Abstract
Background
A myocyte enhancer factor 2A (MEF2A) mutation that segregated with coronary artery disease/myocardial infarction (CAD/MI) in a large family has recently been described. Missense mutations in sporadic coronary artery disease patients were also reported. These data suggest that mutations in exons 7 and 11 of MEF2A cause CAD/MI, though the association was refuted by another study.
Objective
To analyse the genetic variation of exons 7 and 11 in a large cohort of Spanish CAD/MI patients and controls.
Methods and results
A rare polymorphism, P279L, was detected both in patients and controls. Carriers of the 279Leu allele had a threefold risk of suffering CAD/MI compared with controls (p = 0.009; odds ratio = 3.06 (95% confidence interval, 1.17 to 8.06)). In the controls the allele was found only in those under 50 years of age. Exon 11 showed a high degree of heterogeneity caused by a polyglutamine (CAG)n polymorphism, but no significant differences in genotype or allelic frequencies were found.
Conclusions
The 279Leu allele appears to be a genetic risk factor for CAD/MI in the population studied. This effect could be the result of a reduced transcriptional activity on MEF2A with 279Leu.
Keywords: myocardial infarction, risk factors, gene, MEF2A
Myocyte enhancer factor 2 (MEF2) is a family of transcription factors composed of four members: MEF2A, MEF2B, MEF2C, and MEF2D. The MEF2 transcription factors bind to their cognate DNA sequence CAT(A/T)4TAG/A present in the regulatory regions of several genes.1 The promoter regions of various cardiac genes contain MEF2 binding sequences, and MEF2 could regulate inducible gene expression in muscle and endothelium of coronary arteries, among other cell types.1,2,3
Several acquired and inherited risk factors for coronary artery disease (CAD) and its most frequent complication, myocardial infarction (MI), have been described. Coronary artery disease is caused by the development of atherosclerotic lesions in the walls of coronary arteries, and factors such as high cholesterol levels in blood, obesity, diabetes, and high blood pressure contribute to the origin and progression of atherosclerosis, are risk factors for CAD/MI.4 A family history of coronary artery disease is also a significant risk factor for the disease, and in most of the patients the genetic susceptibility lies in common DNA polymorphisms in candidate genes.4,5 In addition, coronary artery disease segregates as a mendelian trait in several families. Some of the loci for these familial forms have been mapped to human chromosomes, but only recently has a candidate gene been identified.2,6 MEF2A is located on chromosomal 15q26 region, which contains a susceptibility locus for familial CAD/MI; a seven amino acid deletion in exon 11 of MEF2A, which disrupts the transcriptional activation activity of MEF2A, has been identified as the causative mutation in a large CAD/MI family.2 Subsequent analysis of the whole MEF2A gene in sporadic cases of coronary artery disease identified several putative missense mutations in exon 7 in as many as 2% of affected patients. These mutations would reduce the activation activity of MEF2A.7
Although these initial studies supported the involvement of MEF2A variants in the risk for CAD/MI, a recent report identified these variants in healthy individuals, thus raising the possibility that they were rare DNA polymorphisms not directly related to the risk for coronary artery disease.8 In order to clarify this issue, we searched for MEF2‐variants in a large cohort of Spanish CAD/MI patients and healthy controls, and compared the frequencies between the two groups.
Methods
Patients and controls
We studied 483 Spanish patients. These patients attended our cardiology departments between 1998 and 2004, and suffered a first episode of myocardial infarction defined according to the WHO criteria.9 Coronary angiograms were carried out on all patients. According to the angiographic appearance, a vessel was regarded as diseased if it contained at least one stenosis involving more than 30% loss of lumen diameter.
The control group consisted of 1189 healthy Spanish individuals who had not suffered from cardiovascular disease. Data on clinical history were collected directly from controls by a normalised questionnaire. Ischaemic coronary, cerebral, and peripheral vascular diseases were excluded on the basis of clinical history. The clinical characteristics of the subjects are shown in table 1.
