ primers generating a 1000 base pair (bp) product. All mice were bred in the specific‐pathogen‐free facilities of the Institute for Animal Health, Compton, UK and used for experiments at six to eight weeks of age unless otherwise stated. All experiments fully complied with relevant Home Office guidelines and were approved by the animal ethics committee of the Institute for Animal Health.
Flow cytometric analysis
Surface phenotypic analysis was carried out on cryopreserved PBMC from healthy C77G carriers and C77C controls: 2×105 PBMC were stained with either allophycocyanine (APC) conjugated CD4 (S3.3, Caltag, Silverstone, UK) or CD8‐APC (clone RPA/T8), along with fluorescein isothiocyanate (FITC) conjugated CD45RA (clone HI100) and phycoerythrin (PE) conjugated CD45R0 (clone UCHL1) monoclonal antibodies (mAbs) in a single step at 4°C for 20 minutes, and washed with phosphate buffered saline (PBS) containing 0.2% bovine serum albumin. The following reagents and antibodies were also used to stain cell suspensions: CD11a‐FITC (G43‐25B), CD28‐FITC (CD28.2), CD62L‐FITC (Dreg56), CD69‐FITC, CCR7 (2H4). Murine lymphocytes were stained with CD4‐FITC (GK1.5), CD8‐PerCP(53‐6.7), CD44‐PE (IM7), and CD122‐PE (Tm‐β1) all from BD Biosciences, Oxford, UK, and CD62‐L (Mel‐14) from Caltag, Silvertone, UK.
Proliferative responses of human PBMC and murine lymph node cells
Human PBMC were plated at 2×105cells/well into 96‐well flat bottom plates and stimulated with anti‐CD3 (clone UCHT1‐2a, at varying concentrations), PMA (50 ng/ml) and ionomycin (0.5 μg/ml) or 5 μg/ml tetanus toxoid. Lymph node cells of CD45+/+, CD45+/−, and CD45RABC/+ mice were plated at 2×105 cells/well into round bottom 96‐well plates. For TCR‐CD3 cross linking, the plates were coated overnight at 4°C with varying concentrations (from 0 to 10 μg/well) of anti‐mouse CD3ε (clone 145‐2C11, BD Biosciences) in the presence or absence of 2 μg/well of CD28 (clone 37.51, BD Biosciences) and washed three times before incubation with the T cells. Cells were harvested at various intervals (24 to 148 hours) after a 12 hour pulse with 1 μCi of [3H] thymidine per well. Early activation was also assessed following CD3 stimulation by harvesting cells at one to five hours and staining with CD69 antibody.
Western blot analysis
Human PBMC or murine lymph node cells were stimulated with appropriate anti‐CD3 antibodies at 2 μg/ml for varying times. For total protein extraction, cell lysis (2×106 cells/ml) was undertaken in RIPA (radio‐immunoprecipitation assay) buffer (PBS, pH 7.4, 1% NP‐40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate (SDS),10 mM NaF, and 0.5 mM phenylmethanesulphonyl fluoride (PMSF)), containing protease and phosphatase inhibitors (protease inhibitor cocktail and phosphatase inhibitor cocktail 2; Sigma‐Aldrich, Poole, UK), on ice at 4°C for 30 minutes. Insoluble materials were removed by centrifugation (15 000×g for 10 minutes at 4°C) and the total protein concentration was quantified by BCA protein assay (Pierce Biotechnology, Rockford, Illinois, USA). Cell lysate proteins were analysed by 10% SDS‐PAGE (10 μg of total protein per lane), transferred onto a nitrocellulose membrane, and immunoblotted using antibodies specific for lymphocyte specific protein tyrosine kinase (Lck) (Upsate Biotechnology Inc, New York, USA) or pY505Lck (Signalling Technologies, Beverly, Massachusetts, USA). Immunoblotting with monoclonal anti‐β‐Actin antibody (Sigma) was used as a loading control. Blots were developed using ECL™ donkey anti‐rabbit HRP linked F(ab′)2 fragment and ECL™ detection reagents (Amersham Biosciences, Amersham, UK).
Statistical analysis
The distribution of genotype and allele frequencies was analysed using standard χ2 or Fisher exact tests, applying SPSS v.12.0. A value of p<0.05 was considered significant. The odds ratio (OR) was calculated to indicate the risk associated and presented with 95% confidence intervals (CI).
