Abstract
Background
Placental mesenchymal dysplasia (PMD) is a distinct syndrome of unknown aetiology that is associated with significant fetal morbidity and mortality. Intrauterine growth restriction is common, yet, paradoxically, many of the associated fetuses/newborns have been diagnosed with Beckwith‐Wiedemann syndrome (BWS).
Methods
We report two cases of PMD with high levels of androgenetic (complete paternal uniparental isodisomy) cells in the placenta and document, in one case, a likely androgenetic contribution to the fetus as well.
Results
The same haploid paternal complement found in the androgenetic cells was present in coexisting biparental cells, suggesting origin from a single fertilisation event.
Conclusions
Preferential allocation of the normal cells into the trophoblast explains the absence of trophoblast overgrowth, a key feature of this syndrome. Interestingly, the distribution of androgenetic cells appears to differ from that reported for artificially created androgenetic mouse chimeras. Androgenetic mosaicism for the first time provides an aetiology for PMD, and may be a novel mechanism for BWS and unexplained intrauterine growth restriction.
Keywords: Beckwith‐Wiedemann syndrome, hydatidiform mole, imprinting, mosaicism, placenta
Androgenetic (paternal only) development is observed in about 1/800 human pregnancies in the form of complete hydatidiform moles (CHMs).1 These embryos can implant in the uterus and are associated with cystic oedematous chorionic villi (fluid accumulation within the placental villi), trophoblastic hyperplasia (overgrowth of the outer layer of the villi), and generally absent fetal development. About 75% of CHMs are thought to arise from fertilisation of an anucleate egg by a single haploid sperm, which subsequently undergoes endoreduplication, while most of the remaining cases involve fertilisation by two sperm. Their abnormal growth is attributed to the parent‐of‐origin dependent expression (imprinting) of a number of developmentally important genes.2,3 Partial hydatidiform moles (PHMs), which are due to triploidy, show some phenotypic overlap with CHMs. However, PHMs exhibit a range of villi from normal to cystic with focal trophoblastic hyperplasia. Furthermore, development of an abnormal fetus is often seen in PHM.4
Placental mesenchymal dysplasia (PMD) is a distinct placental phenotype that has often been misdiagnosed as a PHM and therefore has also been referred to as a “pseudo partial mole”.5,6,7,8,9 Placentas with PMD are typically larger than average and show cystic areas on ultrasound but can co‐exist with a completely normal fetus; however, intrauterine growth restriction and fetal or neonatal death are common. A key feature that distinguishes PMD from a partial or complete mole is the absence of trophoblast hyperplasia. Furthermore, whereas partial moles are triploid, most reported cases of PMD have a normal female 46,XX karyotype. Although an aetiology for PMD has not been shown, the preponderance of females has raised the suspicion of involvement of an X linked gene.10 In about one third of cases, PMD is associated with fetuses exhibiting features of Beckwith‐Wiedemann syndrome (BWS), including omphalocele, macroglossia, and visceromegaly.6,8,9 BWS generally results from abnormal expression of imprinted genes on 11p15.5 by one of several mechanisms and, thus, the abnormal expression of imprinted genes has also been raised as a possible aetiology for PMD.9
In this report, we present two pregnancies affected by PMD in which androgenetic/biparental whole genome mosaicism was observed. The phenotypic features of PMD, including absence of trophoblastic hyperplasia, association with BWS, and preponderance of females, can all be explained by this novel mechanism. Such pregnancies also offer a rare opportunity to consider the imprinting effects associated with uniparental inheritance in different tissues.
Methods
Clinical details
Case 1
A 38 year old woman was referred for amniocentesis with a twin pregnancy at 16 weeks gestation because of increased maternal age. An ultrasound examination revealed a normal placenta in twin 1B, but twin 1A, the smaller twin, had a placenta that was described as cystic and enlarged, with a “moth eaten” appearance. Throughout gestation, the growth of twin 1A continued to lag behind that of twin 1B (36th and 22nd centiles at 28 and 32 weeks gestation, respectively) and liver cysts were noted at 28 weeks. These liver cysts continued to enlarge, eventually encroaching upon the heart, and in utero fetal death was noted at 34 weeks gestation. At this time twin 1B, who showed normal growth and development throughout gestation (58th centile), was delivered. Postnatal development of twin 1B has since been normal. The parents declined an autopsy of twin 1A, therefore further details on the liver cysts seen on ultrasound are unavailable.