Table 1 Clinical characteristics of the patients with myocardial infarction and the controls.
| Patients (n = 483) | Controls (n = 1189) | |
|---|---|---|
| Age (years) (mean (SD)) | 54 (12) | 52 (12) |
| Smoker | 406 (84%) | 440 (37%) |
| Hypertensive | 188 (39%) | 262 (22%) |
| Diabetes | 58 (12%) | 120 (10%) |
| Male | 430 (89%) | 951 (80%) |
| ⩾1 diseased vessel | 459 (95%) | – |
Values are n (%) unless stated otherwise.
Subjects included in the study were all white and were from two Spanish regions (Asturias in northern Spain and Murcia in southeastern Spain). They all gave their informed consent for their participation in the study, which was approved by the ethics committee of Hospital Universitario Central de Asturias.
Single strand conformation analysis of MEF2A
DNA was obtained from patients and controls following a salting out method.10 Exons 7 and 11 of MEF2A were polymerase chain reaction (PCR) amplified with primers obtained from the MEF2A Ensembl Transcript (ID ENST 00000346108): GTTTGTGCCAAAGTATTTTAA and AAGAATGAAGTTGAAGAAAGG (exon 7, annealing at 58°C), and CTTGCAAGCCATCTGACC and CCATCCTCATTCGCTTTACAG (exon 11, annealing at 58°C). Each reaction contained 100 ng of genomic DNA, 10 pmol of each primer, 2mM MgCl2, and 2mM of each dNTP, in a final volume of 20 μl. After 32 PCR cycles of 95° for 30 seconds, 58°C for 40 seconds, and 72°C for 40 seconds, 3 μl of each reaction were mixed with 15 μl of deionised formamide and denatured at 95°C; 10 μl were then electrophoresed on 12% polyacrylamide gels (29:1 acrylamide:bisacrylamide; 30 cm length). Electrophoresis was for 18 hours at 6 W and room temperature. Gels were silver stained to visualise the electrophoretic pattern of each sample.
The nucleotide sequences corresponding to all the different electrophoretic patterns identified through single strand conformation analysis (SSCA) were characterised by direct sequencing of purified PCR fragments on an automated ABI310 system, using the PCR primers and BigDye chemistry.
Genotyping of the MEF2A (CAG)n polymorphism
The number of repeats of the polymorphic polyglutamine (CAG) tract in exon 11 of MEF2A was determined in each patient and control through PCR, followed by capillary electrophoresis in an automated system. Briefly, 50 ng of genomic DNA were amplified in a final volume of 15 μl with primers CTTGCAAGCCATCTGACC and CCGATCACTGCCATCATAGG (annealing at 58°C). The forward primer was 5′‐end fluorescence labelled with FAM. After 30 PCR cycles, 5 μl of each reaction were mixed with 50 μl of formamide, heated at 95°C for five minutes, size fractioned through capillary electrophoresis for 20 minutes in an ABI 310 genetic analyser, and analysed with the GENESCAN software.
Statistical analysis
Allele and genotype frequencies in patients and controls were compared with a χ2 test. The χ2 test was also used to determine whether the observed genotype frequencies in patients and controls differed from those expected under the Hardy–Weinberg equilibrium. Odds ratios (OR) with 95% confidence intervals (CI) were obtained to calculate the relative risk of myocardial infarction associated with the genotypes. The power (per cent chance of detection) was also calculated. All statistical analyses were done using the SPSS statistical package (v.11.0) and EpiInfo v.3.3 software.
Results
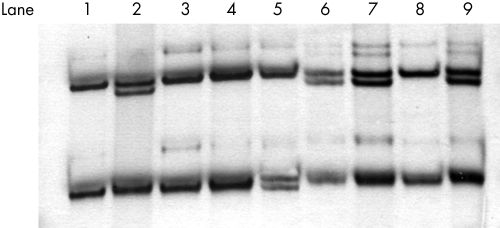
We used SSCA and direct sequencing to analyse exon 7 of the MEF2 gene in 483 CAD/MI patients. We found a rare exon 7 SSCA pattern in 11 patients (fig 1). After sequencing, we found that these were heterozygous for c.1250C→T (nucleotide number according to Genbank accession NM005587), which results in a missense change (P279L). The same genotype was found in nine of the 1189 controls. The frequency of 279Leu carriers was significantly higher in CAD/MI patients than in the controls (2.3% v 0.8%; p = 0.009; OR = 3.06 (95% CI, 1.17 to 8.06)) (table 2). The 279L allele was only found in controls under 50 years of age (9/890; range 30 to 47 years) (table 2). The power of the study for this polymorphism at significance level of 0.05 was 70%.