Results
Frequency of the C77G variant in hepatitis C
As we have previously shown an increased frequency of C77G individuals among HIV patients, we analysed the frequency of the C77G allele in a cohort of 775 white patients with HCV infection and found 28 C77G heterozygotes (allele frequency, 1.8%)—twice as many as in the healthy UK control population (0.9%, p = 0.035, OR = 2.1 (95% CI, 0.99 to 4.52)) (table 1). No G77G homozygotes were found in any group. We next compared the frequency of C77G in patients with chronic HCV infection (that is, HCV RNA positive) with resolvers (anti‐HCV positive, HCV RNA negative). We found 25 C77G heterozygotes among 616 chronic carriers (allele frequency, 2.0%) compared with three of 159 in the resolver group (allele frequency 0.9%), although this difference was not significant. However, more patients carrying the G allele presented with severe fibrosis (7 of 67) compared with those with mild fibrosis (8 of 294), which was statistically significant (p = 0.01: OR = 4.2 (95% CI, 1.3 to 13.3)). Neither the severity of necroinflammation nor the response to interferon treatment was affected by the presence of C77G. Nevertheless these results show a significant effect of the C77G allele on HCV incidence and the severity of the disease.
Table 1 Frequency of CD45 exon 4 C77G polymorphism in control and hepatitis C infected individuals.
| Total | C77C (%) | C77G (%) | p Value/ | OR (95% CI) | |
|---|---|---|---|---|---|
| number | F‐exact | ||||
| Healthy controls | 627 | 626 (99.1) | 11 (0.9) | 0.035 | 2.1 |
| HCV patients | 775 | 747 (98.2) | 28 (1.8) | (1.0 to 4.5) | |
| Chronic HCV | 616 | 591 (98.0) | 25 (2.0) | NS | NS |
| Resolved HCV | 159 | 156 (99.1) | 3 (0.9) | ||
| Fibrosis | |||||
| Mild (<2) | 294 | 286 (97.3) | 8 (2.7) | 0.01 | 4.1 |
| Severe (>3) | 67 | 60 (96.4) | 7 (10.5) | (1.3 to 13.3) | |
| Necroinflammation | |||||
| Mild (<5) | 250 | 241 (96.4) | 9 (3.6) | NS | NS |
| Severe (>5) | 128 | 120 (93.7) | 8 (6.3) | ||
| IFN response | |||||
| Responders | 164 | 160 (97.6) | 4 (2.4) | NS | NS |
| Non‐responders | 45 | 42 (93.3) | 3 (6.7) |
CI, confidence interval; HCV, hepatitis C virus; IFN, interferon; OR, odds ratio.
Phenotypic analysis of PBMC in C77G individuals
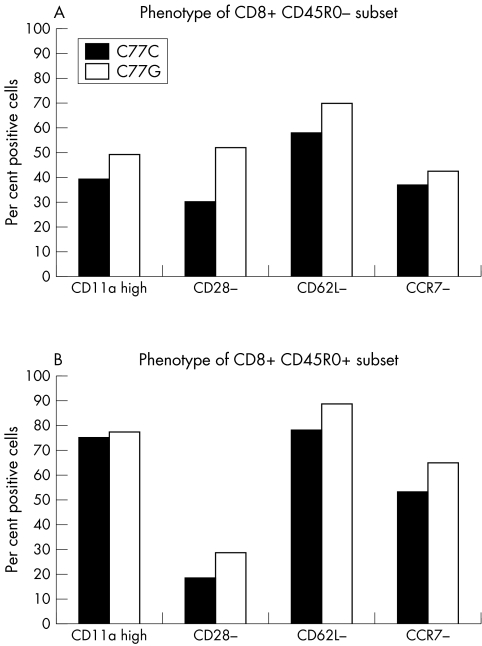
CD45RA and CD45R0 expression defines naïve and memory T cell subsets.3 The proportions of these subsets are altered in C77G individuals, and more CD8 cells express high levels of CD11a and less CD62L.11 Analysis of PBMC from 10 healthy C77G heterozygotes and C77C controls samples showed an increased proportion of single positive CD45RA+ T cells in C77G individuals,11 more apparent among CD8 than CD4 cells (mean of 78.2% single CD8+CD45RA+ cells compared with 44.3% in controls) (p = 0.001) (fig 1). Because of this, we analysed the expression of CD11a, CD28, CD62L, and CCR7 in the CD45RO+ and CD45RO− subsets of CD8 cells. C77G individuals have an increased proportion of CD28−, CD62L−, and CCR7−, and more CD11hi cells, indicating an expansion of the highly differentiated primed CD8+CD45RA+ subset26 (fig 2A). There is also an increase in the proportion of effector memory cells in the CD8+CD45RO+ subset (CD11ahi+, CD28−, CD62L−, and CCR7−) (fig 2B).
