Abstract
Background
Most non‐syndromic congenital heart defects (CHD) are caused by a complex interaction between maternal lifestyle factors, environmental exposures, and maternal and fetal genetic variants. Maternal periconceptional intake of folic acid containing vitamin supplements is reported to decrease the risk of CHD. The 677C→T and 1298A→C polymorphisms in the methylenetetrahydrofolate reductase (MTHFR) gene decrease enzyme activity.
Objective
To examine the relation between CHD and maternal and fetal MTHFR polymorphisms
Methods
375 nuclear families were studied. The transmission/disequilibrium test was used to test for transmission distortion in complete triads. A log‐linear approach was used to test for associations between CHD and maternal and offspring polymorphisms, and to estimate independently the contributions of maternal and fetal variants to relative risks. Haplotype frequencies were estimated and a haplotype transmission disequilibrium test carried out.
Results
The 1298C allele was transmitted less often than expected (p = 0.0013). There was no distortion in the transmission of the 677T allele, neither was there evidence of a parent of origin effect in the transmission of either of the single nucleotide polymorphisms. The 677C–1298C haplotype was also transmitted less often than expected (p = 0.0020). The relative risk associated with inheriting one copy of the 1298C allele was 0.64 (95% confidence interval, 0.48 to 0.87) and the that associated with inheriting two copies of the 1298C allele, 0.38 (0.21 to 0.70).
Conclusions
The apparent protective effect of the MTHFR 1298C allele against CHD could have several explanations and further study is needed.
Keywords: congenital heart defect, MTHFR, polymorphism, folic acid
Congenital heart defects (CHD) are the most common structural birth defects, affecting approximately 8 to 10 of every 1000 live births. The aetiology of non‐syndromic CHD is complex, involving both genetic, epigenetic, and environmental risk factors.1 Periconceptional exposure to folic acid and multivitamins containing folic acid may modulate the maternal risk of having a CHD affected pregnancy.2
It is widely accepted that the impact of folic acid intake on pregnancy outcome is modified by variants in both maternal and fetal genes that code for critical enzymes in the folate and homocysteine pathways.3 The methylenetetrahydrofolate reductase (MTHFR, NM 005957) gene encodes for the enzyme that catalyses the conversion of 5,10‐methylenetetrahydrofolate to 5‐methyltetrahydrofolate. Two single nucleotide polymorphisms (SNPs) in MTHFR, 677C→T (rs1801133) and 1298A→C (rs1801131), are associated with decreased enzyme activity.4
We conducted a case–parent analysis of children with CHD and their parents, ascertained through a population based birth defect registry. In the present study, we examined the independent and interactive impact of the maternal and fetal MHTFR 677C→T and 1298A→C SNPs and their haplotypes on the risk of CHD.
Methods
Study population
The study population consisted of liveborn CHD cases and their parents. Complete triads (case and both parents), and incomplete triads (case with only one participating parent), were included in the study. Cases were identified and ascertained through the Arkansas Reproductive Health Monitoring System, a statewide birth defects registry. Inclusion criteria for cases were as follows:
resident of Arkansas at the time of the completion of the index pregnancy and at the time of enrolment in the study;
pregnancy ended with a live birth between March 1998 and August 2004;
a physician diagnosis of a non‐syndromic septal, conotruncal, or right or left sided obstructive CHD that was confirmed by antenatal or postnatal echocardiogram, surgery, and/or necropsy;
English or Spanish speaking;
cases completed participation in the National Birth Defects Prevention Study (NBDPS).5
Infants with a known single gene disorder, chromosomal abnormality, or syndrome were excluded; only cases of non‐syndromic CHD were included in this study. Further details regarding the NBDPS have been published previously.5
After written consent had been obtained, a blood or cheek cell sample was collected. The protocol and provisions for informed consent were reviewed and approved by the institutional review board at the University of Arkansas for Medical Sciences.
