Abstract
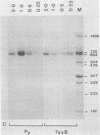
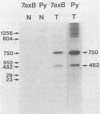
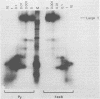
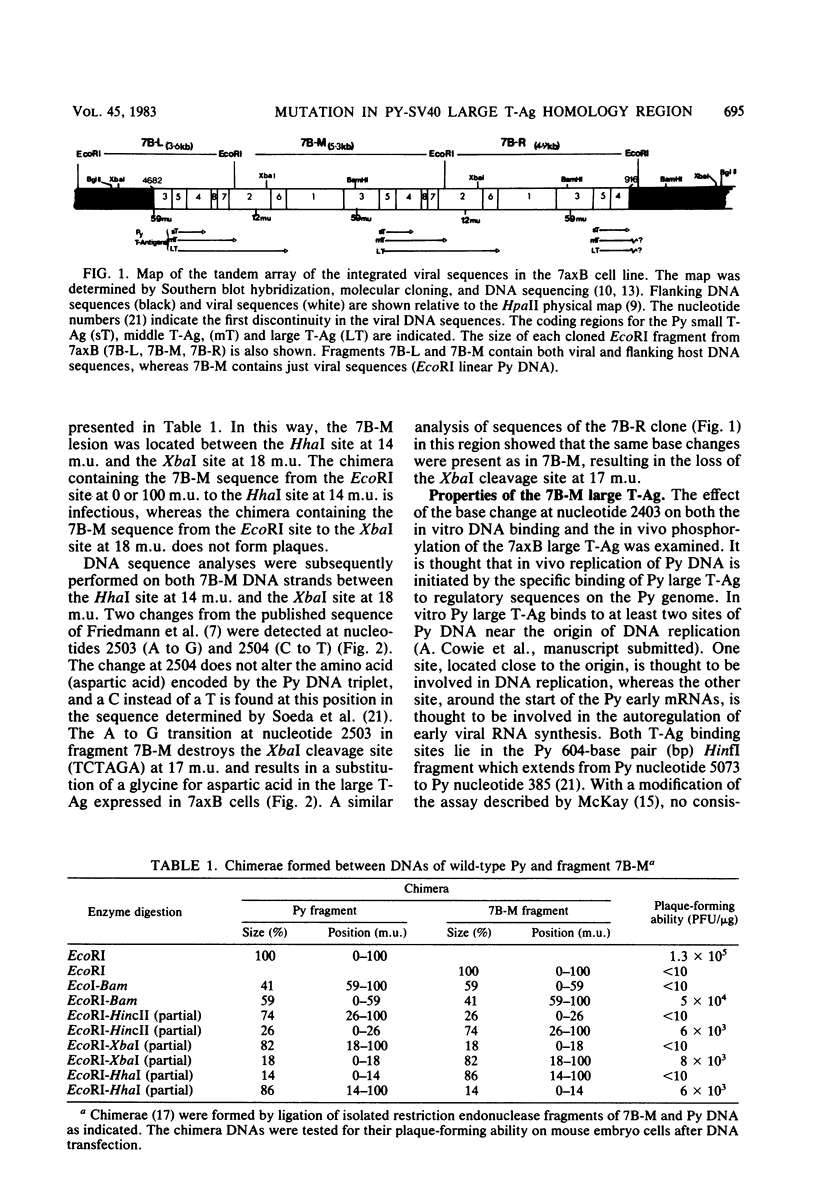
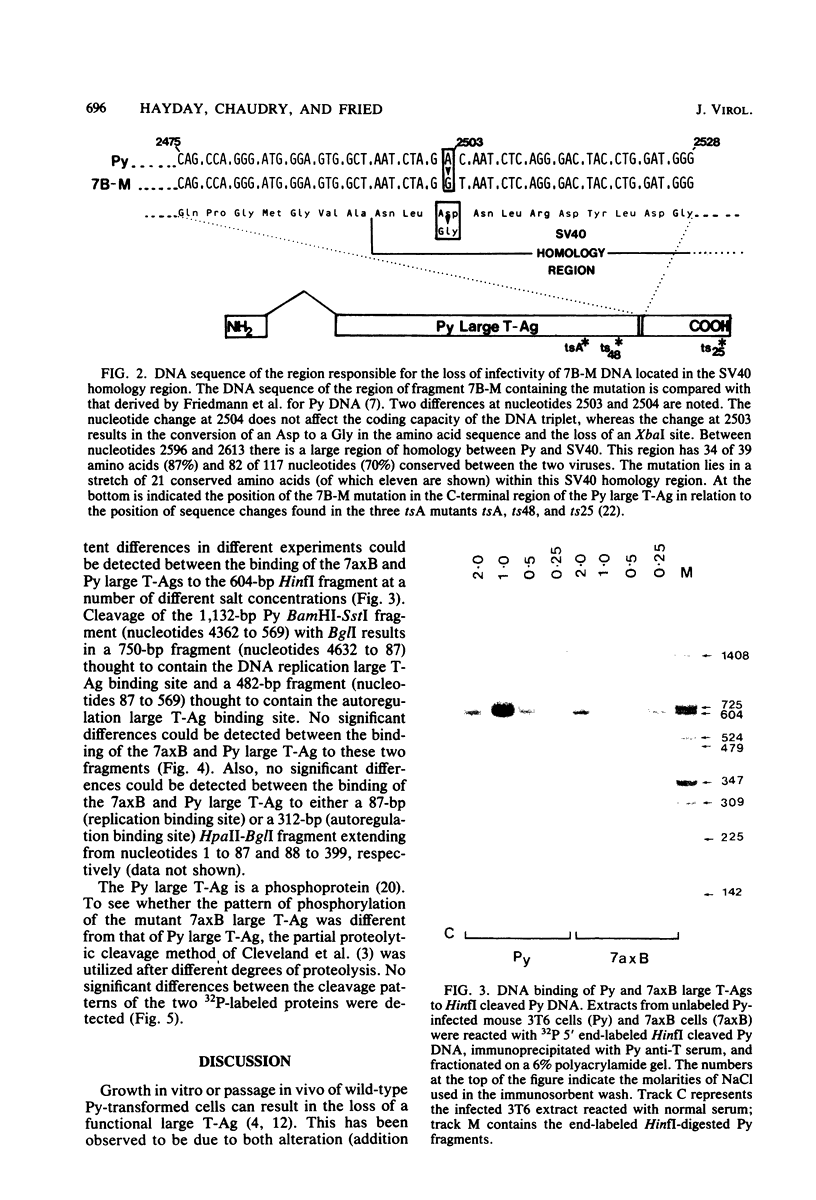
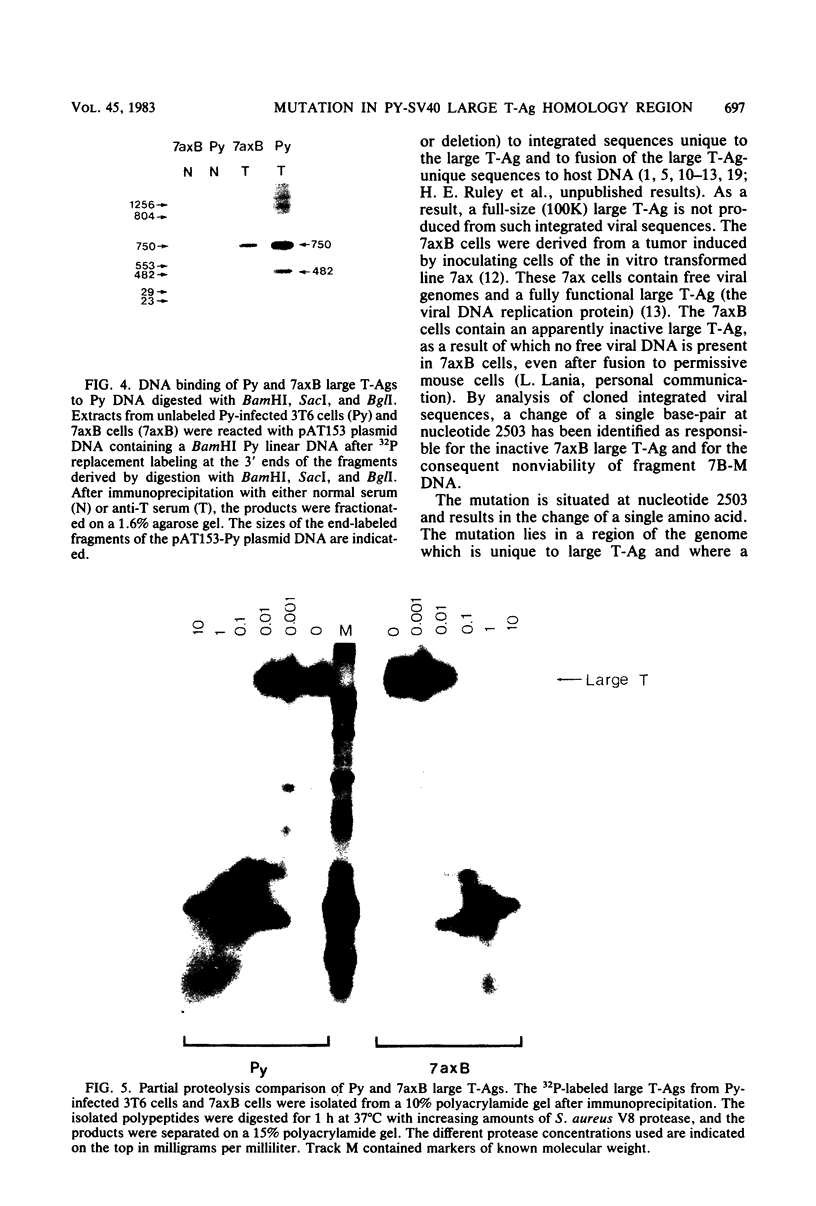
The polyoma virus (Py) transformed cell line 7axB, selected by in vivo passage of an in vitro transformed cell, contains an integrated tandem array of 2.4 genomes and produces the large, middle, and small Py T-antigen species, with molecular weights of 100,000, 55,000, and 22,000, respectively (Hayday et al., J. Virol. 44:67-77, 1982; Lania et al., Cold Spring Harbor Symp. Quant. Biol. 44:597-603, 1980). The integrated viral and adjacent host DNA sequences have been molecularly cloned as three EcoRI fragments (Hayday et al.). One of these fragments (7B-M), derived from within the tandem viral sequences, is equivalent to an EcoRI viral linear molecule. Fragment 7B-M has been found to be transformation competent but incapable of producing infectious virus after DNA transfection (Hayday et al.). By constructing chimerae between 7B-M and Py DNA and by direct DNA sequencing, the mutation responsible for the loss of infectivity has been located to a single base change (adenine to guanine) at nucleotide 2503. This results in a conversion of an aspartic acid to a glycine in the C-terminal region of the Py large T-antigen but does not appear to affect the binding of the Py large T-antigen to Py DNA at the putative DNA replication and autoregulation binding sites. The mutation is located within a 21-amino acid homology region shared by the simian virus 40 large T-antigen (Friedmann et al., Cell 17:715-724, 1979). These results suggest that the mutation in the 7axB large T-antigen may be involved in the active site of the protein for DNA replication.