Figure 1 Flow cytometric analysis of C77G variant individuals. Peripheral blood mononuclear cells were stained with CD45RO‐PE, CD45RA‐FITC, and either CD4 or CD8 allophycocyanin (APC) antibodies before (A) and after (B) stimulation with phytohaemagglutinin for 10 days. Analysis was carried out on CD4 or CD8 gated cells. Normal expression is characterised by the loss of CD45RA and gain of CD45RO expression upon activation. Variant C77G expression is characterised by the absence of a single CD45RO+ population. Even after stimulation for 10 days cells remained double CD45RA+/CD45RO+.
Figure 2 Expression of activation markers on CD8 cells from 10 healthy C77G and 10 C77C control individuals. Proportions of cells that are CD11ahi, CD28−, CD62L−, and CCR7− in the CD8+CD45RO− (A) and CD8+CD45RO+ (B) cell subsets are shown. Although there is a trend, none of the differences is statistically significant.
Thus the most prominent effect in C77G individuals is an increase in the proportion of single positive CD45RA+ primed CD8 T cells.
Proliferative and signalling responses of PBMC in C77G individuals
In order to determine whether the aberrant CD45 isoform expression in C77G (and the presence of more activated T cells) influences the function of PBMC, we analysed proliferative responses and early phosphorylation events following T cell receptor (TcR) stimulation. No differences in the response of PBMC to CD3 or PMA (phorbol 12‐myristate 13‐acetate) and ionomycin were seen between C77G and controls (data not shown). As C77G individuals lack the (classical) single positive memory CD45RA−CD45RO+ cell subset, we analysed the responses to tetanus toxoid. No differences between the C77G and control groups were detected (fig 3A).
Figure 3 Functional responses by peripheral blood mononuclear cells (PBMC) from C77G and control C77C individuals. (A) Mean and standard errors of proliferative responses of 10 C77G and 10 control C77C individuals to 5 μg/ml of tetanus toxoid at various time points. Western blot analysis of lymphocyte specific protein tyrosine kinase (Lck) and pY505Lck of healthy (B) and hepatitis C infected (C) PBMC stimulated with 2 μg/ml CD3 for the times indicated. An equal amount of cell lysate protein (10 μg) was loaded on to each lane. The ratio of pLck to Lck at each time point is shown. Data are representative of five healthy C77G, five healthy C77C, three hepatitis C infected C77G, and three hepatitis control C77C PBMC. (D) Kinetics of CD69 appearance on gated CD4 and CD8 cells in two healthy C77C controls, two hepatitis C C77C, and two C77G patients.
Although no differences in proliferative responses of PBMC were detected, in agreement with previous studies,27 we analysed early signalling events following CD3 stimulation. The dominant role of CD45 is to dephosphorylate the inhibitory tyrosine Y505 of Lck. As shown in fig 3B, pY505Lck in C77G cells shows more rapid and greater dephosphorylation at 0.5 and 1.0 minute following CD3 stimulation (ratio of pLck/Lck 0.54 and 0.66 in C77G v 1.0 and 0.92 in control cells), suggesting increased CD45 phosphatase activity in C77G cells compared with C77C controls. However, this rapid activation of Lck was not seen in three C77G and three C77C control hepatitis C patients (fig 3C), probably because of the well known global immunosuppressive effect of hepatitis C.28 Reduced expression of the early activation marker CD69 following CD3 stimulation of PBMC from hepatitis C C77G and C77C patients compared with healthy controls supports this interpretation (fig 3D).
