Abstract
We have recently developed “monolayer purification” as a rapid and convenient technique to produce specimens of His-tagged proteins or macromolecular complexes for single particle electron microscopy (EM) without prior biochemical purification. Here, we introduce the “Affinity Grid”, a pre-fabricated EM grid featuring a dried lipid monolayer that contains Ni-NTA lipids (lipids functionalized with a Nickel-nitrilotriacetic acid group). The Affinity Grid, which can be stored for several months under ambient conditions, further simplifies and extends the use of monolayer purification. After characterizing the Affinity Grid, we used it to isolate, within minutes, ribosomal complexes from E. coli cell extracts containing His-tagged rpl3, the human homolog of the E. coli 50S subunit rplC. Depending on the way the sample was applied to the Affinity Grid, ribosomal complexes with or without associated mRNA could be prepared. Vitrified Affinity Grid specimens could be used to calculate three-dimensional reconstructions of the 50S ribosomal subunit as well as the 70S ribosome and 30S ribosomal subunit from images of the same sample. In addition, we established that Affinity Grids are stable for some time in the presence of glycerol and detergents. This feature allowed us to isolate His-tagged aquaporin-9 (AQP9) from detergent-solubilized membrane fractions of Sf9 insect cells. The Affinity Grid can thus be used to prepare single particle EM specimens of soluble complexes and membrane proteins.
Keywords: monolayer purification, lipid monolayer, affinity purification, single particle, cryo-electron microscopy
INTRODUCTION
The biochemical purification of proteins and macromolecular complexes has become the rate-limiting step in structure determination by single particle cryo-electron microscopy (EM). A purification protocol of a recombinant protein for structural studies typically consists of an affinity chromatography step followed by size exclusion chromatography. Such a two-step purification scheme requires an appreciable amount of protein, which cannot always be produced, and takes several hours, during which time biological complexes can disintegrate. We have recently introduced “monolayer purification”, a technique that combines protein purification with specimen preparation, as a fast and easy way to prepare specimens suitable for single particle EM, requiring only low expression levels or low concentrations of His-tagged protein 1. In the monolayer purification technique, the extract of cells expressing a His-tagged protein, which can be a subunit of a larger assembly, is overlaid with a lipid monolayer that contains Ni-NTA lipid, a lipid whose head group is functionalized by Nickel-nitrilotriacetic acid (Ni-NTA). The Ni-NTA lipids recruit the His-tagged proteins to the lipid monolayer, which can then be lifted off with an EM grid covered with a continuous or holey carbon support film and prepared by negative staining or vitrification for subsequent imaging in the electron microscope.
Here, we introduce the “Affinity Grid”, a pre-fabricated EM grid with a dried, Ni-NTA lipid-containing monolayer, as a further simplification and extension of the monolayer purification technique. Using His-tagged transferrin-transferrin receptor (Tf-TfR) complex, we show that the Affinity Grid produces specimens equivalent to those obtained by monolayer purification. Although Affinity Grids can be incubated with sample for several hours, an incubation time of only a few minutes usually suffices to produce suitable EM specimens. We also establish that Affinity Grids are resistant to most detergents for a time that depends on the detergent concentration, making them useful for isolating membrane proteins. Finally, we use Affinity Grids to isolate ribosomal complexes from an E. coli extract and the water channel AQP9 from a membrane extract of Sf9 insect cells. Since Affinity Grids can be stored under ambient conditions for several months, they can be pre-fabricated and used whenever needed to prepare specimens for single particle EM – within minutes and with minimum effort.
RESULTS
Characterization of the Affinity Grid
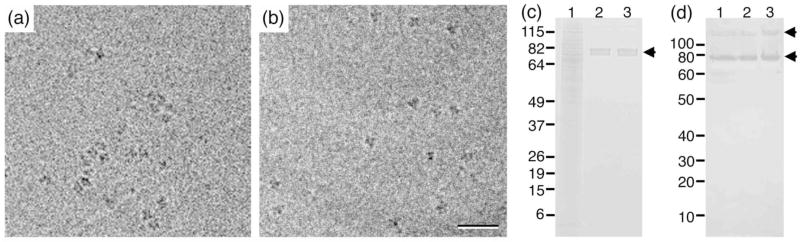
We used His-tagged Tf-TfR complex as a test specimen to characterize the Affinity Grid as a tool to prepare specimens for single particle EM. We first wanted to confirm that Affinity Grids produce samples of the same quality as those prepared with our recently introduced monolayer purification technique 1. We added His-tagged Tf-TfR complex to Sf9 cell extract (6 mg/ml protein, 60 mM imidazole) to a final concentration of 0.15 μg/ml. 3 μl of this mixture was added to a grid covered with a holey carbon film and a dried lipid monolayer containing 20% Ni-NTA lipid, which we refer to as a “20% HC Affinity Grid” (a grid covered with a continuous carbon film and a lipid monolayer containing 2% Ni-NTA lipid would be a “2% CC Affinity Grid”). After a 2-minute incubation, the grid was blotted and vitrified. Images of the Affinity Grid sample (Fig. 1b) looked virtually identical to images taken of Tf-TfR complexes prepared by monolayer purification (Fig. 1a). However, while the Tf-TfR complexes tended to cluster when prepared by monolayer purification (already reported in 1), they were more evenly distributed on the Affinity Grid. Most of the particles attached to the Affinity Grid showed the characteristic shape of the Tf-TfR complex, suggesting the absence of contaminating proteins. The number of particles present on the Affinity Grid can be controlled by adjusting the percentage of Ni-NTA lipid in the lipid mixture used to prepare the Affinity Grid or, more easily, by changing the time the Affinity Grid is incubated with the saple solution. Furthermore, no proteins bound to Affinity Grids when Ni-NTA lipids were omitted from the monolayer (data not shown), consistent with previous results obtained with the monolayer purification technique 1. To confirm the purity of Affinity Grid samples, 20 samples on 20% CC Affinity Grids were eluted into the same 20-μl drop of 300 mM imidazole. For comparison, we also eluted 20 monolayer-purified samples. SDS-PAGE analysis showed a smear over the entire lane for the insect cell extract used as input (Fig. 1c, lane 1), while the samples eluted from the monolayer purification grids (Fig. 1c, lane 2) and the Affinity Grids (Fig. 1c, lane 3) showed only two bands corresponding to Tf and TfR, which have a very similar molecular weight of ~75 kDa. A Western blot of the same samples developed with an anti-His antibody (Fig. 1d) showed two bands for the His-tagged TfR, one at ~75 kDa and one in the higher molecular weight range, presumably representing dimeric TfR. The entire lanes 2 and 3 were excised from the gel shown in Figure 1c and analyzed by mass spectrometry as previously described 1. The results confirmed that only Tf and TfR were present in the samples eluted from grids prepared by monolayer purification and from Affinity Grids (Supplementary Table 1). Affinity Grids reproducibly gave the same results, even if Affinity Grids were used that were stored for up to 6 months under ambient conditions.