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Basilico C., Gattoni S., Zouzias D., Valle G. D. Loss of integrated viral DNA sequences in polyomatransformed cells is associated with an active viral A function. Cell. 1979 Jul;17(3):645–659. doi: 10.1016/0092-8674(79)90272-1. [DOI] [PubMed] [Google Scholar]
- Birg F., Dulbecco R., Fried M., Kamen R. State and organization of polyoma virus DNA sequences in transformed rat cell lines. J Virol. 1979 Feb;29(2):633–648. doi: 10.1128/jvi.29.2.633-648.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Colantuoni V., Dailey L., Basilico C. Amplification of integrated viral DNA sequences in polyoma virus-transformed cells. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3850–3854. doi: 10.1073/pnas.77.7.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey L., Colantuoni V., Fenton R. G., La Bella F., Zouzias D., Gattoni S., Basilico C. The evolution of polyoma-transformed rat cell lines during propagation in vitro. Virology. 1982 Jan 15;116(1):207–220. doi: 10.1016/0042-6822(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Fried M., Griffin B. E., Lund E., Robberson D. L. Polyoma virus--a study of wild-type, mutant and defective DNAs. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):45–52. doi: 10.1101/sqb.1974.039.01.009. [DOI] [PubMed] [Google Scholar]
- Friedmann T., Esty A., LaPorte P., Deininger P. The nucleotide sequence and genome organization of the polyoma early region: extensive nucleotide and amino acid homology with SV40. Cell. 1979 Jul;17(3):715–724. doi: 10.1016/0092-8674(79)90278-2. [DOI] [PubMed] [Google Scholar]