Figure 1 Single strand conformation analysis (SSCA) of exon 7 showing the distinct electrophoretic patterns for 279Pro homozygotes (lanes 1, 3, 4, and 8) and P279L heterozygotes (lanes 2, 6, 7, and 9). Lane 5 is the electrophoretic pattern for a control carrier of a single nucleotide deletion in the flanking intron region (delC+14780/In6; nucleotide number according to ENST 00000346108).
Table 2 Genotype frequencies for the P279L single nucleotide polymorphism.
| Genotype | Total participants∗ | Younger participants† | ||
|---|---|---|---|---|
| Patients (n = 483) | Controls (n = 1189) | Patients (n = 314) | Controls (n = 890) | |
| 279PL | 11 (2.3%) | 9 (0.8%) | 9 (2.9%) | 9 (1%) |
| 279PP | 472 (97.7%) | 1180 (99.2%) | 305 (97.1%) | 881 (99%) |
*p = 0.009; OR = 3.06 (95% CI, 1.17 to 8.06).
†p = 0.01; OR = 2.9 (95% CI, 1.04 to 8.00). Younger participants: male <55 years; female <65 years.
CI, confidence interval; OR, odds ratio.
Exon 11 showed a high degree of SSCA heterogeneity. We sequenced cases representative of all the observed SSCA patterns and found that the variation was caused by a previously described polymorphic trinucleotide CAG repeat (dbSNP rs3138597).11 In addition, two previously described amino acid silent single nucleotide polymorphisms, S417S and G451G (dbSNP rs3730059 and rs325400, respectively) and two length variant alleles in an adjacent poly‐proline repeat (5 and 4 prolines) were identified. The (CAG)n polymorphism was also analysed in 211 patients and 301 controls through a fluorescent capillary automated electrophoresis. Frequencies for the different (CAG)n alleles did not differ between patients and controls (table 3). In addition, the 21 base pair deletion previously found in a large family with CAD/MI was not found among our patients and healthy controls.
Table 3 Allele distribution of the (CAG)n repeat in the patients with myocardial infarction and controls.
| Allele* | |||||
|---|---|---|---|---|---|
| 9 | 10 | 11 | <9 | >11 | |
| Control | 184 (31%) | 97 (16%) | 316 (52%) | 2 | 3 |
| Patients | 136 (32%) | 68 (16%) | 216 (51%) | 2 | 0 |
*Number of (CAG)n repeats: 9, 10, or 11 repeats; <9, alleles with 6, 7, or 8 repeats; >11, alleles with 12 or 13 repeats.
Discussion
MEF2A belongs to a family of four closely related evolutionarily conserved transcriptions factors (MEF2A to MEF2D). The role of MEF2 proteins in cardiovascular physiology has been established from animal models. Most MEF2A knockout mice die in the first week of life, and the few mutants that survive and reach the adulthood are also susceptible to sudden cardiac death. Heterozygous MEF2A+/− mice have a normal phenotype and the cause of the sudden death in homozygous MEF2A−/− mice remains unknown.12 Recently, Wang et al described a MEF2A mutation that segregated with CAD/MI in a large family with several affected members.2 The mutation was a seven amino acid deletion in exon 11, in a region of the protein required for nuclear localisation. Because this mutant protein is sequestered into the cytoplasm and acts in a dominant negative manner, its final effect could be a downregulation of genes that contain the MEF2A binding site in their promoters. MEF2A is expressed in endothelial and smooth muscle cells of coronary arteries, and MEF2A mutations could disrupt the growth or differentiation of these cells, increasing the risk of developing coronary artery disease among mutation carriers. In addition to this deletion in exon 11, a recent report described three putative missense mutations in exon 7 of MEF2A in 2% of sporadic coronary artery disease patients.7 These rare nucleotide changes were absent in healthy controls, were clustered within or close to the major transcriptional activation domain of MEF2A; they significantly reduced the transcriptional activation activity of MEF2A, acting through a loss of function mechanism.7 Together, these data suggested that mutations in the MEF2A gene were involved in the risk of coronary artery disease and myocardial infarction, either familial or sporadic. In a recent report, Weng et al found some of these gene variants among healthy controls, challenging their role in the risk of CAD/MI and suggesting that MEF2A mutations were not a cause of familial or sporadic coronary artery disease.8 However, these investigators did not exclude the presence of coronary artery disease in control group. To clarify this issue, we undertook a screening study of exons 7 and 11 in a large cohort of patients with myocardial infarction and healthy controls from Spain. The only variant identified among patients was P279L, but this was also found in controls. However, carriers of 279Leu were significantly more frequent among the patients, suggesting that this is a rare polymorphism associated with an increased risk for myocardial infarction in our population. According to a previous report, 249Leu significantly reduced the MEF2A transcription activity.7 The (CAG)n repeat polymorphism in exon 11 was highly variable. Polyglutamine expansions are associated with several diseases.13 However, no expansion of the MEF2A‐polyQ repeat in our patients was observed.