Maternal interview data
Mothers of cases were interviewed when they participated in the NBDPS, using a structured computer assisted telephone interview (CATI), which was specifically designed for this ongoing multisite case–control study. The ethnicity of the mother and the father, the maternal periconceptional smoking status, and the use of folic acid or multivitamins containing folic acid were reported by the mothers.
Genotyping
Genomic DNA was extracted from lymphocytes in whole blood or from buccal cells from the cases using the Gentra Puregene protocol (Gentra Systems, Minneapolis, Minnesota, USA) and was stored at −20°C until genotyped. The order of DNA samples in each plate and in each sample group (for example, white controls) was fixed, and included both positive and negative controls. Genotyping assays were carried out for MTHFR 1298A→C using Taqman Custom Genotyping assays from Applied Biosystems (ABI, Foster City, California USA). MTHFR 677C→T primers and probes were designed using Primer Express (ABI). The primer sequences are as follows: forward, 5′ TGGCAGGTTACCCCAAAGG; reverse, 5′ CAC AAAGCGGAAGAATGTGTCA; C‐probe, 5′ 6FAM‐TGATGAAATCGGCTCCCGCA; T‐probe, 5′ VIC‐TGATGATGAAATCGACTCCCGCA. The concentration of each primer and each probe were 900 nM and 200 nM, respectively. Thermal cycling conditions were 50°C for two minutes and 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 62°C for one minute. The polymerase chain reactions (PCRs) for all assays were done on the ABI PRISM 7700 sequence detector or the ABI 7900HT fast real time PCR system (ABI). A post‐PCR plate read was carried out for allelic discrimination, and both the real time and allelic discrimination data were analysed using the ABI sequence detection systems (SDS2.2.1) software.
Statistical analysis
Before the data analysis, genotypic data were checked for Mendelian segregation errors.6 Families with unresolved transmission errors were dropped from the analysis. CHD cases and their parents were tested independently for Hardy–Weinberg (HW) equilibrium using the exact test implemented in Stata's GENHW command.7
The transmission disequilibrium test (TDT) was used to test for allelic associations resulting from linkage disequilibrium between a CHD punitive disease locus and the 677C→T and 1298A→C polymorphisms and also to test for parent of origin effects.8,9 Because the TDT is invalid when data from ambiguous parent–child dyads are excluded and only unambiguous parent–child dyads are included,10 only data from complete triads were analysed. The TDT tests were carried out using the exact TDT implemented in the Stata program SYMMETRY.11
To make use of data from all available families, including mother–child and father–child dyads, the log‐linear based approach and expectation/maximisation (EM) algorithm suggested by Weinberg et al for the analysis of case–parent data were used to test for associations between CHD and the maternal and the offspring MTHFR polymorphisms, and to estimate separately the contributions to the overall genetic relative risks (RR) due to maternal and offspring genetic factors.12,13,14 Additionally, the log‐linear model was used to estimate offspring and maternal RR assuming dominant and recessive models of inheritance. The log‐linear analyses were carried out using the GENECMT program (http://www.biostat‐resources.com/stata).
Haplotype analysis was undertaken using the TRANSMIT program, version 2.5.4.15 TRANSMIT reconstructs haplotypes in the presence of ambiguous phase and missing parental genotype data using an EM algorithm. The program was used to test for haplotype transmission distortion and significance probabilities computed by bootstrap based on 10 000 replicates. The linkage disequilibrium (LD) between the two MTHFR SNPs was estimated using Lewontin's standardised disequilibrium coefficient D, as implemented in the computer program GOLD.16,17
Results
In all, 375 case families were eligible for the study; 92.5% of the families (n = 347) were successfully genotyped for MTHFR 677C→T, 91.5% (n = 343) for 1298A→C, and 84.0% (n = 315) for both SNPs (table 1).
Table 1 Summary of the numbers of case triads/dyads successfully genotyped for MTHFR 677C→T and 1298A→C polymorphisms.
| MTHFR 677C→T | MTHFR 1298A→C | Both polymorphisms | ||||
|---|---|---|---|---|---|---|
| Complete trios | 284 | 268 | 249 | |||
| Mother–child dyads | 57 | 67 | 55 | |||
| Father–child dyads | 6 | 8 | 11 | |||
| Total | 347 | 343 | 315 |
MTHFR, methylenetetrahydrofolate reductase.