Gross placental examination of case 1 revealed a dichorionic twin placenta with fused placental discs. The placental weight was heavy, at 1900 g. The gross and histological features of twin 1B appeared normal. Macroscopically, the twin 1A placental plate contained several large cystic structures, up to 1 cm in size admixed with adherent friable chorionic villi (supplementary figure 1). Microscopic sections demonstrated marked stem villus hydrops with surrounding unremarkable small terminal villi. Subchorionic aneurysmal stem villus vessels and nucleated fetal red blood cells were also identified. No trophoblastic proliferation was present. DNA ploidy studies revealed a diploid population with a minor (<20%) tetraploid population. The histological features were those of PMD.
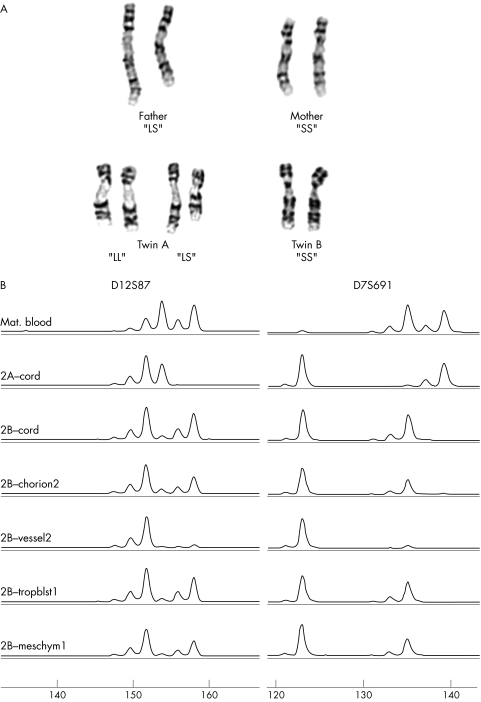
Figure 1 (A) Partial karyotypes from case 1 consistent with upd(9)pat. The paternal chromosome 9 homologues can be distinguished from each other based on the length of the polymorphic region directly beneath the centromere. The father has one large and one small chromosome 9, or an “LS” genotype. Both the mother and twin B have two small 9 homologues or an “SS” genotype. Two cell populations were seen in twin A, one with two large paternally inherited chromosome 9s (LL) and one with both a large paternal and small maternal chromosome 9 (LS). (B) Excess of paternal alleles is seen for case 2, twin B. Molecular analysis of the microsatellite markers D12S87 (left) and D7S691 (right) for (from top to bottom): maternal blood, twin A cord, twin B cord, twin B chorion membrane site no. 2, twin B chorionic vessel site no. 2, twin B trophoblast site no. 1, and twin B chorionic villus mesenchyme site no. 1. A reduced peak size for the maternal alleles is apparent in chorion membrane, vessel, and mesenchyme. Quantification of allele peaks suggests that the trophoblast sample also shows a slight reduction (1.14:1 paternal:maternal ratio for D7S691 and 1.12:1 for D12S87) in the maternal peak.
Case 2
A second trimester triple screen at 15 weeks 6 days gestation in a 28 year old woman showed elevated hCG (5.09 MoM) and normal MSAFP (1.64 MoM). At 15 weeks, ultrasound examination of the pregnancy revealed two normally developed fetuses, however twin 2B was smaller and showed oligohydramnious. Twin 2A had a normal placenta while twin 2B had a cystic placenta that was described as enlarged with a “Swiss cheese” appearance. Amniocentesis performed at 19 weeks showed a 46,XY complement in twin 2A and a 46,XX complement in twin 2B. Amniotic fluid AFP was normal in both twins. Twin 2B lagged slightly behind in growth based on ultrasound estimates. Induction, because of intrauterine growth restriction in twin 2B, was performed at 37 weeks. Twin 2A was a normal boy with APGAR score of 10 and 10, birth weight of 2942 g, head circumference 340 mm, and length 46 cm. Twin 2B was a normal girl, born by breech extraction with APGAR score 10 and 10, birth weight of 2210 g (<5%ile), head circumference 310 mm, length 43 cm, Hb 162 g/l. She was readmitted to the hospital on day 6 for observation due to a short episode of bradycardia noticed on a routine home follow up visit, and was monitored for 48 h. The assessment was normal as were the ECG and echocardiogram. This baby is now 3 months of age and clinical follow up has remained normal with some catch up in growth.