- Gattoni S., Colantuoni V., Basilico C. Relationship between integrated and nonintegrated viral DNA in rat cells transformed by polyoma virus. J Virol. 1980 Jun;34(3):615–626. doi: 10.1128/jvi.34.3.615-626.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B. E., Fried M., Cowie A. Polyoma DNA: a physical map. Proc Natl Acad Sci U S A. 1974 May;71(5):2077–2081. doi: 10.1073/pnas.71.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayday A., Ruley H. E., Fried M. Structural and biological analysis of integrated polyoma virus DNA and its adjacent host sequences cloned from transformed rat cells. J Virol. 1982 Oct;44(1):67–77. doi: 10.1128/jvi.44.1.67-77.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lania L., Gandini-Attardi D., Griffiths M., Cooke B., De Cicco D., Fried M. The polyoma virus 100K large T-antigen is not required for the maintenance of transformation. Virology. 1980 Feb;101(1):217–232. doi: 10.1016/0042-6822(80)90497-3. [DOI] [PubMed] [Google Scholar]
- Lania L., Hayday A., Bjursell G., Gandini-Attardi D., Fried M. Organization and expression of integrated polyoma virus DNA sequences in transformed rodent cells. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):597–603. doi: 10.1101/sqb.1980.044.01.062. [DOI] [PubMed] [Google Scholar]
- Lania L., Hayday A., Fried M. Loss of functional large T-antigen and free viral genomes from cells transformed in vitro by polyoma virus after passage in vivo as tumor cells. J Virol. 1981 Aug;39(2):422–431. doi: 10.1128/jvi.39.2.422-431.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay R. D. Binding of a simian virus 40 T antigen-related protein to DNA. J Mol Biol. 1981 Jan 25;145(3):471–488. doi: 10.1016/0022-2836(81)90540-4. [DOI] [PubMed] [Google Scholar]