Based on the parents of the cases, the frequencies of the MTHFR 677T allele were 32.4% for whites, 12.5% for African‐Americans, and 40.9% for Hispanics. Similarly, allele frequencies for the MTHFR 1298C allele were 29.6% for whites, 17.5% for African‐Americans, and 16.7% for Hispanics. The observed genotype counts did not deviate significantly from HW equilibrium.
Genotype frequencies
The frequencies of each allele, individual genotype, and combined genotype, assuming either a dominant or recessive mode of inheritance for CHD cases and their parents, are presented in table 2. The 677T allele was present in approximately 30% of cases and their parents. The homozygous 677 TT genotype was present in 10.4% of cases, 11.7% of their mothers, and 7.9% of their fathers. Under a dominant model, 51.3% of CHD cases had either the 677 CT or TT genotype, which approximated the frequency of these genotypes in their parents. The 1298C allele was present in 23.8% of CHD cases, 27.5% of mothers, and 28.6% of fathers. A greater percentage of parents had the 1298 AC or CC genotype (47.2% of mothers and 47.1% of fathers) than children (40.8%).
Table 2 Distribution of MTHFR alleles and genotypes among CHD cases and their parents.
| Cases | Mothers | Fathers | ||||
|---|---|---|---|---|---|---|
| 677C→T | (n = 347) | (n = 341) | (n = 290) | |||
| 677C | 480 (69.2%) | 470 (68.9%) | 403 (69.5%) | |||
| 677T | 214 (30.8%) | 212 (31.1%) | 177 (30.5%) | |||
| 677CC | 169 (48.7%) | 169 (49.6%) | 136 (46.9%) | |||
| 677CT | 142 (40.9%) | 132 (38.7%) | 131 (45.2%) | |||
| 677TT | 36 (10.4%) | 40 (11.7%) | 23 (7.9%) | |||
| 677CT/677TT | 178 (51.3%) | 172 (50.4%) | 154 (53.1%) | |||
| 677CC/677CT | 311 (89.6%) | 301 (88.3%) | 267 (92.1%) | |||
| 1298A→C | (n = 343) | (n = 335) | (n = 276) | |||
| 1298A | 523 (76.2%) | 486 (72.5%) | 394 (71.4%) | |||
| 1298C | 163 (23.8%) | 184 (27.5%) | 158 (28.6%) | |||
| 1298AA | 203 (59.2%) | 177 (52.8%) | 146 (52.9%) | |||
| 1298AC | 117 (34.1%) | 132 (39.4%) | 102 (37.0%) | |||
| 1298CC | 23 (6.7%) | 26 (7.8%) | 28 (10.1%) | |||
| 1298AC/1298CC | 140 (40.8%) | 158 (47.2%) | 130 (47.1%) | |||
| 1298AA/1298AC | 320 (93.3%) | 309 (92.2%) | 248 (89.9%) |
CHD, congenital heart defect; MTHFR, methylenetetrahydrofolate reductase.
Haplotype frequencies
The haplotype frequencies of the CHD cases and their parents are shown in table 3. As expected, haplotype analyses indicated near complete linkage disequilibrium between the MTHFR 677 and the 1298 locus (D′ = 0.985). The most common haplotype among cases and their parents was the 677C‐1298A. Fewer cases (24.0%) had the 677C‐1298C haplotype than either mothers (27.0%) or fathers (28.7%).
Table 3 Estimated haplotype frequencies of the MTHFR 677C→T and 1298A→C single nucleotide polymorphisms.
| Haplotype | Cases (%) | Mothers (%) | Fathers (%) | All | ||||
|---|---|---|---|---|---|---|---|---|
| 677C – 1298A | 44.6 | 41.9 | 40.7 | 42.0% | ||||
| 677C – 1298C | 24.0 | 27.0 | 28.7 | 25.5% | ||||
| 677T – 1298A | 31.2 | 31.1 | 30.3 | 30.9% | ||||
| 677T – 1298C | 0.3 | 0.0 | 0.3 | 1.6% |
MTHFR, methylenetetrahydrofolate reductase.
