Abstract
Background
The genetic contribution to pain sensitivity underlies a complex composite of parallel pain pathways, multiple mechanisms, and diverse inter‐individual pain experiences and expectations.
Methods
Variations for genes encoding receptors related to cold and heat sensation, such as transient receptor potential A subtype 1 (TRPA1), M subtype 8 (TRPM8), V subtype 1 (TRPV1), δ opioid receptor subtype 1 (OPRD1), catechol O‐methyltransferase (COMT), and fatty acid amide hydrolyase (FAAH), were investigated in four major ethnic populations.
Results
We defined 13 haplotype blocks in European Americans, seven blocks in African Americans, seven blocks in Hispanic subjects, and 11 blocks in Asian Americans. Further study in European American subjects found significant associations between short duration cold pain sensitivity and variations in TRPA1, COMT, and FAAH in a gender dependent manner. Our observations demonstrate that genetic variations in TRPA1, COMT, and FAAH contribute gender specifically to individual variations in short duration cold pain sensitivity in a European American cohort.
Conclusions
The effects of TRPA1 variations on experimental short duration heat pain sensitivity may contribute to inter‐individual variation in pain sensitivity in humans.
Keywords: cold pain, experimental pain, haplotype, thermal pain
Considering moderate heritability estimates, multiple pain mechanisms, complex networks of neural circuits and pain related molecules and environmental factors, ascertaining the genetic contribution to the individual variance in pain sensitivity is a complex problem. The contribution of each gene likely is a subtle effect on this multiplicity of mechanisms, making its signal difficult to detect. Even though the individual gene effects may be small, interactions among the genes and environment may make a substantial contribution to the final manifestation of pain sensitivity. In spite of recent technological progress, it is still necessary to choose candidate genes within the identified regions based on their biological role1 and examine them for informative polymorphisms. These can range from polymorphisms resulting in an amino acid change, alterations affecting the stability, splicing, or localisation of the mRNA, and regulatory elements for gene expression in upstream regions or in introns or intron derived microRNAs.
Transient receptor potential (TRP) channels are the vanguard of sensory systems, responding to temperature, touch, pain, osmolarity, pheromones, taste, and other stimuli. Unlike most ion channels, TRP channels are identified by their homology rather than by ligand function or selectivity, because their functions are disparate and often unknown. TRP subfamily A member 1 (TRPA1), also known as ankyrin‐like protein with transmembrane domains 1 (ANKTM1), is activated by noxious cold temperature (<15°C),2 although this observation has not been reproduced by others. Among the TRP subfamily M, TRPM member 8 is a non‐selective, outwardly rectifying channel that can be activated by cold (8–28°C).3 Another TRP subfamily V has been proposed to mediate warm and noxious heat sensation with TRP subfamily V member 1 (TRPV1) affecting responses to heat stimuli greater than 43°C.4 In addition to the flexible temperature range of activation,5 response to chemical or mechanical stimuli and heteromultimerisation of many TRP subtypes6 contribute to their complicated interactions in many sensory pathways.
The expression of opioid receptors in skin sensory nerve fibres suggests their potential contribution to direct elicitation of neurogenic inflammation, pain, and pruritus in the skin. Several findings have directly implicated genetic variation in opioid function in pain sensitivity. Basal nociceptive sensitivity is correlated to δ opioid analgesic sensitivity in animals, implying genetic co‐determination.7 Genetic linkage mapping for thermal nociception in mice implicates Oprd1, which encodes the murine δ opioid receptor.7 Murine Oprd1 is orthologous to the human δ opioid receptor subtype 1 (OPRD1) and has been proposed to affect responses to heat stimuli.7,8
Catechol O‐methyltransferase is one of the two enzymes along with monoamine oxidase that account for the initial steps in the elimination of catecholamines. The catechol O‐methyltransferase gene (COMT) contains a common functional polymorphism, COMT G1947A, also known as COMT Val158Met, which substitutes from valine to methionine at amino acid position 158 (or 108 of S‐COMT) and markedly reduces enzyme activity to approximately 20–40% of wild type levels.9,10,11 This amino acid change may regulate the amounts of active dopamine and norepinephrine in various parts of the brain and therefore may be associated with mood and other mental processes.12 The Met158 allele is reported to increase pain report and to decrease brain opioid system activation after an experimental pain challenge involving infusion of hypertonic saline into the masseter muscle.13 Haplotypes including COMT Val158Met were recently identified and an association suggested with experimental pain sensitivity and a chronic pain condition.14
Fatty acid hydrolase (FAAH) is an enzyme that terminates the endogenous signalling activity of a large class of fatty acid amides. Animal studies indicate that FAAH serves as the primary catabolic regulator of fatty acid amide in the nervous system and mice lacking the FAAH gene (FAAH) exhibit less pain responses to thermal stimuli.15 Reduced cellular expression of the enzyme by the common natural mutant FAAH Pro129Thr in human lymphocytes suggests significant functional variability exists in the endocannabinoid system in humans which may influence pain sensitivity.16
We have examined the effects of the human TRPA1, TRPM8, TRPV1, OPRD1, COMT, and FAAH loci on variation in relatively short duration experimental pain responses to investigate the contribution of genetic factors to pain sensitivity in humans. We further examined the characteristics of haploblocks from the above genes in major ethnic populations and the potential role of the genetic variations including haploblocks and single nucleotide polymorphisms (SNPs) in the interindividual variation in cold and heat pain sensitivity in European Americans.
Methods
Subjects
Normal subjects (443 females and 292 males) were evaluated at the National Institute of Dental and Craniofacial Research following informed consent under a human research protocol approved by the NIDCR IRB. Participants were recruited from the greater Washington area by the National Institutes of Health patient recruitment office and through referral from local dental offices. Subjects were not experiencing any clinical pain as symptomatic patients were referred elsewhere at the time of initial screening. Self reported ethnicity in the sample was 50.1% European American, 22.8% African American, 10.6% Hispanic subjects, and 14.1% Asian American. For the genotype linkage, we excluded individuals with mixed race parentage (2.3%).
SNP genotyping
For genotyping, 50 ml of venous blood from each subject was collected. DNA isolation was performed with the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN, USA) following the manufacturer's instructions.