Fig. 1. Affinity Grid and monolayer purification of Tf-TfR complexes from Sf9 cell extract.
(a, b) Images of Tf-TfR complex added to Sf9 cell extract containing 60 mM imidazole and prepared by monolayer purification using a 20% Ni-NTA lipid monolayer (a) or a 20% HC Affinity Grid (b). The images of the vitrified samples show that the Tf-TfR complexes are more evenly distributed on the Affinity Grid. Scale bar is 30 nm. (c) SimplyBlue stained SDS-PAGE gel. Lane 1: cell extract containing Tf-TfR complex; lane 2: protein eluted from 20 monolayer purification samples; lane 3: protein eluted from 20 Affinity Grid samples. (d) Western blot analysis of the gel shown in (c) developed with anti-His antibody to detect His-tagged TfR. The band at ~75 kDa represents monomeric TfR, whereas the band at ~150 kDa most likely represents dimeric TfR.
To test the potential use of Affinity Grids for preparing membrane protein samples, we tested their tolerance for detergents. We prepared His-tagged Tf-TfR complex (0.15 μg/ml) in buffers containing either 1% OG, 0.2% DM, 0.02% DDM, 0.03% Triton X-100, 0.014% Tween 20, 0.5% CHAPS, 0.2% Fos-choline 11 or 0.1% digitonin and applied 3-μl aliquots of these samples to 2% CC Affinity Grids. Samples were incubated for 30 seconds, 1, 2, 5, 10, 20, 30 and 60 minutes prior to negative staining and assessed by visual inspection in the electron microscope. We found that Affinity Grids were stable in the presence of 1% OG, 0.2% DM and 0.02% DDM for up to 30 minutes, producing useful specimens for single particle EM. After 60 minutes of incubation, the lipid monolayer started to show signs of degradation, such as the appearance of small holes in the lipid film or small (~5 – 10 nm), round lipid vesicles, which are easily distinguished from protein due to their very low contrast. This resulted in specimens with little or no Tf-TfR complexes present on the grid. In the presence of 0.03% Triton X-100 and 0.2% Fos-choline 11, the lipid monolayer was stable for up to 10 minutes before it began to dissolve. Samples in 0.5% CHAPS could only be incubated for 2 minutes and samples in 0.014% Tween-20 for 30 seconds before the monolayer degraded. 0.1% digitonin proved incompatible with Affinity Grids, even at incubation times as short as 30 seconds. To determine whether the detergent concentration has an influence on the stability of the Affinity Grid, we prepared Tf-TfR complex in 1, 2, 3, 4 and 5% OG on 2% CC Affinity Grids and incubated them for the same times prior to negative staining. We found that Affinity Grids can tolerate up to 2% OG for 20 – 30 minutes before substantial degradation of the lipid monolayer occurred, while 3% OG already began to destabilize the monolayer after 2 minutes. OG concentrations of 4 – 5% were incompatible with Affinity Grids even for very short incubation times. The results of these experiments (summarized in Table 1) suggest that Affinity Grids are compatible with most detergents, at least for some time at detergent concentrations not too far above the critical micelle concentration.
Table 1.
Stability of Affinity Grids in the presence of detergents as assessed by visual inspection of the specimens in the electron microscope.
| Detergent | CMC | 0.5 min | 1 min | 2 min | 5 min | 10 min | 20 min | 30 min | 60 min |
|---|---|---|---|---|---|---|---|---|---|
| 1% OG | 0.53% * | ++ | ++ | ++ | ++ | ++ | + | + | − |
| 2% OG | ++ | ++ | ++ | ++ | ++ | + | − | − | |
| 3% OG | + | + | + | − | − | − | − − | − − | |
| 4% OG | − | − | − | − − | − − | − − | − − | − − | |
| 5% OG | − − | − − | − − | − − | − − | − − | − − | − − | |
| 0.2% DM | 0.087% * | ++ | ++ | ++ | ++ | ++ | ++ | + | − |
| 0.02% DDM | 0.0087% * | ++ | ++ | ++ | ++ | ++ | ++ | + | − |
| 0.03% Triton X– 100 | 0.015% * | ++ | ++ | ++ | + | + | − | − | − − |
| 0.014% Tween 20 | 0.0072% * | + | − | − | − | − − | − − | − − | − − |
| 0.5% CHAPS | 0.49% * | ++ | + | + | − | − − | − − | − − | − − |
| 0.2% Fos– Choline 11 | 0.062% * | + | + | + | + | + | − | − | − − |
| 0.1% Digitonin | 0.075% ** | − | − − | − − | − − | − − | − − | − − | − − |
according to Anatrace Inc. (Maumee, OH); **according to Sigma Chemical Company (St. Louis, MO)
No apparent degradation of the lipid monolayer (continuous lipid monolayer)
Minor degradation of the lipid monolayer (appearance of some small holes in the lipid monolayer and of some small lipid vesicles)
Major degradation of the lipid monolayer (presence of many small holes in the lipid monolayer and of many small lipid vesicles)
Almost complete degradation of the lipid monolayer (large grid areas without lipid monolayer
We also tested the compatibility of Affinity Grids with glycerol, as glycerol is often used to stabilize protein complexes and is employed in certain cryo-negative staining protocols. We prepared His-tagged Tf-TfR complex (0.15 μg/ml) in buffer solution containing 1, 2, 3, 4 and 5% glycerol and applied 3-μl aliquots of each sample to 2% CC Affinity Grids. Samples were incubated for 1, 5, 10 and 15 minutes prior to extensive washing and negative staining. Affinity Grids were stable in the presence of up to 5% glycerol for at least a 5-minute incubation period. After 10 minutes of incubation, only glycerol concentrations of less than or equal to 3% produced useful samples with only minor lipid layer degradation. By 15 minutes, all glycerol concentrations tested caused substantial degradation of the lipid monolayer (see Table 2 for a summary of these results).