Thus the only functional difference we have detected to date between C77G and C77C individuals is the more rapid dephosphorylation of pY505Lck in healthy C77G subjects.
CD45RABC/+ Tg mice as a model for the human C77G variant
C77G individuals cannot splice out the fourth (A) exon, and their lymphocytes always co‐express exon‐A‐containing CD45 isoforms. We therefore constructed mice with one splicable wild type CD45 allele and a non‐splicing CD45RABC Tg. In these CD45RABC/+ mice all lymphocytes express CD45RABC together with the CD45 isoforms generated by alternative splicing from the wild type allele, mimicking the pattern of expression in C77G heterozygotes.
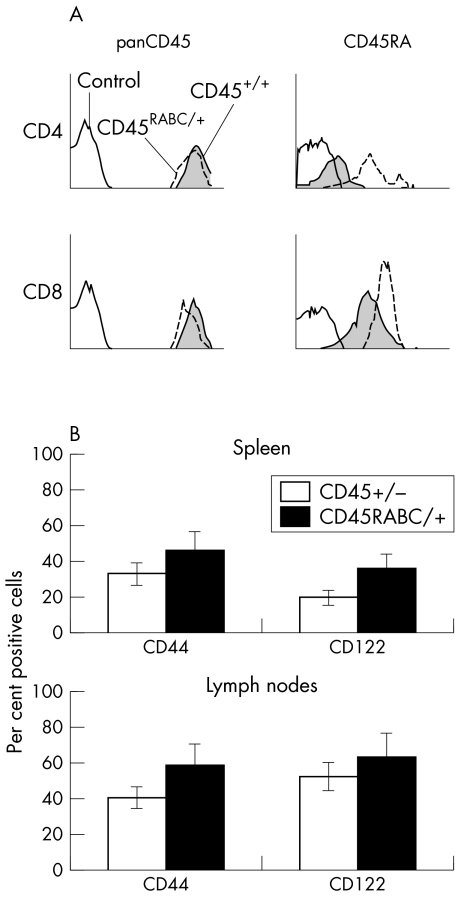
Total CD45 expression on CD4 and CD8 lymphocytes is very similar in CD45+/+ and CD45RABC/+ mice, and these mice have similar numbers of T cells and an identical CD4 to CD8 ratio to CD45+/+ mice (fig 4A). As expected, the CD45RABC/+ mice expressed more A exon, particularly on CD4 cells. The effect on CD4 cells is presumably more pronounced because wild type CD4 cells express very low levels of CD45 RA. However, as there are more highly differentiated T cells among CD8 than CD4 cells in C77G humans, we examined the expression of CD44 and CD122 as markers for activated/memory CD8 cells in CD45RABC/+ mice. Both lymph node and spleen showed an increased (∼20%) proportion of cells expressing CD44 and CD122 compared with control CD45+/+ mice (fig 4B). No change in the proportion of memory/activated CD4 T cells lacking CD62L was detected (data not shown).
Figure 4 Phenotypic analysis in CD45RABC/+ mice. (A) CD45 expression in CD45+/+ and CD45RABC/+ mice. Lymph node cells were stained with pan CD45 or exon specific antibodies against CD45RA, together with CD4 and CD8. Analysis was undertaken on CD4 or CD8 gated cells. The shaded histogram is CD45+/+, the dotted line CD45RABC/+, and the solid line isotype control. (B) CD44 and CD122 expression in CD45+/+ and CD45RABC/+ mice. Lymph node and spleen cells were stained with antibodies against CD44 and CD122 together with CD3, and analysis undertaken on CD3 gated cells. Examples are representative of three mice of each type.
Lymphocytes from peripheral and mesenteric lymph nodes of CD45+/+, CD45+/−, and CD45RABC/+ mice were activated with CD3. An increased proliferative response was detected in CD45RABC/+ Tg at 24 and 48 hours, followed by a reduction at 96 hours compared with CD45+/+ or CD45+/− mice (fig 5A). Following CD3 stimulation there is greater dephosphorylation of pY505Lck at 0.5 and 1.0 minutes in CD45RABC/+ than in CD45+/+ cells (fig 5B). The increased dephosphorylation correlates with the brisk initial proliferative response and suggests increased CD45 phosphatase activity in the CD45RABC/+ mice.