For SNP genotyping, Assays‐on‐Demand or Assays‐by‐Design SNP Genotyping Products (Applied Biosystems, Foster City, CA, USA) were used. Each well contained 2.5 μl of TaqMan universal master mix, 0.25 μl of genotyping assay mix, and 2.25 μl of DNAse free water. Polymerase chain reaction (PCR) was performed under the following conditions: 95°C, 10 min followed by 40 cycles of 92°C, 15 s and 60°C, 1 min in a Perkin‐Elmer 9700 thermocycler (Perkin‐Elmer, Boston, MA, USA). Following PCR, fluorescence of each well was measured using the ABI Prism 7900 Sequence Detection System (Applied Biosystems). From the genomic sequences including their flanking regions, eight SNPs (average distance 6.8 kb) from TRPA1 (genomic size 54 kb), 12 SNPs (average distance 8.4 kb) from TRPM8 (genomic size 102 kb), 13 SNPs (average distance 4.4 kb) from TRPV1 (genomic size 44 kb), 11 SNPs (average distance 5.5 kb) from OPRD1 (genomic size 52 kb), 12 SNPs (average distance 3.0 kb) from COMT (genomic size 36.4 kb), and seven SNPs (average distance 2.8 kb) from FAAH (genomic size 19.8 kb) were screened. Detailed information on genotyped SNPs is given in table 1.
Table 1 Information on genotyped SNPs.
| Gene | Order | SNP ID | Location from transcription site (bp) | Nucleotide variation | Rarer allele frequency |
|---|---|---|---|---|---|
| TRPA1 | 1 | rs1443952 | 7176 | G/A | 0.38 |
| 2 | rs3735943 | 21 817 | T/C | 0.50 | |
| 3 | rs3735942 | 21 846 | C/T | 0.33 | |
| 4 | rs1025928 | 24 561 | G/A | 0.40 | |
| 5 | rs13255063 | 28 284 | A/T | 0.20 | |
| 6 | rs1198795 | 38 218 | G/A | 0.41 | |
| 7 | rs13279503 | 48 193 | C/G | 0.32 | |
| 8 | rs1947913 | 60 806 | A/T | 0.21 | |
| TRPM8 | 1 | rs6431648 | 7415 | G/A | 0.41 |
| 2 | rs10803666 | 12 905 | G/C | 0.12 | |
| 3 | rs12472151 | 26 650 | G/A | 0.05 | |
| 4 | rs2215173 | 29 129 | G/A | 0.37 | |
| 5 | rs6740118 | 34 064 | C/T | 0.35 | |
| 6 | rs4663995 | 66 084 | C/T | 0.25 | |
| 7 | rs1016062 | 68 850 | G/A | 0.17 | |
| 8 | rs2362294 | 74 470 | G/A | 0.41 | |
| 9 | rs2362295 | 80 483 | T/C | 0.23 | |
| 10 | rs10490018 | 91 689 | C/T | 0.27 | |
| 11 | rs2052029 | 101 645 | G/C | 0.28 | |
| 12 | rs663924 | 108 143 | A/G | 0.30 | |
| TRPV1 | 1 | rs150854 | −6355 | A/C | 0.41 |
| 2 | rs465563 | −1014 | T/C | 0.41 | |
| 3 | rs182637 | 2873 | G/A | 0.43 | |
| 4 | rs161381 | 9178 | C/A | 0.15 | |
| 5 | rs733080 | 12 094 | A/G | 0.35 | |
| 6 | rs150846 | 17 837 | C/T | 0.49 | |
| 7 | rs222748 | 18 344 | C/T | 0.11 | |
| 8 | rs222747 | 19 505 | C/G | 0.25 | |
| 9 | rs222745 | 23 834 | C/T | 0.12 | |
| 10 | rs8065080 | 32 258 | A/G | 0.36 | |
| 11 | rs224547 | 37 686 | C/T | 0.35 | |
| 12 | rs4790522 | 42 851 | G/T | 0.42 | |
| 13 | rs322957 | 50 575 | A/T | 0.26 | |
| OPRD1 | 1 | rs1042114 | 322 | G/T | 0.08 |
| 2 | rs533123 | 2502 | A/G | 0.24 | |
| 3 | rs204048 | 6367 | A/G | 0.20 | |
| 4 | rs678849 | 6535 | C/T | 0.45 | |
| 5 | rs2236857 | 22 956 | T/C | 0.27 | |
| 6 | rs421300 | 30 940 | A/G | 0.36 | |
| 7 | rs12749204 | 37 560 | A/G | 0.15 | |
| 8 | rs2234918 | 50 944 | T/C | 0.46 | |
| 9 | rs204076 | 51 737 | A/T | 0.27 | |
| 10 | rs175969 | 59 905 | T/C | 0.09 | |
| 11 | rs379944 | 61 346 | C/T | 0.10 | |
| COMT | 1 | rs5746846 | −8663 | G/C | 0.39 |
| 2 | rs2020917 | −425 | C/T | 0.32 | |
| 3 | rs933271 | 2099 | T/C | 0.29 | |
| 4 | rs5993882 | 8225 | T/G | 0.20 | |
| 5 | rs740603 | 15 869 | G/A | 0.46 | |
| 6 | rs4646312 | 19 029 | T/C | 0.33 | |
| 7 | rs165722 | 19 705 | T/C | 0.48 | |
| 8 | rs6269 | 20 644 | A/G | 0.44 | |
| 9 | rs4633 | 20 927 | T/C | 0.49 | |
| 10 | rs4818 | 21 899 | C/G | 0.37 | |
| 11 | rs4680 | 21 963 | G/A | 0.42 | |
| 12 | rs174699 | 25 150 | T/C | 0.07 | |
| 13 | rs165728 | 27 715 | T/C | 0.08 | |
| FAAH | 1 | rs932816 | −239 | G/A | 0.19 |
| 2 | rs4141964 | 5053 | T/C | 0.49 | |
| 3 | rs3766246 | 5684 | G/A | 0.41 | |
| 4 | rs324420 | 10 774 | C/A | 0.27 | |
| 5 | rs324419 | 11 999 | C/T | 0.11 | |
| 6 | rs2295633 | 14 396 | G/A | 0.43 | |
| 7 | rs2295632 | 19 575 | C/T | 0.38 |
Genotype discrimination was performed using TaqMan Sequence Detector version 2.1 software. Samples that failed to amplify were not included in the final analysis.