Table 2.
Stability of Affinity Grids in the presence of glycerol as assessed by visual inspection of the specimens in the electron microscope.
| % Glycerol | 1 min | 5 min | 10 min | 15 min |
|---|---|---|---|---|
| 1 | ++ | ++ | + | − |
| 2 | ++ | ++ | + | − |
| 3 | ++ | + | + | − |
| 4 | ++ | + | − | −− |
| 5 | + | + | − | −− |
No apparent degradation of the lipid monolayer (continuous lipid monolayer)
Minor degradation of the lipid monolayer (appearance of some small holes in the lipid monolayer and of some small lipid vesicles)
Major degradation of the lipid monolayer (presence of many small holes in the lipid monolayer and of many small lipid vesicles)
Almost complete degradation of the lipid monolayer (large grid areas without lipid monolayer)
Affinity Grid purification of ribosomal complexes
We previously demonstrated that native 50S ribosomal subunits containing His-tagged human rpl3 can be purified from E. coli extracts using monolayer purification 1. We now tested the Affinity Grid with the same system. We added a 3-μl drop of E. coli extract (~3 mg/ml) to a 2% CC Affinity Grid (Fig. 2e). After an incubation of 2 minutes, the sample was negatively stained and imaged in the electron microscope (Fig. 2a). The images showed the same kind of complexes, 20 to 30 nm in size, that we previously observed with specimens produced by conventional Ni-affinity chromatography and monolayer purification 1. To assess the composition of the complexes seen in the images, protein was eluted from 20 samples on 20% CC Affinity Grids and analyzed by mass spectrometry. The same proteins were present as previously identified in monolayer-purified samples 1; all of them were known ribosomal subunits and no contaminating proteins could be identified (Supplementary Table 2).
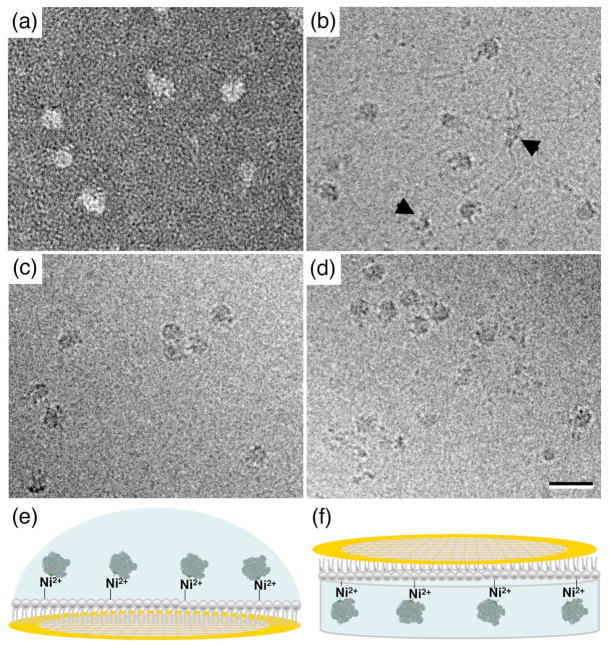
Fig. 2. Affinity Grid preparation of ribosomal complexes.
(a) Image of negatively stained ribosomal complexes adsorbed to a 2% CC Affinity Grid from E. coli cell lysate containing 60 mM imidazole. (b) Image of vitrified ribosomal complexes adsorbed to a 20% HC Affinity Grid from the same E. coli cell lysate. The image shows mRNA emanating from the ribosomal complexes. The arrowheads point to 30S ribosomal subunits. (c) Image of vitrified ribosomal complexes prepared by lipid monolayer purification using a 20% Ni-NTA lipid monolayer. No mRNA strands are visible. (d) Image of vitrified ribosomal complexes adsorbed to a 20% HC Affinity Grid from E. coli cell lysate that was treated with RNAse A. No mRNA strands are visible. Scale bar is 30 nm. (e, f) Illustrations of the two different methods used to prepare Affinity Grids of ribosomal complexes. Either a drop of cell extract was pipetted onto an Affinity Grid (e) or the Affinity Grid was placed on top of cell extract contained in a well of a teflon block (f).
To produce vitrified specimens, we added a 3-μl drop of E. coli extract to a 20% HC Affinity Grid. After a 2-minute incubation, the sample was blotted, quick-frozen in liquid ethane and imaged in the electron microscope (Fig. 2b). A comparison with images of vitrified samples prepared by monolayer purification (Fig. 2c) revealed distinct differences. First, as previously observed, clustering of the particles often seen in samples prepared by monolayer purification did not occur on the Affinity Grid. Second, while both samples contained mostly particles consistent in size with the 50S ribosomal subunit or the 70S ribosome, the Affinity Grid specimen also contained smaller particles that would be consistent in size and shape with a 30S ribosomal subunit (black arrows in Fig. 2b). Third, and most notably, the images obtained with the Affinity Grid sample showed that many of the ribosomal complexes were attached to ~15 Å thick, string-like densities, which we thought most likely represented mRNA strands. To test this interpretation, we incubated the extract with RNAse A for 30 minutes prior to applying it to an Affinity Grid. The images from this preparation no longer showed the string-like densities associated with the ribosomal complexes (Fig. 2d), confirming that they represented mRNA. mRNA was consistently present in Affinity Grid preparations of ribosomal complexes (Fig. 2b), but was always missing from preparations produced by monolayer purification (Fig. 2c). To obtain an Affinity Grid sample in a manner more resembling the way a monolayer purification sample is prepared, we placed 25 μl of extract into a well in a Teflon block and placed a 20% HC Affinity Grid on top of the extract (as illustrated in Fig. 2f). After a 30-minute incubation, the Affinity Grid was lifted off, blotted and vitrified. Images of this preparation did not show mRNA, while mRNA was visible on an Affinity Grid specimen prepared by pippetting a drop of the same extract onto an Affinity Grid (as illustrated in Fig. 2e) at the same time (data not shown). These results indicate that the presence of mRNA in Affinity Grid preparations depends on the exact way the sample was applied to the Affinity Grid.