Transmission disequilibrium
Table 4 presents the transmission of the 677T allele and the 1298C allele from heterozygous case parents to CHD affected offspring. Of the 241 informative transmissions, there was no evidence of distortion of the 677T allele. In contrast, among the 205 informative transmissions in cases, the 1298C allele was transmitted less frequently than would be expected based on a Mendelian inheritance (p = 0.0013).
Table 4 Transmission of MTHFR variant alleles from heterozygous case parents to CHD‐affected offspring (TDT results using completed triads only).
| Allele | Transmitted | Not transmitted | Exact p value | |||
|---|---|---|---|---|---|---|
| 677T* | 124 (51.5%) | 117 (48.5%) | 0.6992 | |||
| 1298C† | 79 (38.5%) | 126 (61.5%) | 0.0013 |
*241 informative transmissions.
†205 informative transmissions.
MTHFR, methylenetetrahydrofolate reductase.
The 677C‐1298A haplotype was transmitted significantly more often than expected (p = 0.0159). In contrast, the 677C‐1298C haplotype was transmitted significantly less often than expected (p = 0.0020; table 5).
Table 5 Haplotype frequencies and tests of association for haplotypes defined by the MTHFR gene variants 677C→T and 1298A→C.
| Haplotype | Estimated haplotype frequency | Observed | Expected | Var(O−E) | χ2(1 df) | p Value* | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 677C‐1298A | 41.6% | 333.54 | 312.72 | 74.87 | 5.79 | 0.0159 | ||||||
| 677C‐1298C | 27.7% | 183.78 | 208.32 | 62.90 | 9.57 | 0.0020 | ||||||
| 677T‐1298A | 30.4% | 233.32 | 228.32 | 68.85 | 0.36 | 0.5365 | ||||||
| 677T‐1298C | 0.4% | 1.35 | 2.64 | 0.98 | 1.69 | 0.1947 |
Global χ2 test on 3 degrees of freedom = 12.17, p = 0.0079.
*Significant probabilities derived from the haplotype based TDT (TRANSMIT) based on 10 000 bootstrap samples and robust standard errors.
E, expected; MTHFR, methylenetetrahydrofolate reductase; O, observed; Var, variance.
Impact of variant alleles on relative risk of CHD
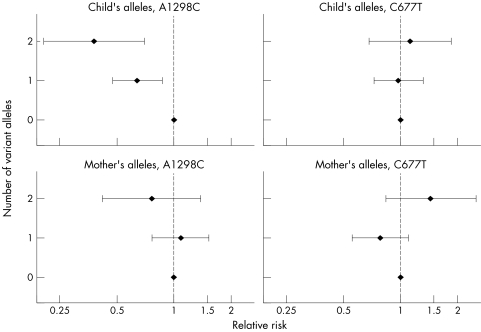
Figure 1 shows the relative risks of CHD associated with both the maternal and fetal variant of each of the two alleles. The child's 1298C allele was associated with a decreased risk of CHD (p<0.001), and the protective effect appeared greater when the child inherited two copies of the variant allele compared with inheriting only one: the relative risk among offspring who inherited one copy of the 1298C allele was 0.64 (95% CI, 0.48 to 0.87) and for those inheriting two copies it was 0.38 (0.21 to 0.70). Neither the fetal MTHFR 677T allele nor either of the mother's variant alleles was significantly associated with the risk of CHD.
Figure 1 Estimated relative risk of congenital heart defects associated with the child (top panel) or mother (bottom panel) carrying zero, one, or two copies of the variant allele (results from log‐linear model using all available families).