Experimental pain sensitivity measurements
We measured pain sensitivity in response to experimental painful thermal stimuli and cold stimuli with separate visual analogue scale (VAS) ratings for pain intensity. Individuals were trained to use a sliding VAS by rating a visual grey scale. This procedure also provides an indication of each subject's comprehension of the rating process using a VAS.17 For cold stimuli, we recorded cold pain intensity (CPI) VAS ratings every 30 s following submersion of the subject's hand up to the wrist into an insulated bucket filled with iced water (2–4°C). We instructed subjects to keep their hand submerged while repeatedly clenching and unclenching to prevent local warming in the water until the pain reached an “unbearable level” or 180 s, whichever occurred soonest. The temperature of iced water was maintained by ice cubes separated from the subject's hand by a wire mesh. Subjects rated CPI at 30 s; if they withdrew their hand prior to 30 s, CPI was rated at their cold withdrawal time (CWT).
Individuals rated heat pain intensity (HPI) using a VAS following application of 35, 43, 44, 45, 46, 47, and 49°C thermal stimuli for 5 s. The thermode probe area was 1 cm in diameter and mounted in a housing to maintain constant pressure with the skin. The probe was self applied at six different sites on the volar forearm within an area of ∼40×100 mm. Four iterations were completed for each temperature by moving the probe from spot to spot to eliminate the possibility of sensitisation or tissue damage. The order of each temperature was pre‐determined for each trial, but the order was quasi‐random to prevent subjects from anticipating each subsequent stimulus. Subjects were blinded with regard to the temperature of the stimulus.
Data analysis
For the linkage disequilibrium (LD) block evaluation, we included SNPs with minor allele frequency >0.1, genotype success rate >0.9, and p>0.05 in the Hardy‐Weinberg equilibrium test. The Hardy‐Weinberg equilibrium analysis was performed for the observed genotypes to remove non‐polymorphic markers in populations using GeneStat software and the chi square (χ2) test value was used to determine whether there was a significant deviation from Hardy‐Weinberg equilibrium. To resolve phase unknown genotypes and estimate population frequencies in unrelated individuals, we employed the PHASE method, a probability based Bayesian algorithm. Haploblocks based on the confidence interval rule18 were generated by Haploview version 3.11. To quantify LD (allelic association between marker and genes), the widely used D′ and r2 were calculated. Both measures are built on the basic pairwise‐disequilibrium coefficient, D, which is the difference between the probability of observing two marker alleles on the same haplotype and observing them independently in the population. A value of 0 implies independence, while 1.0 means complete co‐transfer.
Due to the smaller sample size when subdivided into ethnic groups, second association analysis was carried out only in European Americans. For the associations between haplotypes, individual SNPs, and cold/heat pain sensitivity, statistical evaluation was performed with one way analysis of variance (ANOVA) models. The total number of statistical comparisons evaluating the association between genetic variation and pain responses in this study was 96.
Results
The minor allelic frequencies of each locus in the TRPA1, TRPM8, TRPV1, OPRD1, COMT, and FAAH genes are shown in table 1. Ten SNPs from European Americans, six SNPs from African Americans, two SNPs from Hispanic subjects, and two SNPs from Asian Americans deviated from Hardy‐Weinberg equilibrium and were excluded for LD block evaluation in each population.
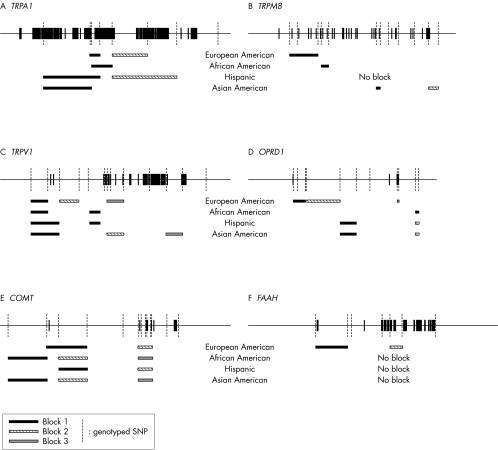
We next determined what haplotypes are formed by SNPs following Hardy‐Weinberg equilibrium. Thirteen haploblocks were found in European Americans (average minimum span 7.0 kb), seven in African Americans (average minimum span 5.1 kb), seven in Hispanic subjects (average minimum span 9.3 kb), and 11 in Asian Americans (average minimum span 6.4 kb). The positions of those SNPs and haploblocks within the loci of TRPA1, TRPM8, TRPV1, OPRD1, COMT, and FAAH are shown in fig 1. Their D′ with confidence intervals and r2 matrices in each ethnic population are given in supplement 1 (available from http://www.jmedgenet.com/supplemental).
Figure 1 LD blocks in genomic structures. (A) TRPA1; (B) TRPM8; (C) TRPV1; (D) OPRD1; (E) COMT; (F) FAAH.
Further haploblock analysis was performed with European Americans only. We found two haploblocks (three SNPs spanning ∼2.7 kb and two SNPs spanning ∼9.9 kb) from TRPA1, one haploblock (three SNPs spanning ∼19.2 kb) from TRPM8, three haploblocks (two SNPs spanning ∼5.3 kb, two SNPs spanning ∼6.3 kb, three SNPs spanning ∼5.5 kb) from TRPV1, three haploblocks (three SNPs spanning ∼6.0 kb, two SNPs spanning ∼16.4 kb, two SNPs spanning ∼1.0 kb) from OPRD1, two haploblocks (three SNPs spanning ∼8.6 kb, five SNPs spanning ∼2.9 kb) from COMT, and two haploblocks (two SNPs spanning ∼5.3 kb, two SNPs spanning ∼2.4 kb) from FAAH.
In TRPA1 haploblock 1, a total of six haplotypes were detected for the three SNPs, with the most frequent haplotype (43%) composed of C_C_A. The four most frequent haplotypes constituted 99% of total haplotypes. In TRPA1 haploblock 2, a total of four haplotypes were detected for the two chosen SNPs, with the most frequent haplotype (40%) composed of the most frequent alleles for both markers (A_G). Three of the four haplotypes were frequent (>5%), and these three haplotypes constituted 99% of the total haplotypes. In TRPM8 haploblock 1, a total of five haplotypes were detected for the three SNPs, with the most frequent haplotype (80%) composed of G_G_G. Three frequent haplotypes constituted 95% of the total haplotypes. In TRPV1 haploblock 1, a total of three haplotypes were detected for the two SNPs, with the most frequent haplotype (44%) composed of C_T. Three frequent haplotypes constituted 100% of the total haplotypes. In TRPV1 haploblock 2, a total of four haplotypes were detected for the two SNPs. The most frequent haplotype (47%) was composed of A_C and three frequent haplotypes constituted 99% of the total haplotypes. In TRPV1 haploblock 3, a total of six haplotypes were detected for the three SNPs. The most frequent haplotype (65%) was composed of C_C_C and three frequent haplotypes constituted 99% of the total haplotypes.