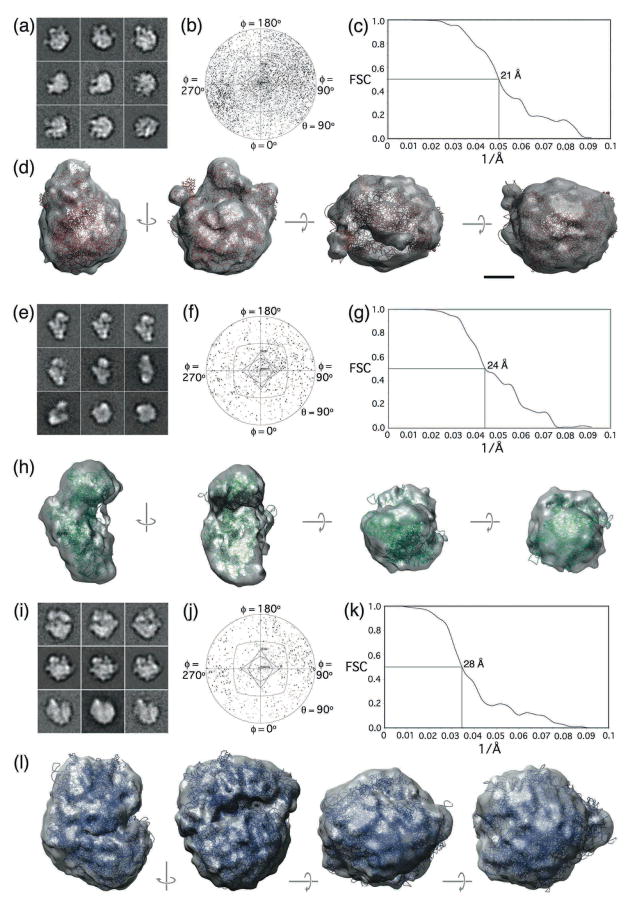
To calculate structures of ribosomal complexes, we selected 52,507 particles from 274 images of vitrified Affinity Grid specimens containing mRNA and classified them into 200 classes (Supplementary Fig. 1). To account for the heterogeneity in the class averages due to the presence of different ribosomal complexes, we created 30-Å density maps of the 30S, 50S and 70S ribosomal complexes based on the crystal structure of the E. coli 70S ribosome (pdb code: 1ML5; 2). We used these reference volumes to sort the class averages into groups representing the 50S ribosomal subunit (representative class averages shown in Fig. 3a), the 30S ribosomal subunit (representative class averages shown in Fig. 3e) and the 70S ribosome (representative class averages shown in Fig. 3i) using the normalized cross-correlation routine implemented in SPIDER 3. We then used FREALIGN 4 to calculate 3D reconstructions of the 50S subunit (Fig. 3d; 23,680 particles in the final map) at a resolution of 21 Å (Fig. 3c), the 30S subunit (Fig. 3h; 7,226 particles in the final map) at a resolution of 24 Å (Fig. 3g), and the 70S ribosome (Fig. 3l; 3,178 particles in the final map) at a resolution of 28 Å (Fig. 3k). The Euler angle distribution for each reconstruction showed that the particle orientations sampled the entire 3D space (Figs. 3b, f and j). Manual placement of the atomic models for each ribosomal complex into the corresponding density map demonstrated that the structural features of the density maps were consistent with the crystal structures (Fig. 3d, h and l).
Fig. 3. 3D reconstructions of vitrified ribosomal complexes purified on Affinity Grids.
(a–d) 3D reconstruction of the 50S ribosomal subunit showing (a) representative class averages, (b) the angular distribution plot, (c) the FSC curve indicating a resolution of 21 Å and (d) different views of the final density map with the fitted atomic model in red (pdb code: 1ML5, chains a - x 2). (e–h) 3D reconstruction of the 30S ribosomal subunit showing (e) representative class averages, (f) the angular distribution plot, (g) the FSC curve indicating a resolution of 24 Å and (h) different views of the final density map with the fitted atomic model in green (pdb code: 1ML5, chains A and C - X 2). (i–l) 3D reconstruction of the 70S ribosome showing (i) representative class averages, (j) the angular distribution plot, (k) the FSC curve indicating a resolution of 28 Å and (l) different views of the final density map with the fitted atomic model in blue (pdb code: 1ML5, all chains 2). The side length of individual panels in (a), (e) and (i) is 38 nm. The scale bar in (d) is 5 nm.
Affinity Grid purification of AQP9
Since the Affinity Grid proved to be sufficiently resistant to OG, we tested whether we could use it to isolate AQP9 from a membrane extract of Sf9 insect cell. Sf9 cells over-expressing His-tagged AQP9 were lysed and the membrane fraction was solubilized with 2% OG. Imidazole was added to a final concentration of 60 mM and the detergent-solubilized membranes were applied to a 2% CC Affinity Grid. In parallel, a conventional purification was performed for the same membrane fraction using Ni-affinity and gel filtration chromatography. Negatively stained specimens were prepared for the two samples and examined in the electron microscope.