Figure 5 Activation of T cells. (A) Mesenteric and peripheral lymph node cells from CD45+/+, CD45+/−, and CD45RABC/+ mice were activated in the presence of varying amounts of plate bound CD3 and 1 μg/ml of CD28 antibodies. Cells were harvested at 24, 48, 72, and 96 hours after a 12 hour pulse with [3H]‐thymidine. Means and standard deviations of triplicate cultures from three mice are shown. Background counts were less than 500 cpm and have been subtracted. Data are representative of five experiments. (B) Western blot of Lck and pY505Lck of lymph node T cells stimulated with 2 μg/ml CD3 for the times indicated. An equal amount of cell lysate protein (10 μg) was loaded on each lane. Data are representative of three experiments.
Taken together these results suggest that changing the balance of CD45 isoforms in CD45RABC/+ mice alters the phenotype of CD8 cells, the proliferative responses, and Lck activation, as in human CD45 variants.
Discussion
HCV infected individuals show an increased frequency of C77G carriers compared with healthy controls (p = 0.035, OR = 2.1). The presence of C77G also influences the severity of disease, as there are more C77G carriers among patients with severe fibrosis. In the light of these observations we tried to understand how altered CD45 expression in C77G influences immune responses and disease. Our results indicate that lymphocytes from C77G carriers have an altered phenotype, particularly of CD8 cells. Although they lack single positive CD45RA−CD45RO+ memory cells, they have an increased proportion and absolute number of single positive primed CD8+CD45RA+ cells (CD11hi, CD28−, CD62L−, CCR7−). Development of this subset of CD8 cells has been associated with CMV seroconversion29 and in elderly individuals it contains large clones of CMV specific cells,30,31 although these may be functionally compromised.32,33 Less effective anti‐viral responses in C77G individuals might lead to an increase in the CD8+CD45RA+ primed subset. Whether this is the cause of the increased number of activated/memory CD8+ cells in CD45RABC/+ mice remains to be investigated.
In both C77G humans and CD45RABC/+ mice, there is more rapid dephosphorylation of the inhibitory Y505Lck, suggesting a more vigorous initial response to TcR stimulation. The apparent paradox of a more vigorous response and increased disease severity might be because a non‐protective immune response is generated (for example, Th2 rather than Th1) or because the more intense immune response contributes to more severe fibrosis. A more vigorous response and lowered TcR threshold for activation may also lead to increased autoreactivity and underlie the association of C77G with multiple sclerosis or other autoimmune diseases. CD45RABC/+ Tg T cells show similar increased dephosphorylation of Lck and interestingly these mice have increased susceptibility to experimental autoimmune encephalomyelitis. Disease progression in HIV has also been associated with immune activation, but whether this is a cause or consequence of progression is not clear.
Proliferative responses of C77G and C77C PBMC to CD3 or tetanus toxoid do not differ, in agreement with earlier studies.27 However alloreactive T cell lines from C77G carriers show enhanced proliferation and increased interleukin 2 production following stimulation with CD3 or antigen.34 Murine CD45RABC/+ Tg T cells also show more rapid and vigorous proliferative responses. This discrepancy may be because only half of the C77G PBMCs have an abnormal CD45RA/RO phenotype while all of the alloreactive cells are CD45RA/RO and all the transgenic cells express CD45RA as well as other isoforms.
Although phenotypic changes in CD8 T cells are prominent in C77G individuals, effects on HCV susceptibility and disease progression may be caused by altered function of other cell types such as dendritic cells, B cells, or NK cells. The latter have been shown to be involved in HCV susceptibility.35 The fact that disease susceptibility as well as progression may be affected supports the idea that early (innate) immune responses might affected by C77G.
In summary C77G individuals show increased susceptibility and more severe fibrosis following HCV infection. Healthy C77G individuals have an altered T cell phenotype and earlier Lck dephosphorylation following TcR stimulation. It is intriguing that the only common CD45 variant allele described, exon 6 A138G, has a protective effect in hepatitis C in the Far East (unpublished data). These two polymorphisms in exon 4 (C77G) and exon 6 (A138G) therefore have contrasting phenotypes and disease associations. The exon 4 77G allele causes increased CD45RA expression and is present at a higher frequency in autoimmune and infectious diseases such as HIV and HCV, as reported here. In contrast the exon 6 138G variant promotes splicing towards low molecular weight isoforms (CD45RO) and is associated with protection against Graves' disease, hepatitis B, and hepatitis C. Transgenic mice expressing a modified combination of CD45 isoforms also show altered immune function. These data provide convincing evidence that CD45 expression patterns can have profound effects on immune function and human diseases.