In OPRD1 haploblock 1, a total of six haplotypes were detected for the three SNPs, with the most frequent haplotype (59%) composed of G_A_A. The three most frequent haplotypes constituted 99% of the total haplotypes. In OPRD1 haploblock 2, a total of four haplotypes were detected for the two SNPs, with the most frequent haplotype (52%) composed of T_T. The three most frequent haplotypes constituted 99% of the total haplotypes. In OPRD1 haploblock 3, a total of four haplotypes were detected for the two SNPs, with the most frequent haplotype (56%) composed of T_A. The three most frequent haplotypes constituted 99% of the total haplotypes. In COMT haploblock 1, a total of five haplotypes were detected for the three SNPs, with the most frequent haplotype composed of C_T_T. The three most frequent haplotypes constituted 95.3% of the total haplotypes. In COMT haploblock 2, three frequent haplotypes constituted 98.3% of the total of 10 haplotypes and T_T_A_T_C_A was the most frequent (49.4%). In FAAH haploblock 1, a total of four haplotypes were detected and G_T (36.7%) was the most frequent of three frequent haplotypes (99.8%). In FAAH haploblock 2, three frequent haplotypes constituted 99.8% of the total of four haplotypes and C_G was the most frequent haplotype (62.9%) (table 2).
Table 2 Frequent haplotypes in European Americans (frequency >5%).
| Frequency | |
|---|---|
| TRPA1 block 1 (SNP 2, 3, 4) | |
| C_C_A | 0.43 |
| T_T_G | 0.30 |
| T_C_G | 0.16 |
| C_C_G | 0.11 |
| Sum | 0.99 |
| TRPA1 block 2 (SNP 5, 6) | |
| A_G | 0.40 |
| A_A | 0.36 |
| T_G | 0.23 |
| Sum | 0.99 |
| TRPM8 block 1 (SNP 1, 2, 3) | |
| G_G_G | 0.80 |
| A_G_G | 0.09 |
| A_C_G | 0.06 |
| Sum | 0.95 |
| TRPV1 block 1 (SNP 1, 2) | |
| C_T | 0.44 |
| A_C | 0.31 |
| A_T | 0.25 |
| Sum | 1.00 |
| TRPV1 block 2 (SNP 3, 4) | |
| A_C | 0.47 |
| G_C | 0.35 |
| G_A | 0.17 |
| Sum | 0.99 |
| TRPV1 block 3 (SNP 7, 8, 9) | |
| C_C_C | 0.65 |
| C_G_C | 0.23 |
| T_C_T | 0.11 |
| Sum | 0.99 |
| OPRD1 block 1 (SNP 1, 2, 3) | |
| G_A_A | 0.59 |
| G_A_G | 0.20 |
| T_G_A | 0.11 |
| G_G_A | 0.10 |
| Sum | 0.99 |
| OPRD1 block 2 (SNP 4, 5) | |
| T_ T | 0.52 |
| C_C | 0.27 |
| C_T | 0.20 |
| Sum | 0.99 |
| OPRD1 block 3 (SNP 8, 9) | |
| T_A | 0.56 |
| C_T | 0.33 |
| C_A | 0.11 |
| Sum | 0.99 |
| COMT block 1 (SNP 2, 3, 4) | |
| C_T_T | 0.42 |
| T_T_T | 0.33 |
| C_C_G | 0.20 |
| Sum | 0.95 |
| COMT block 2 (SNP 6, 7, 8, 9, 10, 11) | |
| T_T_A_T_C_A | 0.49 |
| C_C_G_C_G_G | 0.42 |
| T_C_A_C_C_G | 0.07 |
| Sum | 0.98 |
| FAAH block 1 (SNP 1, 2) | |
| G_T | 0.37 |
| G_C | 0.35 |
| A_C | 0.28 |
| Sum | 0.99 |
| FAAH block 2 (SNP 5, 6) | |
| C_G | 0.63 |
| C_A | 0.22 |
| T_A | 0.15 |
| Sum | 0.99 |
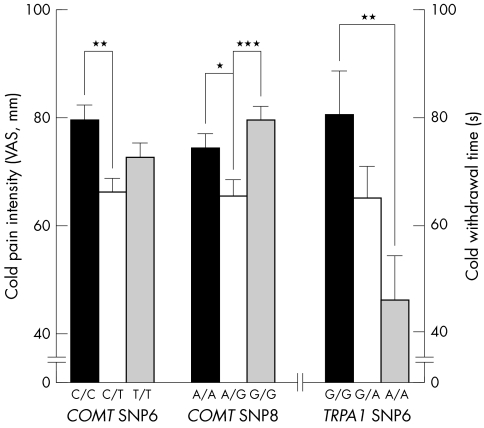
Considering the relatively dominating effect of gender on pain, male and female subjects were analysed separately for the association between combination of haplotypes and cold/heat pain sensitivity. In European Americans, there are five common combinations of haploblock 2 in TRPA1. We found a significant association between common combinations (combination of T_G+A_G, T_G+A_A, A_G+A_G, A_G+A_A, and A_A+A_A) of haploblock 2 in TRPA1 and CWT accounting for 7% of variation (ANOVA, df = 146, F = 2.43, p = 0.050) and HPI at 49°C (HPI49) accounting for 10.5% of variation (ANOVA, df = 145, F = 3.76, p = 0.006) in females. Further analysis of individual SNPs of haploblock 2 of TRPA1 showed significant association only between SNP 6 (rs11988795) and CWT (ANOVA, df = 166, F = 3.85, p = 0.023) (fig 2). There are four common combinations of haploblock 2 in COMT. We also found significant associations between CPI and common combinations (combinations C_C_G_C_G_G+C_C_G_C_G_G, C_C_G_C_G_G+T_T_A_T_C_A, T_T_A_T_C_A+T_T_A_T_C_A, and T_T_A_T_C_A+T_C_A_C_C_G) of haploblock 2, accounting for 5.3% (ANOVA, df = 160, F = 2.94, p = 0.035) of variation in CPI in females. SNP 6 (rs4646312) and SNP 8 (rs6269) from this haploblock showed association with CPI (ANOVA, df = 179, F = 4.00, p = 0.021 and df = 178, F = 5.15, p = 0.007, respectively) (fig 2).