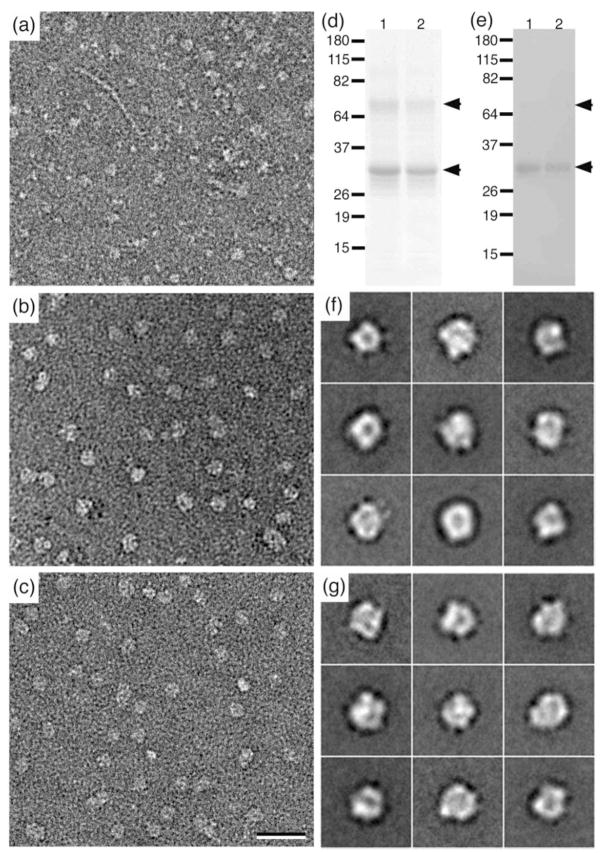
Negatively stained specimens of the Sf9 membrane extract prepared in the conventional way showed a variety of proteins, making it impossible to identify individual AQP9 tetramers (Fig. 4a). In contrast, specimens prepared by conventional Ni-affinity and gel filtration chromatography (Fig. 4b) and using an Affinity Grid (Fig. 4c) showed largely homogeneous particle populations. To assess the purity of the sample produced with the Affinity Grid, the protein was eluted from 20 samples on 40% CC Affinity Grids and analyzed by SDS-PAGE (Fig. 4d, lane 2) and Western blotting (Fig. 4e, lane 2) as well as by mass spectrometry. For comparison, AQP9 purified by conventional Ni-affinity chromatography was subjected to the same analyses (lane 1 in Figs. 4d and 4e). The SDS-PAGE gel and Western blot of the two samples show the two bands representing unglycosylated (~32 kDa) and glycosylated AQP9 (~35 kDa) that have been previously observed 5 as well as dimeric AQP9 (~68 kDa). In addition, the mass spectrometry results confirmed that AQP9 was the only protein present in the two samples (Supplementary Table 3). We then collected 15 images each of the chromatographically and Affinity Grid purified samples and selected 9,526 and 10,292 particles, respectively, that were each classified into 50 classes. The class averages of OG-solubilized AQP9 obtained with the Affinity Grid specimen (Fig. 4g) appeared to be of the same quality as those obtained with conventionally prepared sample (Fig. 4f). In both cases we observed particles about ~10 nm in size with a central stain accumulation, which looked like those previously reported for single particles of AQP9 5. Because of the small size of the AQP9 tetramer (~130 kDa), we did not attempt to prepare vitrified specimens of AQP9, but we are optimistic that it should be possible, with sufficiently large membrane proteins, to prepare vitrified specimens using Affinity Grids.
Fig. 4. Affinity Grid purification of His-tagged AQP9.
(a) Image of negatively stained Sf9 membrane extract. (b) Image of negatively stained AQP9 purified by Ni-affinity and gel filtration chromatography. (c) Image of negatively stained AQP9 adsorbed to a 2% CC Affinity Grid from Sf9 membrane extract containing 60 mM imidazole. Scale bar is 30 nm. (d, e) SimplyBlue stained SDS-PAGE gel (d) and Western blot (e) detecting His-tagged AQP9 (lanes 1: AQP9 purified by conventional chromatographic methods, lanes 2: AQP9 eluted from Affinity Grids). (f, g) Representative class averages of negatively stained AQP9 purified by conventional chromatographic methods (f) or by adsorption to an Affinity Grid (g). Side length of individual panels is 26 nm.
DISCUSSION
Affinity Grids in comparison to monolayer purification
We have recently introduced monolayer purification as an easy and rapid technique to purify a His-tagged protein or macromolecular complex from cell extract while simultaneously preparing a specimen suitable for single particle EM 1. While it is easy to cast a monolayer over a cell extract and to then pick it up with an EM grid, it does require this procedure be performed for every sample, potentially creating a threshold for the use of monolayer purification. We therefore set out to test whether we could produce an Affinity Grid featuring a dried, pre-deposited monolayer containing Ni-NTA lipids on a carbon-coated EM grid. If successful, the Affinity Grid would make it possible to simply apply a drop of extract to the pre-fabricated grid, which would specifically adsorb only His-tagged proteins while other proteins would be removed during the washing of the grid. Our test with His-tagged Tf-TfR and ribosomal complexes showed that the Affinity Grid indeed worked and produced samples that were of the same purity as those produced with monolayer purification. However, preparing an Affinity Grid sample takes much less time and is as convenient as preparing a normal EM grid while not requiring any plasma cleaning or glow discharging of the grid prior to sample application. Particularly useful is the fact that Affinity Grids are stable and can be stored under ambient conditions for at least six months. Thus, Affinity Grids can be produced at any time and used when needed.
Affinity Grids extend the applicability of monolayer purification
Monolayer purification depends on the formation of a lipid monolayer at the air-water interface. Since detergents and glycerol generally dissolve lipid monolayers (personal experience), these substances are incompatible with monolayer purification. Detergents are, however, used in the standard protocols to prepare yeast extracts and they are occasionally added in low concentrations to protein solutions to minimize aggregation through hydrophobic interactions. Detergents are also used to solubilize membrane proteins from cell membranes. Glycerol is another common additive that stabilizes proteins and complexes in solution, and it is also employed in certain cryo-negative staining procedures 6. The incompatibility of lipid monolayers with detergents and glycerol thus limits the use of monolayer purification. Since the lipid monolayer on the Affinity Grid is adsorbed to a carbon film, we propose that this interaction may be the reason why the lipid monolayer is more resistant to detergents and glycerol. With the exception of digitonin, which is known to be particularly potent in solubilizing lipids 7 and destroys the Affinity Grid almost immediately, the Affinity Grid is stable in most detergents for at least some period of time. The time of stability increases with decreasing detergent concentration. Similarly, Affinity Grids are only useful for a limited time if the sample contains glycerol, with the lifetime of the Affinity Grid again increasing with decreasing glycerol concentrations. As Affinity Grids were sufficiently stable in the presence of 2% OG, they could be used to directly adsorb His-tagged AQP9 from an insect cell membrane extract. For samples containing more aggressive detergents and/or a high concentration of glycerol, the best strategy would presumably be to minimize the required incubation time by using an Affinity Grid with a high percentage of Ni-NTA lipid. An alternative solution to this problem would be to prepare Affinity Grids with a lipid monolayer that contains fluorinated lipids as the filler lipid. Fluorinated lipids are more resistant to detergents 8 and have already been used to form two-dimensional (2D) crystals on lipid monolayers 9. Although not yet tested, fluorinated lipids, which are not commercially available at this time, may also be more resistant to glycerol.