Acknowledgements
We thank the Trent and HENCORE Study Groups for providing the samples. We are also grateful to Cathy De Lara, Vjera Pandol‐Kaljevic, Yan Zhang, and Hala Ghattas for assistance in recruiting subjects for functional analysis.
Abbreviations
HCV - hepatitis C virus
HENCORE - Hepatitis C European Network for Cooperative Research
Lck - lymphocyte specific protein tyrosine kinase
PBMC - peripheral blood mononuclear cells
SNP - single nucleotide polymorphism
TcR - T cell receptor
Footnotes
Conflicts of interest: none declared
References
- 1.Hermiston M L, Xu Z, Weiss A. CD45: a critical regulator of signaling thresholds in immune cells. Annu Rev Immunol 200321107–137. [DOI] [PubMed] [Google Scholar]
- 2.Penninger J M, Irie‐Sasaki J, Sasaki T, Oliveira‐dos‐Santos A J. CD45: new jobs for an old acquaintance. Nat Immunol 20012389–396. [DOI] [PubMed] [Google Scholar]
- 3.Akbar A N, Terry L, Timms A, Beverley P C, Janossy G. Loss of CD45R and gain of UCHL1 reactivity is a feature of primed T cells. J Immunol 19881402171–2178. [PubMed] [Google Scholar]
- 4.Okumura M, Matthews R J, Robb B, Litman G W, Bork P, Thomas M L. Comparison of CD45 extracellular domain sequences from divergent vertebrate species suggests the conservation of three fibronectin type III domains. J Immunol 19961571569–1575. [PubMed] [Google Scholar]
- 5.Kung C, Pingel J T, Heikinheimo M, Klemola T, Varkila K, Yoo L I, Vuopala K, Poyhonen M, Uhari M, Rogers M, Speck S H, Chatila T, Thomas M L. Mutations in the tyrosine phosphatase CD45 gene in a child with severe combined immunodeficiency disease. Nat Med 20006343–345. [DOI] [PubMed] [Google Scholar]
- 6.Tchilian E Z, Wallace D L, Wells R S, Flower D R, Morgan G, Beverley P C. A deletion in the gene encoding the CD45 antigen in a patient with SCID. J Immunol 20011661308–1313. [DOI] [PubMed] [Google Scholar]
- 7.Jacobsen M, Schweer D, Ziegler A, Gaber R, Schock S, Schwinzer R, Wonigeit K, Lindert R B, Kantarci O, Schaefer‐Klein J, Schipper H I, Oertel W H, Heidenreich F, Weinshenker B G, Sommer N, Hemmer B. A point mutation in PTPRC is associated with the development of multiple sclerosis. Nat Genet 200026495–499. [DOI] [PubMed] [Google Scholar]
- 8.Tchilian E Z, Wallace D L, Dawes R, Imami N, Burton C, Gotch F, Beverley P C. A point mutation in CD45 may be associated with HIV‐1 infection. AIDS 2001151892–1894. [DOI] [PubMed] [Google Scholar]
- 9.Stanton T, Boxall S, Hirai K, Dawes R, Tonks S, Yasui T, Kanaoka Y, Yuldasheva N, Ishiko O, Bodmer W, Beverley P C, Tchilian E Z. A high‐frequency polymorphism in exon 6 of the CD45 tyrosine phosphatase gene (PTPRC) resulting in altered isoform expression. Proc Natl Acad Sci USA 20031005997–6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanton T, Boxall S, Bennett A, Kaleebu P, Watera C, Whitworth J, French N, Dawes R, Hill A V, Bodmer W, Beverley P C, Tchilian E Z. CD45 variant alleles: possibly increased frequency of a novel exon 4 CD45 polymorphism in HIV seropositive Ugandans. Immunogenetics 200456107–110. [DOI] [PubMed] [Google Scholar]
- 11.Boxall S, McCormick J, Beverley P, Strobel S, De Filippi P, Dawes R, Klersy C, Clementi R, De Juli E, Ferster A, Wallace D, Arico M, Danesino C, Tchilian E. Abnormal cell surface antigen expression in individuals with variant CD45 splicing and histiocytosis. Pediatr Res 200455478–484. [DOI] [PubMed] [Google Scholar]