Figure 2 Association between cold pain response and SNPs of haploblocks from TRPA1 and COMT in European American females.
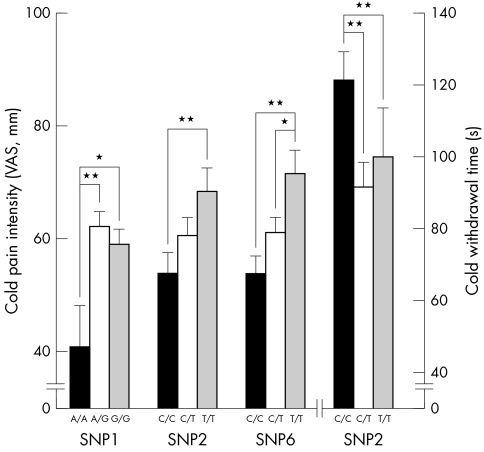
In contrast, in males the common combinations (G_C+G_C, G_C+A_C, G_C+G_T, A_C+G_T, and G_T+G_T) of haploblock 1 in FAAH accounted for 8.6% of variation in CPI (ANOVA, df = 150, F = 2.89, p = 0.025) and SNP 1 and 2 (rs932816 and rs4141964) were both individually associated with CPI (ANOVA, df = 165, F = 4.18, p = 0.017 and df = 166, F = 3.94, p = 0.021, respectively), while only SNP 2 was associated with CWT (ANOVA, df = 166, F = 4.07, p = 0.019) (fig 3). Common combinations (C_G+C_G, C_G+C_A, and C_G+T_A) of haploblock 2 of FAAH accounted for 7.4% of variation in CWT (ANOVA, df = 134, F = 5.24, p = 0.006) in males. SNP 6 (rs2295633) from haploblock 2 was associated with CPI (ANOVA, df = 161, F = 5.14, p = 0.007) (fig 3). Common combinations of haplotypes in TRPM8, TRPV1, and OPRD1 showed no association with cold and heat pain sensitivity in this cohort of European American females and males.
Figure 3 Association between cold pain response and SNPs of haploblocks from FAAH in European American males.
Discussion
We have examined genetic variations such as SNPs and haplotypes in genes related to thermosensation in various ethnic populations, which provide a basis for genetic association studies in painful conditions. Significant associations found in European Americans in our study suggest not only the role of variations of those genes in responses to short duration heat and cold painful stimuli in humans but also the necessity of further studies including other ethnic populations.
Because the vast majority of heterozygosity in the human population is attributable to common variants, one promising approach is to test common genetic variations for association to pain sensitivity. However, it is not easy to choose the candidate genes from the more than 20 000 human genes. One suggested approach is to choose candidate genes based on a plausible biological role for the relevant gene in the mechanisms of the phenotype and ideally with a functional change in physiology by variation of the gene.19 Reducing the number of candidate genes is also important to avoid type I error. Thus rather than query the entire genome, we systematically examined SNPs and haplotypes for several major ion channels that transduce the physical stimuli (cold and heat) we used for experimental pain stimulation. We also examined three other important molecules that modulate nociceptive processing at many levels of the nervous system, the delta opioid receptor and two enzymes involved in clearance of neuroactive signalling molecules. All have been demonstrated to have a biological role or relationship with pain sensitivity.
The search for predictive SNPs within genes may require examination of not only exons but also promoters and/or the area of intron‐exon boundaries and mRNA processing signals. Any portion of genomic DNA can be meaningful for the phenotype because of the complicated dynamics of DNA structure, gene expression, and interchromosomal associations. Therefore, it may be appropriate to evaluate the association between a common set of SNPs and pain phenotypes to find a candidate region as the first step. This higher order of genetic organisation was used to probe genetic and behavioural associations. The analyses of individual SNPs within the candidate region may be the second step to narrow the responsible genetic variation for the phenotypes. This approach can also decrease the risk of false positive findings by reducing the number of comparisons. Considering the sample size, it is reported that samples of 45 unrelated individuals should be sufficient to find 99% of haplotypes with a frequency of ⩾5% in a population.20
We generated haploblocks using three different algorithms from Haploview 3.11: (1) confidence intervals for D′, (2) the 4 gamete rule, and (3) solid spine. These methods are known to generate haplotype block partition quite differently and we also obtained different results with the different methods. There is currently no consensus on the definition that best generates haplotype block structures. Therefore, we chose confidence intervals for D′ to define haplotype blocks because it generated haploblocks most conservatively in our data set (see supplement 2, available from http://www.jmedgenet.com/supplemental). The general properties of haplotypes including average span, number of markers needed for identifying a region as a block, and block characteristics across populations in the present human cohort are consistent with previously reported properties with the same definition of haplotype block, supporting the generalisability of the findings.18 Each haploblock has three to five common haplotypes (>5%) and those accounted for more than 95% of the total haplotypes. No gender difference was found either in individual marker SNP allelic frequencies or haplotypes in the present human cohort.
The ethnicity of the subjects was determined by self determined ethnicity instead of using ancestry informative markers. It is generally acceptable to use self identified ethnicity based on the geographic ancestry because it is very highly correlated with genetically defined clusters,21 though it is less reliable than using explicit genetic data.22 The results of haplotypes from our sample are consistent with the results of the International HapMap Project Database (http://www.hapmap.org/) in the comparable regions. Haplotype patterns in African Americans showed similarity with Hispanic subjects who are an admixed group that includes white and Native American ancestry, as well as African ancestry.23 These consistent findings suggest the generalisability of our sample.