When monolayer purification is used to prepare a specimen, the particles adsorbed to the lipid monolayer often seem to have a tendency to cluster (Figs. 1a and 2c). Interestingly, such clustering was not observed with samples prepared on Affinity Grids (Figs. 1b and 2b). Lipid monolayers are fluid and proteins adsorbing to them can easily diffuse, a feature that is exploited in the 2D crystallization of proteins on lipid monolayers 10. By contrast, the lipids constituting the monolayer on the Affinity Grid are attached to the carbon film. We hypothesize that this attachment makes the monolayer not only more resistant to detergents and glycerol, but also prevents or at least substantially reduces diffusion of the lipids. Clustering of proteins on monolayers thus appears to be due to affinity of the proteins for each other, with the lipids enabling the proteins to find each other by diffusion. Since the lipids in the monolayer are attached to the carbon film of the Affinity Grid, they are presumably prevented from diffusing thus also preventing particle clustering.
In summary, the Affinity Grid makes monolayer purification compatible for use with membrane proteins and protein solutions containing glycerol, and it is superior to monolayer purification in most cases because it prevents particle clustering. Once the technique for producing Affinity Grids has been mastered, the preparation of the grids is highly reproducible, and the grids are usually almost completely covered with the lipid monolayer.
Affinity Grid samples of ribosomal complexes differ depending on the preparation method
Unexpectedly, Affinity Grid samples of ribosomal complexes looked quite different from samples prepared by monolayer purification. Most notably, the ribosomal complexes were still associated with mRNA. The possibility that the RNA was simply degraded in the extract used to prepare the sample by monolayer purification was ruled out by preparing monolayer and Affinity Grid samples from the same extract. mRNA was again present in the Affinity Grid sample but not in the monolayer purification sample. Since preparing a monolayer purification sample takes about 20 minutes while preparing an Affinity Grid requires only about 2 minutes, another possibility was that the RNA was degraded in the additional time it took to prepare the grid by monolayer purification. However, when the Affinity Grid sample was prepared once the preparation of the monolayer purification grid was completed, the outcome did not change. Finally, we prepared an Affinity Grid sample by placing it onto the top of cell extract (Fig. 2f) rather than to place a drop of extract onto the Affinity Grid (Fig. 2e). In this preparation, mRNA was no longer visible. The reason why the two different procedures produce different samples is currently not clear, but the results were reproducible. It may be that placing extract onto the Affinity Grid is more gentle and thus allows the mRNA to remain on the monolayer, while lifting the Affinity Grid from the surface of the extract may be harsher and rip the mRNA off the monolayer. An alternative explanation may be that the surface tension at the air-water interface disrupts the RNA strands present in the sample when the specimen is prepared by placing the Affinity grid on top of the sample and then lifting it off from the aqueous solution.
Affinity Grid samples produced by placing cell extract on the grid showed not only the presence of mRNA, in addition to 50S and 70S complexes, but also revealed 30S ribosomal complexes. Since the 30S complex does not contain a His-tagged subunit (and accordingly was absent in samples prepared by monolayer purification 1), the only possibility for it to be present in Affinity Grid samples is that they were bound to mRNAs that, in turn, were adsorbed to the lipid monolayer through His-tagged ribosomal complexes. The gentleness of the Affinity Grid preparation thus seems to allow the preparation of large, functional, macromolecular assemblies that usually disintegrate during specimen preparation. The Affinity Grid may thus open a new avenue to the visualization of complex biological assemblies by cryo-EM or electron tomography that were not possible to isolate with previously available techniques.
MATERIALS AND METHODS
Expression of rpl3, AQP9, TfR and production of Tf-TfR complex
Rpl3 was expressed in E. coli, TfR was expressed in 293-T cells, and the Tf-TfR complex was produced as described in 1. AQP9 was expressed in Sf9 cells as described in 5.
Preparation of cell and membrane extracts
Sf9 and E. coli cell extracts were prepared as described in 1. Membrane extract from Sf9 cells was produced by centrifugation of 50 ml Sf9 cell extract at 100,000g for 30 minutes at 4°C. The pellet containing the cell membranes was homogenized in 100 ml buffer (20 mM Tris, pH 8.0, 300 mM NaCl) containing 2% octyl-β,D-glucoside (OG) (Anatrace, Inc., Maumee, OH). The homogenate was centrifuged at 100,000g for 30 minutes at 4°C, and the supernatant was used for Affinity Grid experiments.
Conventional purification of His-tagged Tf-TfR complex
Conventional Ni-affinity purification of His-tagged Tf-TfR complex was performed as described in 1.