Interaction between genetic variants and folic acid
We investigated the interaction between MTHFR 677C→T and 1298A→C and the periconceptional use of folic acid or multivitamins containing folic acid. The periconceptional period was defined as approximately one month before conception through the second month of pregnancy. Of the 375 mothers in the study, 301(80%) reported taking either folic acid or a multivitamin containing folic acid during this folic acid period. The effect of the variant allele was calculated separately for each stratum of periconceptional folic acid use, assuming a dominant effect of the variant allele and HW equilibrium. The relative risks associated with each maternal variant allele remained non‐significant, with point estimates near 1 for those who used folic acid periconceptionally and those who did not. The apparent protective effect of the 1298C allele was similar between children whose mothers used folic acid periconceptionally (RR = 0.62 (95% CI, 0.44 to 0.86)) and children whose mothers did not use folic acid (RR = 0.64 (0.31 to 1.33)).
Discussion
In this population based case–parent study, we examined the impact of two polymorphisms in the MTHFR gene that codes for a critical enzyme in the folate pathway. The frequency of the genotypes and haplotypes in the parents in our sample are consistent with findings of other investigators.18
We did not observe an effect of the MTHFR 677T allele on CHD frequency even after controlling for use of folic acid supplements. Our study was carried out after the introduction of folate food fortification in the USA, which resulted in a doubling of serum folate among women of childbearing age in the general population.19 Increased homocysteine and decreased serum folate are most pronounced in individuals with the MTHFR TT genotype who have a low folate intake.20 We cannot exclude a detrimental effect of the MTHFR 677C→T polymorphism among women with low dietary folate intake.
Our results indicate that the MTHFR 1298C allele was transmitted to infants with CHD less often than would be expected based on independent segregation. The 677C‐1298C haplotype was also transmitted less often than would be expected. The results of the log‐linear analysis, which used data from all of the available families, were consistent with the results of the TDT and provide direct estimates of disease risk associated with allelic variants. The estimated relative risk of CHD was lower in children with one or two copies of the 1298C allele than in children with the homozygous wild type.
There are at least four possible interpretations of our findings. First, the fetal 1298C allele in each of these analyses appears to protect against CHD. Other case–parent studies have examined the association between MTHFR 677C→T and 1298A→C and orofacial clefts and neural tube defects.18,21 In two of these studies, one on neural tube defects,21 and one on orofacial clefts,18 the 1298C appeared to have a protective effect. Both groups of investigators explained these findings by noting the strong linkage disequilibrium between the 677T allele and the 1298A allele. Because the normal allele at one marker (1298A) almost always travels with the variant allele at the other marker (677T), the investigators of these two studies postulate that the adverse effect of the MTHFR 677T allele caused an apparent, but spurious, protective effect of 1298C.18,21
We provide three alternative explanations for the observed protective effect of the MTHFR 1298C allele. First, it is possible that the 1298A→C polymorphism could be in linkage disequilibrium with another nearby yet undetected functional polymorphism in the MTHFR gene. Second, as is the case for some cancers, the MTHFR 677T and 1298C alleles could be protective. Multiple studies have shown that the MTHFR 677T allele and the 1298C allele are associated with a decreased activity of the enzyme, which results in more methylene‐THF at the expense of methyl‐THF.22 The MTHFR enzyme is at a metabolic branch point that directs folate towards homocysteine remethylation of methionine at the expense of DNA and RNA biosynthesis.22 Cancer researchers postulate that the protective effect of the MTHFR 677T allele is related to the abundant purines and pyrimidines available for DNA synthesis leading to error‐free DNA synthesis.23 It is plausible that a decrease in fetal MTHFR activity associated with 1298C allele exerts a true protective effect during cardiogenesis when rapid cell proliferation and error‐free DNA synthesis are critical. It might be that the slightly lower activity associated with the C allele provides a better balance between methylation and DNA synthesis. The protective effect may be even greater when folic acid is taken periconceptionally, because the additional folic acid protects maternal homocysteine remethylation of methionine while the fetal 1298C allele enhances error‐free DNA synthesis.