Principal temperature sensors belong to the transient receptor potential (TRP) family of non‐selective cation channels.24 The large number of TRP subtypes, their overlapping electrophysiological characteristics, broad expression patterns, heteromultimerisation, lack of specific blockers, and poorly understood mechanisms of activation have made their study difficult. A/A homozygote variants at TRPA1 SNP 6, which is in the intron, showed less pain tolerance to the cold stimuli compared to G/G homozygotes. The association between genetic variations of TRPA1 and CWT is consistent with previous in vitro finding,2 although there still remains debate about the role of TRPA1 in pain.25 Different cell types (Chinese hamster ovary cells versus rat trigeminal neuronal cell, human embryonic kidney cell), or different experimental environments used in these studies may explain the inconsistency.2,25,26 Even genetically identical cells exposed to the same environmental conditions can show significant variation in molecular content and marked differences in phenotypic characteristics.
A paradoxical relationship between TRPs and pain sensation has been suggested previously based on the observation that long term topical application of capsaicin affects cold sensation longer than any other sensations including heat and mechanical sensations.27 The inhibition of capsaicin induced current by cooling and burning sensation by noxious cold also suggests a cross‐relationship between cold and heat pain sensation. It was recently reported that TRPA1 is activated not only by cold but also by mustard oil and bradykinin which suggests a role in various painful sensations. It is co‐expressed in 97% of primary afferent nerves with TRPV12 and the colocalisation of TRPV1 and TRPM8 is also reported, although it is still controversial. The co‐expression of TRPV1 and TRPA1 or TRPM8 and the flexibility of the TRP family channels raise the possibility that these channels might interact to influence the properties of one another. In this regard, recently reported heteromultimerisation among the TRP channels is suggestive of the mechanism for interactions.6
TRPA1 is also suggested as a mechanically gated transduction channel which mediates the perception of sound in the ear. Drosophila orthologues of TRPA1 show an activation threshold at 24–29°C suggesting flexibility in TRP family channel response to different temperature ranges. TRPA1 channels inactivate in hyperpolarised cells but remain open in depolarised cells. This property provides a mechanism for the lack of desensitisation, coincidence detection, and allodynia that characterise pain by allowing a sensory neuron to respond constantly to sustained stimulation that is noxious and yet permitting the same cell to ignore sustained stimulation that is innocuous.28 Nevertheless, the association between HPI at 49°C and the TRPA1 haploblock is somewhat surprising, although it was not confirmed in the individual SNP analysis. It is not clear yet whether this finding was by chance or other factors such as the three‐dimensional structure of TRPA1 haploblock 1 affect on heat pain sensitivity. When we analysed European Americans with these specific SNPs, we could not find significant association either in female or males. This negative association requires further studies to confirm the potentially significant effect of TRPA1 genetic variation on heat pain sensitivity in humans.
The cold receptor TRPM8 is expressed in a separate set of primary afferent nerves, suggesting that the sensation of cold versus cold pain may be transmitted by two different populations of sensory nerves.2 However, we could not find any association between TRPM8 and cold or heat pain sensitivity in our sample. Considering the relatively higher range of threshold temperature of TRPM8 (8–28°C),3 it is likely that the TRPM8 receptor was not activated by the painful cold temperature we used (2–4°C).
Among the markers, SNP 8 (rs222747) and 10 (rs8065080) in TRPV1 induce amino acid changes (methionine to isoleucine at codon 315 and isoleucine to valine at codon 585, respectively) in its coding sequence. However, we could not find any significant association with TRPV1 and cold/heat pain sensitivity in European Americans. Our result is consistent with previous in vitro findings.29 In spite of the significant association between oprd1 and heat pain sensitivity in vivo data,7OPRD1 haploblocks did not show significant association with cold and/or heat pain sensitivity. Because the significant effect of oprd1 on heat pain sensitivity was strain dependent as well as gender dependent,7 additional ethnic populations should be investigated to confirm our results.
Although the missense SNP 11 (rs4680) of COMT is included in the cold pain associated haploblock, this substitution from valine to methionine at codon 158 cannot explain the association because individual SNP analysis showed no differences among val/val, val/met, and met/met populations. It is not consistent with previous findings reporting higher sensory ratings of pain in met/met homozygotes.13 We also failed to reproduce the existence of previously reported haplotypes inducing low, moderate, and high pain sensitivity,14 in spite of the similar haplotype frequencies. Our data even demonstrated opposite trends in that the haplotypes, claimed to be related to higher pain sensitivity, are actually included in the lower pain sensitive populations in European Americans, although this is not statistically significant (data not shown). Because the observed association between human diseases and the met allele (low enzyme activity) is relatively modest and occasionally produces opposite results,14 it is not surprising to find this kind of inconsistency. Even with SNP 6 and 8 showing significant association with CPI, both of them are located in introns and the heterozygote group showed a lower response compared to homozygotes. The observed relationship between this COMT haploblock with two SNPs and CPI suggests that another mechanism including different, unidentified but significant genetic factors may play a role in cold pain perception. Considering the multiple pathways involved in different types of pain sensitivity, including different dimensions of pain such as unpleasantness, it is important to note that our results apply only to briefly induced cold and heat pain.
In FAAH, SNP 4 (rs324420) induces amino acid change from proline to threonine at amino acid 129 and is known to lower enzymatic activity. Based on its biological role and knock out mice data,15,30 it is probable that Thr/Thr homozygotes show less pain response compared to Pro/Pro homozygotes. Because humans homozygous for the Thr129 may possess constitutively lower levels of FAAH activity in both central and peripheral tissues, it is tempting to speculate that these subjects may display a corresponding increase in endocannabinoid signalling that contributes, at least in part, to decreased basal pain sensitivity. However, we could not find an association. Instead, among four SNPs contributing haploblocks in FAAH, three SNPs in introns showed significant association with cold pain sensitivity. Four additional SNPs whose heterozygosity was greater than 0.25 were reported in FAAH and two non‐synonymous SNPs are close to SNP 6 and 7 showing significant association with CPI. Given the significant findings between FAAH variations and CPI, additional high density genotyping of the FAAH region is necessary. Like FAAH, genes showing association between their variations and pain sensitivity, such as TRPA1 and COMT, contain other SNPs located within or close to them. Some of these SNPs induce important functional change such as non‐synonymous amino acid substitution, splicing variants, etc. Although their heterozygosity is low or not known, future studies including higher density genotyping around the candidate regions are needed.