Preparation of Affinity Grids
1,2-dilauryl-sn-glycero-3-phosphatidylcholine (DLPC) and 1,2-dioleoyl-sn-glycero-3-[N(5-amino-1-carboxypentyl)iminodi acetic acid] succinyl-nickel salt (Ni-NTA lipid) were purchased from Avanti Polar lipids (Alabaster, AL). Each lipid was reconstituted in chloroform to 1 mg/ml. A 25-μl buffer aliquot (20 mM Hepes, pH 7.9, 150 mM NaCl) was placed into the well of a Teflon block, and 1 μl of a lipid mixture (DLPC containing the desired percentage of Ni-NTA lipid in chloroform) was added on top of the aqueous solution to form a lipid monolayer at the air-water interface. The Teflon block was incubated in a sealed humid environment at 4°C for 15 minutes. A copper EM grid (400 mesh, Ted Pella, Redding, CA) covered with a continuous carbon film or a Quantifoil 2/1 holey carbon grid (400 mesh, Quantifoil Micro Tools GmbH, Germany) was placed onto the lipid monolayer. The grid was gently lifted off with forceps, blotted from the side (perpendicular to the grid) with Whatman #1 filter paper (Whatman International Ltd, Middlesex, England) and allowed to air-dry. In the case of Quantifoil 2/1 holey carbon grids, a thin layer of carbon was evaporated onto the carbon side of the grid for stabilization during storage. Affinity Grids prepared in this way were stored for up to 6 months in a closed grid box (Ted Pella) under ambient conditions of room temperature (~25°C) and humidity (not controlled) before use. To specify the type of Affinity Grids used for each set of experiments, we adapt the following nomenclature: X% CC/HC Affinity Grid, where the percentage denotes the proportion of Ni-NTA lipid in the monolayer and CC or HC denotes continuous or holey carbon film.
Affinity Grid purification of His-tagged Tf-TfR complex, ribosomal complexes and AQP9
Tf-TfR complex
2 μl of Tf-TfR complex (0.03 mg/ml) in 20 mM Hepes, pH 7.4, 150 mM NaCl was added to 38 μl of Sf9 cell extract (6 mg/ml). This mixture was diluted 1:10 using the same buffer, and a 3 μl aliquot was applied to a 20% HC Affinity Grid. The sample was incubated on the grid for 2 minutes prior to blotting and vitrification. Likewise, 2 μl of Tf-TfR complex was added to 38 μl of buffer solution (20 mM Hepes, pH 7.4, 150 mM NaCl) containing either glycerol (1 – 5% final concentration) or one of the following detergents: 1 – 5% OG (Anatrace), 0.2% n-decyl-β,D-maltoside (DM) (Anatrace), 0.02% n-dodecyl-β,D-maltoside (DDM) (Anatrace), 0.03% Triton X-100 (EMD Bioscsciences, San Diego, CA), 0.014% Tween 20 (EMD Biosciences), 0.5% CHAPS (Anatrace), 0.2% Fos-choline 11 (Anatrace) and 0.1% digitonin (Sigma-Aldrich, St. Louis, MO). 3 μl of a 1:10 dilution of Tf-TfR complex in solutions containing these detergents or glycerol was added to a 2% CC Affinity Grid and incubated for various times prior to negative staining.
Ribosomal complexes
Based on previous results 1, 60 mM imidazole and 20 mM MgCl2 (final concentrations) were added to 1 ml of E. coli extract containing His-tagged rpl3. 3 μl of this mixture was placed on a 2% CC Affinity Grid (for negative staining) or a 20% HC Affinity Grid (for vitrification). Samples were incubated for 2 minutes, blotted from the side and either negatively stained or vitrified.
In addition, 25-μl aliquots of the same mixture were placed into two tubes, and 1 μl of RNAse A (Ambion, Inc., Austin, TX) (~ 0.5 units) was added to one tube while 1 μl of buffer was added to the other tube. The samples were incubated for 30 minutes at 4ºC. Two 10-μl aliquots from each tube were placed into wells of a teflon block. One of the two wells from each sample was overlayed with a 20% Ni-NTA lipid monolayer while 20% HC Affinity Grids were placed on top of the other two wells and later recovered. In parallel, 3 μl aliquots of the mixtures with and without RNAse A were added to two 20% HC Affinity Grids. All samples were incubated for 30 minutes. For the samples in the teflon block, monolayers were recovered using Quantifoil holey carbon grids while the Affinity Grids were simply lifted off the wells. All samples were vitrified.
AQP9
60 mM imidazole (final concentration) was added to 1 ml solubilized Sf9 membranes. 3 μl of this mixture was placed on a 2% CC Affinity Grid and incubated for 5 minutes. The grid was washed with 7 drops of MilliQ water prior to negative staining.
Sample elution from Affinity Grids
A sample was prepared on a 20% CC Affinity Grid, excess solution was blotted off with filter paper and the grid was then incubated for 2 minutes with 20 μl of 300 mM imidazole. The 20-μl drop was recovered using a pipetteman and added to the next Affinity Grid sample. In this way, proteins adsorbed to 20 Affinity Grids were eluted into the same 20-μl volume of 300 mM imidazole.
SDS-PAGE, Western blotting and mass spectrometry
SDS-PAGE
Samples were run on 10% SDS-PAGE gels and stained either with Coomassie blue or SimplyBlue stain (Invitrogen Corporation, Carlsbad, CA). Western blotting: His-tagged proteins were detected with anti-His antibody (GE Healthcare, Buckinghamshire, UK) and developed by the alkaline phosphatase method using the Sigma Fast system (Sigma-Aldrich, St. Louis, MO).
Protein assay
Protein concentrations were determined using the BCA protein assay kit (Pierce, Rockford, IL).
Mass spectrometry
Entire lanes were excised from SimplyBlue stained SDS-PAGE gels and analyzed by liquid chromatography tandem mass spectrometry (LC/MS/MS) in the Taplin Biological Mass Spectrometry Facility at Harvard Medical School.
Specimen preparation
Negative staining
For conventionally purified protein samples, grids were negatively stained with 0.75% uranyl formate as described in 6 and grids of monolayer-purified samples were stained as described in 1. For Affinity Grids, 3-μl sample aliquots were placed on the Affinity Grids and incubated for various times. The grids were blotted from the side, washed with one drop of 0.75% uranyl formate and stained for 20 seconds with another drop of 0.75% uranyl formate. Affinity Grids of specimens in detergent solution were washed with 7 drops of MilliQ water before staining and grids of specimens in glycerol solution with 15 drops of MilliQ water.
Vitrification
Grids of monolayer-purified samples were vitrified as described in 1. For Affinity Grids, 3-μl sample aliquots were placed on Affinity Grids, which were blotted for 3 seconds and plunged into liquid ethane using a Vitrobot (FEI Company, Hillsboro, Oregon) operating at 22°C and 65% relative humidity.