A third explanation for the apparent protective effect of the 1298C allele is the selective survival of those with the 1298A allele. The 1298C allele is more common in fetal tissue than in neonatal tissue.24 Thus our observation of a decreased transmission could be an artefact caused by decreased viability of fetuses with this variant allele.
Further investigation using large scale population based investigations of genetic variants and environmental modifiers that affect DNA synthesis and methylation is indicated. Both DNA synthesis and methylation are essential for normal fetal growth and development.25 A microenvironment that provides the most favourable balance between DNA synthesis and cellular methylation would be expected to promote optimal fetal development and survival.
Acknowledgements
We thank Macy Guan for DNA extraction, Melissa Nicholas for database management, William J Gabello, Mary Dornhoffer, and the Office of Grants and Scientific Publications at the University of Arkansas for Medical Sciences, and Cynthia Bond for editorial assistance during the preparation of this manuscript. We also thank Sadia Malik MD, MPH for interpreting and classifying cardiovascular defects. We appreciate and acknowledge the generous participation of the many study families who made this work possible.
Abbreviations
CHD - congenital heart defect
LD - linkage disequilibrium
MTHFR - methylenetetrahydrofolate reductase
NBDPS - National Birth Defects Prevention Study
SNP - single nucleotide polymorphism
TDT - transmission disequilibrium test
Footnotes
This study was supported by grants from the National Institute of Child Health and Human Development No 5R01 HD39054‐05, National Center for Research Resources No 1C06 RR16517‐01 and 3C06 RR16517‐01S1, and Cooperative Agreement No U50/CCU613236‐08 from the Centers for Disease Control and Prevention (CDC). Funding was also provided by the Arkansas Biosciences Institute (ABI); a partnership of scientists from Arkansas Children's Hospital; Arkansas State University; the University of Arkansas Division of Agriculture; the University of Arkansas, Fayetteville; and the University of Arkansas for Medical Sciences. The contents of this letter to JMG are solely the responsibility of the authors and do not necessarily represent the official views of the CDC, National Institutes of Health, or ABI.
Conflicts of interest: none declared
References
- 1.Botto L D, Correa A. Decreasing the burden of congenital heart anomalies: an epidemiologic evaluation of risk factors and survival. Prog Pediatr Cardiol 200318111–121. [Google Scholar]
- 2.Botto L D, Mulinare J, Erickson J D. Do multivitamin or folic acid supplements reduce the risk for congenital heart defects? Evidence and gaps. Am J Med Genet 2003121A95–101. [DOI] [PubMed] [Google Scholar]
- 3.Doolin M T, Barbaux S, McDonnell M, Hoess K, Whitehead A S, Mitchell L E. Maternal genetic effects, exerted by genes involved in homocysteine remethylation, influence the risk of spina bifida. Am J Hum Genet 2002711222–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botto L D, Yang Q. 5,10‐Methylenetetrahydrofolate reductase gene variants and congenital anomalies: a HuGE review. Am J Epidemiol 2000151862–877. [DOI] [PubMed] [Google Scholar]
- 5.Yoon P W, Rasmussen S A, Lynberg M C, Moore C A, Anderka M, Carmichael S L, Costa P, Druschel C, Hobbs C A, Romitti P A, Langlois P H, Edmonds L D. The National Birth Defects Prevention Study. Public Health Rep 200111632–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu J, Turner A, Little J, Bleecker E R, Meyers D A. Positive results in association studies are associated with departure from Hardy‐Weinberg equilibrium: hint for genotyping error? Hum Genet 200211573–574. [DOI] [PubMed] [Google Scholar]
- 7.Cleves M A. Hardy‐Weinberg equilibrium tests and allele frequency estimation. STATA Technical Bull 19994834–37. [Google Scholar]