Even though we found some significant association between TRPA1, COMT, and FAAH genetic variations and cold pain sensitivity, for modest genetic effects and for identification of genotypic subgroups, much larger sample sizes may be required. Furthermore, the risk of a chance finding should be considered carefully. In general, SNPs showing association with pain sensitivity in these candidate genes are in introns or synonymous. Again, more complicated unidentified mechanisms rather than the simple amino acid change in the coding sequence may play a major role. However, the results are promising especially for TRPA1 and FAAH because we obtained relatively low p values (data not shown) and reasonable data profiles, that is, homozygote 1>heterozygote>homozygote 2, and similar results from different statistical strategies. A sample drawn from only one ethnic group may increase the ability to study phenotype‐genotype association. Therefore, larger samples from other ethnic populations including African Americans, Hispanic subjects, and Asian Americans are needed to generalise the association between these genetic predictors and cold pain sensitivity. It is also necessary to take account of the substructure of the examined population even in a relatively homogenous genetic group such as Icelanders. Icelanders cannot be considered to be a single, randomly interbreeding population.31 It is interesting to note that the subpopulation with red hair colour is more sensitive to painful stimuli than other hair colour subpopulations in Caucasians.32,33 This will probably be more important in larger populations such as European Americans or African Americans. Again, independent analysis of each gender population is needed. Along with its binding affinity to DNA and gene expression regulation, estradiol is also thought to influence various brain functions by acting on receptors localised to the neuronal membrane surface.34 However, we still do not know the mechanism of gender difference on pain sensitivity.
Although our results do not suggest that heat sensitive and cold sensitive subgroups are identical as previously reported in animals, this does not imply that heat and cold pain sensitivities are genetically different types of nociception. On the contrary, the potential association between TRPA1 haplotypes and heat pain suggests a possible relationship between cold pain and heat pain. The value of estimating heritability is to establish the likely success of specific strategies to detect the action of individual genes. Considering the low heritability estimate of heat pain sensitivity in humans, it is less likely the same effects will be found though a specific genetic polymorphism that is responsible for both heat pain sensitivity and cold pain sensitivity. Consistent with the heritability estimates, we found a few haploblocks and their SNPs had a significant effect on pain responses, mostly to cold stimuli.
Because haploblocks can arise from several causes, simply identifying them does not ensure either their conservation within or between populations or their utility for mapping genes associated with the phenotypes of interest. Identification of causal variations including the three‐dimensional structure of genomic or complementary DNA, and SNPs predicting amino acid change may be the first step for investigating the genetic role in pain sensitivity. Functional tests should be performed in vitro to directly measure the effect of the SNP to confirm the association and provide a stronger rationale for the relationship to pain sensitivity. Our association analyses have identified several loci associated with cold and heat pain sensitivity, but their functional importance has yet to be confirmed.
Supplements 1 and 2 are available from http://www.jmedgenet.com/supplemental
Acknowledgements
We thank Dr Yin Yao for consultation on statistical analyses used in this manuscript.
Abbreviations
ANOVA - one way analysis of variance
CPI - cold pain intensity
CWT - cold withdrawal time
FAAH - fatty acid hydrolase
HPI - heat pain intensity
LD - linkage disequilibrium
SNP - single nucleotide polymorphism
TRP - transient receptor potential
VAS - visual analogue scale
Footnotes
This research was supported by the Division of Intramural Research, NIDCR, NIH
Competing interests: none declared
Supplements 1 and 2 are available from http://www.jmedgenet.com/supplemental
References
- 1.Hirschhorn J N, Daly M. Genome‐wide association studies for common diseases and complex traits. Nat Rev Genet 2005695–108. [DOI] [PubMed] [Google Scholar]
- 2.Story G M, Peier A M, Reeve A J, Eid S R, Mosbacher J, Hricik T R, Earley T J, Hergarden A C, Andersson D A, Hwang S W, McIntyre P, Jegla T, Bevan S, Patapoutian A. ANKTM1, a TRP‐like channel expressed in nociceptive neurons, is activated by cold temperatures. Cell 2003112(6)819–829. [DOI] [PubMed] [Google Scholar]
- 3.Peier A M, Moqrich A, Hergarden A C, Reeve A J, Andersson D A, Story G M, Earley T J, Dragoni I, McIntyre P, Bevan S, Patapoutian A. A TRP channel that senses cold stimuli and menthol. Cell 2002108(5)705–715. [DOI] [PubMed] [Google Scholar]
- 4.Caterina M, Schumacher M, Tominaga M, Rosen T A, Levine J D, Julius D. The capsaicin receptor: a heat‐activated ion channel in the pain pathway. Nature 1997389(23)816–824. [DOI] [PubMed] [Google Scholar]
- 5.Viswanath V, Story G M, Peier A M, Petrus M J, Lee V M, Hwang S W, Patapoutian A, Jegla T. Opposite thermosensor in fruitfly and mouse. Nature 2003423(6942)822–823. [DOI] [PubMed] [Google Scholar]
- 6.Clapham D E, Runnels L W, Strubing C. The TRP ion channel family. Nat Rev Neurosci 20012(6)387–396. [DOI] [PubMed] [Google Scholar]
- 7.Mogil J S, Richards S P, O'Toole L A, Helms M L, Mitchell S R, Belknap J K. Genetic sensitivity to hot‐plate nociception in DBA/2J and C57BL/6J inbred mouse strains: possible sex‐specific mediation by δ2‐opioid receptors. Pain 199770267–277. [DOI] [PubMed] [Google Scholar]
- 8.Kim H, Neubert J K, San Miguel A, Xu K, Krishnaraju R K, Iadarola M J, Goldman D, Dionne R A. Genetic influence on variability in human pain sensitivity associated with gender, ethnicity and psychological temperament. Pain 2004109488–496. [DOI] [PubMed] [Google Scholar]