- 12.Boxall S, Stanton T, Hirai K, Ward V, Yasui T, Tahara H, Tamori A, Nishiguchi S, Shiomi S, Ishiko O, Inaba M, Nishizawa Y, Dawes R, Bodmer W, Beverley P C, Tchilian E Z. Disease associations and altered immune function in CD45 138G variant carriers. Hum Mol Genet 2004132377–2384. [DOI] [PubMed] [Google Scholar]
- 13.Thude H, Hundrieser J, Wonigeit K, Schwinzer R. A point mutation in the human CD45 gene associated with defective splicing of exon A. Eur J Immunol 1995252101–2106. [DOI] [PubMed] [Google Scholar]
- 14.Lynch K W, Weiss A. A CD45 polymorphism associated with multiple sclerosis disrupts an exonic splicing silencer. J Biol Chem 200127624341–24347. [DOI] [PubMed] [Google Scholar]
- 15.Barcellos L F, Caillier S, Dragone L, Elder M, Vittinghoff E, Bucher P, Lincoln R R, Pericak‐Vance M, Haines J L, Weiss A, Hauser S L, Oksenberg J R. PTPRC (CD45) is not associated with the development of multiple sclerosis in US patients. Nat Genet 20012923–24. [DOI] [PubMed] [Google Scholar]
- 16.Ballerini C, Rosati E, Salvetti M, Ristori G, Cannoni S, Biagioli T, Massacesi L, Sorbi S, Vergelli M. Protein tyrosine phosphatase receptor‐type C exon 4 gene mutation distribution in an Italian multiple sclerosis population. Neurosci Lett 2002328325–327. [DOI] [PubMed] [Google Scholar]
- 17.Vorechovsky I, Kralovicova J, Tchilian E, Masterman T, Zhang Z, Ferry B, Misbah S, Chapel H, Webster D, Hellgren D, Anvret M, Hillert J, Hammarstrom L, Beverley P C. Does 77C→G in PTPRC modify autoimmune disorders linked to the major histocompatibility locus? Nat Genet 20012922–23. [DOI] [PubMed] [Google Scholar]
- 18.Vogel A, Strassburg C P, Manns M P. 77 C/G mutation in the tyrosine phosphatase CD45 gene and autoimmune hepatitis: evidence for a genetic link. Genes Immun 2003479–81. [DOI] [PubMed] [Google Scholar]
- 19.Schwinzer R, Witte T, Hundrieser J, Ehlers S, Momot T, Hunzelmann N, Krieg T, Schmidt R E, Wonigeit K. Enhanced frequency of a PTPRC (CD45) exon A mutation (77C→G) in systemic sclerosis. Genes Immun 20034168–169. [DOI] [PubMed] [Google Scholar]
- 20.Gomez‐Lira M, Liguori M, Magnani C, Bonamini D, Salviati A, Leone M, Andreoli V, Trojano M, Valentino P, Savettieri G, Quattrone A, Pignatti P F, Momigliano‐Richiardi P, Giordano M. CD45 and multiple sclerosis: the exon 4 C77G polymorphism (additional studies and meta‐analysis) and new markers. J Neuroimmunol 2003140216–221. [DOI] [PubMed] [Google Scholar]
- 21.Mohsen A H, Group T H. The epidemiology of hepatitis C in a UK health regional population of 5.12 million. Gut 200148707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hennig B J, Hellier S, Frodsham A J, Zhang L, Klenerman P, Knapp S, Wright M, Thomas H C, Thursz M, Hill A V. Association of low‐density lipoprotein receptor polymorphisms and outcome of hepatitis C infection. Genes Immun 20023359–367. [DOI] [PubMed] [Google Scholar]
- 23.Thursz M, Yallop R, Goldin R, Trepo C, Thomas H C. Influence of MHC class II genotype on outcome of infection with hepatitis C virus. The HENCORE group. Hepatitis C European Network for Cooperative Research. Lancet 19993542119–2124. [DOI] [PubMed] [Google Scholar]