Electron microscopy
Negatively stained specimens were imaged in an FEI Tecnai 12 electron microscope (FEI, Hillsboro, OR) equipped with a LaB6 filament and operated at an acceleration voltage of 120 kV. Images were recorded on imaging plates under low-dose conditions at a nominal magnification of 67,000x and a defocus value of about −1.5 μm. Imaging plates were scanned with a Ditabis scanner (Pforzheim, Germany) using a step size of 15 μm, a gain setting of 20,000 and a laser power setting of 30%. The images were binned over 2 x 2 pixels for a final sampling of 4.5 Å/pixel at the specimen level.
Grids of vitrified specimens were transferred into an FEI F20 electron microscope equipped with a field emission gun using an Oxford cryo-specimen holder, maintaining a temperature of −180°C. Samples were examined at an acceleration voltage of 200 kV and images were recorded on Kodak SO-163 film at a nominal magnification of 50,000x using low-dose procedures and a defocus ranging from −2 to −4 μm. Film negatives were developed for 12 minutes with full-strength Kodak D-19 developer at 20°C. Micrographs were digitized with a Zeiss SCAI scanner (Carl Zeiss Inc., Oberkochen, Germany) using a step size of 7 μm and binned over 3 x 3 pixels for a final sampling of 4.2 Å/pixel at the specimen level.
Image processing
Ribosomal complexes
WEB, the display program associated with the SPIDER software package 3, was used to select 52,507 particles from 274 images of vitrified specimens of ribosomal complexes, in which mRNA could also be seen. The particles were windowed into individual images of 90 x 90 pixels. Using the SPIDER software package, the particles were low-pass filtered to 20 Å, rotationally and translationally aligned and subjected to 10 cycles of multi-reference alignment. Each round of multi-reference alignment was followed by K-means classification into 200 classes. The references used for the first multi-reference alignment were randomly chosen from the raw images.
The EMAN software package 11 was used to calculate reference volumes filtered to 30 Å resolution based on the atomic model of the 70S ribosome (pdb code: 1ML5) 2 for the 70S ribosome (all chains) and the 50S (chains a - x) and 30S (chains A and C - X) ribosomal subunits. Re-projections from the reference volumes were calculated at 4° intervals and cross-correlated with the 200 experimental class averages. Class averages were assigned either to the 70S, 50S or 30S ribosomal complexes depending on a normalized cross-correlation coefficient of 0.8 or higher with re-projections of the respective reference volume. The particles belonging to the class averages were then used to create 3 image stacks corresponding to the 70S (3,973 particles from 10 classes), 50S (29,600 particles from 81 classes) or 30S (9,032 particles from 24 classes) ribosomal complexes.
Three independent reconstructions were calculated with FREALIGN version 7.05 4, which was used to determine and refine the orientation parameters for each particle and to correct for the contrast transfer function (CTF) of the microscope. The correct defocus value for each particle image was deduced from the position of each particle in the image and the tilt angles and defocus values of the images, which were determined with CTFTILT 12. FREALIGN was first run for one round using mode 3 (systematic parameter search) with an angular step of 7° to determine initial orientation parameters for each particle relative to the reference model. The resulting parameters were iteratively refined over 15 additional cycles running in mode 1 (local parameter refinement) including data in the 200 − 10 Å resolution range. Only particles with a weighted cross-correlation coefficient better than 0.8 were included in the final reconstructions, which were 23,680 particles for the 50S subunit, 7,226 particles for the 30S subunit, and 3,178 particles for the 70S ribosome. The final density maps were low-pass filtered according to their respective resolutions, which were estimated by Fourier shell correlation (FSC) with the FSC = 0.5 cut-off criterion 13.
AQP9
9,526 particles from the conventionally purified sample and 10,292 particles from the Affinity Grid sample were selected from 15 images of each specimen and windowed into 64 x 64 pixel images. The particles in each data set were classified into 50 classes as described above.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant GM62580 (to S. C. Harrison). The molecular EM facility at Harvard Medical School was established with a generous donation from the Giovanni Armenise Harvard Center for Structural Biology. We thank Matthias Wolf for discussions and advice on image processing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kelly DF, Dukovski D, Walz T. Monolayer purification: a rapid method for isolating protein complexes for single-particle electron microscopy. Proc Natl Acad Sci U S A. 2008;105:4703–4708. doi: 10.1073/pnas.0800867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klaholz BP, Pape T, Zavialov AV, Myasnikov AG, Orlova EV, Vestergaard B, Ehrenberg M, van Heel M. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature. 2003;421:90–94. doi: 10.1038/nature01225. [DOI] [PubMed] [Google Scholar]
- 3.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 4.Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–25. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Viadiu H, Gonen T, Walz T. Projection map of aquaporin-9 at 7 Å resolution. J Mol Biol. 2007;367:80–88. doi: 10.1016/j.jmb.2006.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification - powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore RJ, Wilson JD. Extraction of the reduced nicotinamide adenine dinucleotide phosphate:delta 4-3-ketosteroid-5-α-oxidoreductase of rat prostate with digitonin and potassium chloride. Biochemistry. 1974;13:450–456. doi: 10.1021/bi00700a009. [DOI] [PubMed] [Google Scholar]
- 8.Lebeau L, Lach F, Venien-Bryan C, Renault A, Dietrich J, Jahn T, Palmgren MG, Kühlbrandt W, Mioskowski C. Two-dimensional crystallization of a membrane protein on a detergent-resistant lipid monolayer. J Mol Biol. 2001;308:639–647. doi: 10.1006/jmbi.2001.4629. [DOI] [PubMed] [Google Scholar]
- 9.Levy D, Chami M, Rigaud JL. Two-dimensional crystallization of membrane proteins: the lipid layer strategy. FEBS Lett. 2001;504:187–193. doi: 10.1016/s0014-5793(01)02748-x. [DOI] [PubMed] [Google Scholar]
- 10.Uzgiris EE, Kornberg RD. Two-dimensional crystallization technique for imaging macromolecules, with application to antigen-antibody-complement complexes. Nature. 1983;301:125–129. doi: 10.1038/301125a0. [DOI] [PubMed] [Google Scholar]
- 11.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 12.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 13.Bottcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.