- 9.Xie T, Ho S, Li L, Ma O C. G/A1947 Polymorphism in catechol‐O‐methyltransferase (COMT) gene in Parkinson's disease. Mov Disord 199712(3)426–427. [DOI] [PubMed] [Google Scholar]
- 10.Lachman H M, Papolos D F, Saito T, Yu Y M, Szumlanski C L, Weinshilboum R M. Human catechol‐O‐methyltransferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics 19966(3)243–250. [DOI] [PubMed] [Google Scholar]
- 11.Shield A J, Thomae B A, Eckloff B W, Wieben E D, Weinshilboum R M. Human catechol O‐methyltransferase genetic variation: gene resequencing and functional characterization of variant allozymes. Mol Psychiatry 20049(2)151–160. [DOI] [PubMed] [Google Scholar]
- 12.Männistö P, Kaakkola S. Catechol‐O‐methyltransferase (COMT): biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective COMT inhibitors. Pharmacol Rev 199951(4)593–628. [PubMed] [Google Scholar]
- 13.Zubieta J K, Heitzeg M M, Smith Y R, Bueller J A, Xu K, Xu Y, Koeppe R A, Stohler C S, Goldman D. COMT val158met genotype affects mu‐opioid neurotransmitter responses to a pain stressor. Science 2003299(5610)1240–1243. [DOI] [PubMed] [Google Scholar]
- 14.Diatchenko L, Slade G D, Nackley A G, Bhalang K, Sigurdsson A, Belfer I, Goldman D, Xu K, Shabalina S A, Shagin D, Max M B, Makarov S S, Maixner W. Genetic basis for individual variations in pain perception and the development of a chronic pain condition. Hum Mol Genet 200514(1)135–143. [DOI] [PubMed] [Google Scholar]
- 15.Lichtman A H, Shelton C C, Advani T, Cravatt B F. Mice lacking fatty acid amide hydrolase exhibit a cannabinoid receptor‐mediated phenotypic hypoalgesia. Pain 2004109(3)319–327. [DOI] [PubMed] [Google Scholar]
- 16.Chiang K P, Gerber A L, Sipe J C, Cravatt B F. Reduced cellular expression and activity of the P129T mutant of human fatty acid amide hydrolase: evidence for a link between defects in the endocannabinoid system and problem drug use. Hum Mol Genet 200413(18)2113–2119. [DOI] [PubMed] [Google Scholar]
- 17.Rosier E M, Iadarola M J, Coghill R C. Reproducibility of pain measurement and pain perception. Pain 200298205–216. [DOI] [PubMed] [Google Scholar]
- 18.Gabriel S B, Schaffner S F, Nguyen H, Moore J M, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu‐Cordero S N, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander E S, Daly M J, Altshuler D. The structure of haplotype blocks in the human genome. Science 2002296(5576)2225–2229. [DOI] [PubMed] [Google Scholar]
- 19.Bird T D, Jarvik G P, Wood N W. Genetic association studies: genes in search of diseases. Neurology 200157(7)1153–1154. [DOI] [PubMed] [Google Scholar]
- 20.The International HapMap Consortium The International HapMap Project. Nature 2003426(6968)789–796. [DOI] [PubMed] [Google Scholar]
- 21.Risch N, Burchard E, Ziv E, Tang H. Categorization of humans in biomedical research: genes, race and disease. Genome Biol 20023(7)comment2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bamshad M, Wooding S, Salisbury B A, Stephens J C. Deconstructing the relationship between genetics and race. Nat Rev Genet 20045(8)598–609. [DOI] [PubMed] [Google Scholar]
- 23.Burchard E G, Ziv E, Coyle N, Gomez S L, Tang H, Karter A J, Mountain J L, Perez‐Stable E J, Sheppard D, Risch N. The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med 2003348(12)1170–1175. [DOI] [PubMed] [Google Scholar]
- 24.Voets T, Droogmans G, Wissenbach U, Janssens A, Flockerzi V, Nilius B. The principle of temperature‐dependent gating in cold‐ and heat‐sensitive TRP channels. Nature 2004430(7001)748–754. [DOI] [PubMed] [Google Scholar]
- 25.Jordt S E, Bautista D M, Chuang H H, McKemy D D, Zygmunt P M, Hogestatt E D, Meng I D, Julius D. Mustard oils and cannabinoids excite sensory nerve fibres through the TRP channel ANKTM1. Nature 2004427(6971)260–265. [DOI] [PubMed] [Google Scholar]
- 26.Chesler E J, Wilson S G, Lariviere W R, Rodriguez‐Zas S, Mogil J S. Influences of laboratory environment on behavior. Nat Neurosci 20025(11)1101–1102. [DOI] [PubMed] [Google Scholar]
- 27.Nolano M, Simone D, Wedelschafer‐Crabb G, Johnson T, Hazen E, Kennedy W R. Topical capsaicin in humans: parallel loss of epidermal nerve fibers and pain sensation. Pain 199981135–145. [DOI] [PubMed] [Google Scholar]
- 28.Nagata K, Duggan A, Kumar G, Garcia‐Anoveros J. Nociceptor and hair cell transducer properties of TRPA1, a channel for pain and hearing. J Neurosci 200525(16)4052–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayes P, Meadows H, Gunthorpe M, Harries M H, Duckworth D M, Cairns W, Harrison D C, Clarke C E, Ellington K, Prinjha R K, Barton A J, Medhurst A D, Smith G D, Topp S, Murdock P, Sanger G J, Terrett J, Jenkins O, Benham C D, Randall A D, Gloger I S, Davis J B. Cloning and functional expression of a human orthologue of rat vanilloid receptor‐1. Pain 200088205–215. [DOI] [PubMed] [Google Scholar]
- 30.Cravatt B F, Saghatelian A, Hawkins E G, Clement A B, Bracey M H, Lichtman A H. Functional disassociation of the central and peripheral fatty acid amide signaling systems. Proc Natl Acad Sci U S A 2004101(29)10821–10826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helgason A, Yngvadottir B, Hrafnkelsson B, Gulcher J, Stefansson K. An Icelandic example of the impact of population structure on association studies. Nat Genet 200537(1)90–95. [DOI] [PubMed] [Google Scholar]
- 32.Liem E B, Lin C M, Suleman M I, Doufas A G, Gregg R G, Veauthier J M, Loyd G, Sessler D I. Anesthetic requirement is increased in redheads. Anesthesiology 2004101(2)279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liem E B, Joiner T V, Tsueda K, Sessler D I. Increased sensitivity to thermal pain and reduced subcutaneous lidocaine efficacy in redheads. Anesthesiology 2005102(3)509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boulware M I, Weick J P, Becklund B R, Kuo S P, Groth R D, Mermelstein P G. Estradiol activates group I and II metabotropic glutamate receptor signaling, leading to opposing influences on cAMP response element‐binding protein. J Neurosci 200525(20)5066–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]