The association between P279L and myocardial infarction was based on a comparison between 483 patients with myocardial infarction and 1189 controls. Interesting, all the controls who were 289Leu carriers were under 50 years of age. The lack of 279Leu among elderly controls could reflect the predisposition to develop CAD/MI conferred by this MEF2A allele. However, because we did not undertake coronary angiography to confirm the lack of diseased vessels among the controls, our study could underestimate the risk conferred by 279Leu if this allele was associated with an increased risk of developing atherosclerotic lesions.
In conclusion, we found a significantly greater frequency of 279Leu among patients with myocardial infarction. This allele could be a genetic risk factor for myocardial infarction in our population.
Acknowledgements
This work was supported by grants from Spanish Fondo de Investigaciones Sanitarias (FIS 03/05) and Red Temática de Centros de Genética Molecular (FIS C03/07).
Abbreviations
CAD/MI - coronary artery disease/myocardial infarction
MEF2A - myocyte enhancer factor 2A
SSCA - single strand conformation analysis
Footnotes
Conflicts of interest: none declared
References
- 1.Akanaza H, Komuro I. Roles of cardiac transcription factors in cardiac hypertrophy. Circ Res 2003921079–1088. [DOI] [PubMed] [Google Scholar]
- 2.Wang L, Fan C, Topol E S, Topol E J, Wang Q. Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science 20033021578–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Firulli B, Miano J M, Bi W, Johnson W, Casscells W, Olson N, Schwarz J J. Myocyte enhancer binding factor‐2 expression and activity in vascular smooth muscle cells. Circ Res 199678196–204. [DOI] [PubMed] [Google Scholar]
- 4.Lusis A J. Atherosclerosis. Nature 2000407233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cambien F. Coronary heart disease and polymorphisms in gene affecting lipid metabolism and inflammation. Curr Atheroscler Rep 20057188–195. [DOI] [PubMed] [Google Scholar]
- 6.Wang Q. Advances in the genetics basis of coronary artery disease. Curr Atheroscler Rep 20057235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhagavatula M R, Fan C, Shen G Q, Cassano J, Plow E F, Topol E J, Wang Q. Transcription factor MEF2A mutations in patients with coronary artery disease. Hum Mol Genet 2004133181–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weng L, Kavalar N, Ustaszewska A, Doelle H, Schackwitz W, Hebert S, Cohen J, McPherson R, Pennacchio L A. Lack of MEF2A mutations in coronary artery disease. J Clin Invest 20051151016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO MONICA Project MONICA manual, revised edition. Geneva: WHO Cardiovascular Diseases Unit, 1990
- 10.Miller S A, Dykes D D, Polesky H F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988161215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachinski L L, Abchee A, Durand J B, Roberts R, Krahe R, Hobson G M. Polymorphic trinucleotide repeat in the MEF2A gene at 15q26 is not expanded in familial cardiomyopathies. Mol Cell Probes 19971155–58. [DOI] [PubMed] [Google Scholar]
- 12.Naya F J, Black B L, Wu H, Bassel‐Duby R, Richardson J A, Hill J A, Olson E N. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat Med 200281303–1309. [DOI] [PubMed] [Google Scholar]
- 13.Morfini G, Pigino G, Brady S T. Polyglutamine expansion diseases: failing to deliver. Trends Mol Med 20051164–70. [DOI] [PubMed] [Google Scholar]