- 8.Spielman R S, McGinnis R E, Ewens W J. Transmission test for linkage disequilibrium: The insulin gene region and insulin‐dependent diabetes mellitus (IDDM). Am J Hum Genet 199352506–516. [PMC free article] [PubMed] [Google Scholar]
- 9.Ewens W J, Spielman R S. The transmission/disequilibrium test: history, subdivision, and admixture. Am J Hum Genet 199557455–464. [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis D, Sham P C. A note on the application of the transmission disequilibrium test when a parent is missing. Am J Hum Genet 199556811–812. [PMC free article] [PubMed] [Google Scholar]
- 11.Cleves M A, Olson J M, Jacobs K B. Exact transmission‐disequilibrium tests with multiallelic markers. Genet Epidemiol 199714337–347. [DOI] [PubMed] [Google Scholar]
- 12.Weinberg C R, Wilcox A J, Lie R T. A log‐linear approach to case‐parent‐triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am J Hum Genet 199862969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilcox A J, Weinberg C, Lie R. Distinguishing the effects of maternal and offspring genes through studies of “Case‐Parent Triads”. Am J Epidemiol 1998148893–901. [DOI] [PubMed] [Google Scholar]
- 14.Weinberg C R. Allowing for missing parents in genetic studies of case‐parent triads. Am J Hum Genet 1999641186–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clayton D. A generalization of the transmission/disequilibrium test for uncertain‐haplotype transmission. Am J Hum Genet 1999651170–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewontin R C. On measures of gametic disequilibrium. Genetics 1988120849–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abecasis G R, Cookson W O. GOLD – graphical overview of linkage disequilibrium. Bioinformatics 200016182–183. [DOI] [PubMed] [Google Scholar]
- 18.Jugessur A, Wilcox A J, Lie R T, Murray J C, Taylor J A, Ulvik A, Drevon C A, Vindenes H A, Abyholm F E. Exploring the effects of methylenetetrahydrofolate reductase gene variants C677T and A1298C on the risk of orofacial clefts in 261 Norwegian case‐parent triads. Am J Epidemiol 20031571083–1091. [DOI] [PubMed] [Google Scholar]
- 19. Folate status in women of childbearing age, by race/ethnicity‐‐United States, 1999–2000. MMWR 200251808–810. [PubMed] [Google Scholar]
- 20.de Bree A, Verschuren W M, Bjorke‐Monsen A L, van der Put N M, Heil S G, Trijbels F J, Blom H J. Effect of the methylenetetrahydrofolate reductase 677C→T mutation on the relations among folate intake and plasma folate and homocysteine concentrations in a general population sample. Am J Clin Nutr 200377687–693. [DOI] [PubMed] [Google Scholar]
- 21.Parle‐McDermott A, Mills J L, Kirke P N, O'Leary V B, Swanson D A, Pangilinan F, Conley M, Molloy A M, Cox C, Scott J M, Brody L C. Analysis of the MTHFR 1298A→C and 677C→T polymorphisms as risk factors for neural tube defects. J Hum Genet 200348190–193. [DOI] [PubMed] [Google Scholar]
- 22.Ueland P M, Hustad S, Schneede J, Refsum H, Vollset S E. Biological and clinical implications of the MTHFR C677T polymorphism. Trends Pharmacol Sci 200122195–201. [DOI] [PubMed] [Google Scholar]
- 23.Fang J Y, Xiao S D. Folic acid, polymorphism of methyl‐group metabolism genes, and DNA methylation in relation to GI carcinogenesis. J Gastroenterol 200338821–829. [DOI] [PubMed] [Google Scholar]
- 24.Zetterberg H, Regland B, Palmer M, Ricksten A, Palmqvist L, Rymo L, Arvanitis D A, Spandidos D A, Blennow K. Increased frequency of combined methylenetetrahydrofolate reductase C677T and A1298C mutated alleles in spontaneously aborted embryos. Eur J Hum Genet 200210113–118. [DOI] [PubMed] [Google Scholar]
- 25.Hobbs C A, Cleves M A, Lauer R M, Burns T L, James S J. Preferential transmission of the MTHFR 677 T allele to infants with Down syndrome: implications for a survival advantage. Am J Med Genet 20021139–14. [DOI] [PubMed] [Google Scholar]