- Miller L. K., Cooke B. E., Fried M. Fate of mismatched base-pair regions in polyoma heteroduplex DNA during infection of mouse cells. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3073–3077. doi: 10.1073/pnas.73.9.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. K., Fried M. Construction of infectious polyoma hybrid genomes in vitro. Nature. 1976 Feb 19;259(5544):598–601. doi: 10.1038/259598a0. [DOI] [PubMed] [Google Scholar]
- Miller L. K., Fried M. Construction of the genetic map of the polyoma genome. J Virol. 1976 Jun;18(3):824–832. doi: 10.1128/jvi.18.3.824-832.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruley H. E., Lania L., Chaudry F., Fried M. Use of a cellular polyadenylation signal by viral transcripts in polyoma virus transformed cells. Nucleic Acids Res. 1982 Aug 11;10(15):4515–4524. doi: 10.1093/nar/10.15.4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffhausen B. S., Benjamin T. L. Phosphorylation of polyoma T antigens. Cell. 1979 Dec;18(4):935–946. doi: 10.1016/0092-8674(79)90206-x. [DOI] [PubMed] [Google Scholar]
- Thomas T., Vollmer P., Folk W. R. Nucleotide sequence changes in polyoma virus A gene mutants. J Virol. 1981 Mar;37(3):1094–1098. doi: 10.1128/jvi.37.3.1094-1098.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson V. G., Tevethia M. J., Lewton B. A., Tegtmeyer P. DNA binding properties of simian virus 40 temperature-sensitive A proteins. J Virol. 1982 Nov;44(2):458–466. doi: 10.1128/jvi.44.2.458-466.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]