- 24.Jurinke C, van den Boom D, Cantor C R, Koster H. The use of MassARRAY technology for high throughput genotyping. Adv Biochem Eng Biotechnol 20027757–74. [DOI] [PubMed] [Google Scholar]
- 25.Tchilian E Z, Dawes R, Hyland L, Montoya M, Le Bon A, Borrow P, Hou S, Tough D, Beverley P C. Altered CD45 isoform expression affects lymphocyte function in CD45 Tg mice. Int Immunol 2004161323–1332. [DOI] [PubMed] [Google Scholar]
- 26.Hamann D, Kostense S, Wolthers K C, Otto S A, Baars P A, Miedema F, van Lier R A. Evidence that human CD8+CD45RA+CD27− cells are induced by antigen and evolve through extensive rounds of division. Int Immunol 1999111027–1033. [DOI] [PubMed] [Google Scholar]
- 27.Schwinzer R, Schraven B, Kyas U, Meuer S C, Wonigeit K. Phenotypical and biochemical characterisation of a variant CD45R expression pattern in human leucocytes. Eur J Immunol 1992221095–1098. [DOI] [PubMed] [Google Scholar]
- 28.Lucas M, Vargas‐Cuero A L, Lauer G M, Barnes E, Willberg C B, Semmo N, Walker B D, Phillips R, Klenerman P. Pervasive influence of hepatitis C virus on the phenotype of antiviral CD8+ T cells. J Immunol 20041721744–1753. [DOI] [PubMed] [Google Scholar]
- 29.Kuijpers T W, Vossen M T, Gent M R, Davin J C, Roos M T, Wertheim‐van Dillen P M, Weel J F, Baars P A, van Lier R A. Frequencies of circulating cytolytic, CD45RA+CD27−, CD8+ T lymphocytes depend on infection with CMV. J Immunol 20031704342–4348. [DOI] [PubMed] [Google Scholar]
- 30.Ouyang Q, Wagner W M, Walter S, Muller C A, Wikby A, Aubert G, Klatt T, Stevanovic S, Dodi T, Pawelec G. An age‐related increase in the number of CD8+ T cells carrying receptors for an immunodominant Epstein‐Barr virus (EBV) epitope is counteracted by a decreased frequency of their antigen‐specific responsiveness. Mech Ageing Dev 2003124477–485. [DOI] [PubMed] [Google Scholar]
- 31.Khan N, Shariff N, Cobbold M, Bruton R, Ainsworth J A, Sinclair A J, Nayak L, Moss P A. Cytomegalovirus seropositivity drives the CD8 T cell repertoire toward greater clonality in healthy elderly individuals. J Immunol 20021691984–1992. [DOI] [PubMed] [Google Scholar]
- 32.Ouyang Q, Wagner W M, Wikby A, Walter S, Aubert G, Dodi A I, Travers P, Pawelec G. Large numbers of dysfunctional CD8+ T lymphocytes bearing receptors for a single dominant CMV epitope in the very old. J Clin Immunol 200323247–257. [DOI] [PubMed] [Google Scholar]
- 33.Ouyang Q, Wagner W M, Zheng W, Wikby A, Remarque E J, Pawelec G. Dysfunctional CMV‐specific CD8(+) T cells accumulate in the elderly. Exp Gerontol 200439607–613. [DOI] [PubMed] [Google Scholar]
- 34.Do H T, Baars W, Schwinzer R. Functional significance of the 77C→G polymorphism in the human CD45 gene: enhanced T‐cell reactivity by variantly expressed CD45RA isoforms. Transplant Proc 20053751–52. [DOI] [PubMed] [Google Scholar]
- 35.Khakoo S I, Thio C L, Martin M P, Brooks C R, Gao X, Astemborski J, Cheng J, Goedert J J, Vlahov D, Hilgartner M, Cox S, Little A M, Alexander G J, Cramp M E, O'Brien S J, Rosenberg W M, Thomas D L, Carrington M. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science 2004305872–874. [DOI] [PubMed] [Google Scholar]