Examination of the placenta in case 2 showed a dichorionic twin placenta with fused placental discs, together weighing 690 g. Approximately 60% of the fetal surface was associated with twin 2B. The placenta for twin 2A was grossly and histologically normal. The placenta for twin 2B was grossly abnormal with abnormal chorionic vessels that were large, dilated, and had a tortuous course (supplementary figure 1). There were thrombi in the dilated subchorionic vessels. Grossly, there were some cysts that, histologically, were in stem villi. The stem villi had abundant myxoid stroma and oedema with abnormal vessels that extended from the fetal to maternal surface. The chorionic villi were largely unremarkable, with only focal hydropic change. There was no trophoblast hyperplasia. The features were those of PMD.
Cytogenetics
Amniotic fluid samples were cultured, harvested, and GTG banded according to standard procedures.
CGH
Comparative genomic hybridisation (CGH) was performed on trophoblast samples following the labelling and hybridisation protocol detailed previously by Lestou et al.11 DNA extracted from the trophoblast samples from case 2, twin A and twin B were each labelled with FITC‐12‐dUTP and the 46,XX reference DNA with TRITC‐6‐dUTP. Diploid metaphase spreads were obtained from peripheral blood lymphocyte cultures of a normal male donor by use of standard protocols. Digital image analysis was performed using a Zeiss Axioplan epifluorescence microscope with selective filters, a COHU charge coupled device camera (COHU, San Diego, CA), and PathVysion Smart Capture VP software (Vysis, Downers Grove, IL) as described in Lomax et al.12 A shift in the average red‐to‐green ratio value above 1.1 or below 0.9 (excluding known heterochromatic and polymorphic regions) is considered to be evidence for a dosage difference.
FISH
Fluorescence in situ hybridisation (FISH) studies were performed on the mesenchyme‐1 sample from case 2, twin B as described by Henderson.32 A probe specific for the alpha satellite region of chromosome 4 was used (CEP4, Vysis). A total of 500 nuclei were scored and cut offs for significant levels of trisomy were taken from tissue matched diploid controls as described by Lomax et al.12
Molecular investigation
Placental samples were taken from the fetal side of the placenta, and the amnion and chorionic plate were isolated from the remaining villi and thoroughly washed with saline solution. The remaining chorionic villi were subjected to treatment with collagenase IA (350 U/mg; Sigma, St Louis, MO) to separate the chorionic villus mesenchyme (mostly stromal core) from the trophoblast. A sample from one of the enlarged chorionic vessels was also taken from case 2B. DNA was extracted from each sample using a salting out method.
PCR amplification was performed using standard conditions with primer pairs for microsatellite markers for chromosomes 7, 9, 12, 14, 16, and X. Most markers were genotyped using fluorescently labelled primers with PCR products quantified on an ABI 310 Prism Genetic Analyzer (ABI, Foster City, CA). The PCR products for the markers D9S286, D9S157, D9S162, D9S165, D9S152, and D9S176 were resolved on a polyacrylamide gel and the relative dosage of parental alleles was estimated following silver staining (for example, allele ratios were scored as approximately equal, clear excess of paternal peak, or extreme excess of paternal peak). Linearity of amplification was demonstrated for AR13 and D7S691 (data not shown) by mixing samples in different ratios (ranging from 1:9 to 9:1). Allele ratios using multiple markers consistently showed excess of paternal alleles in the same tissues. Peak areas for D7S691 were used to estimate the proportion of the androgenetic cell line, as this marker was informative in both cases, clearly amplified distinct alleles, and showed no evidence of an allele amplification bias. The estimated frequency of the androgenetic cell line (fAnd) was calculated as: fAnd = Pp−Pm, where Pp and Pm represent the fractions of the total peak area contributed by the paternal and maternal peaks, respectively.
Results
Cytogenetic analysis of amniotic fluid obtained in the second trimester of pregnancy of case 1 twin A revealed two different diploid female cell populations that could be distinguished based on a structural polymorphism in the proximal long arm of chromosome 9 (fig 1A). In half of the cells, both chromosome 9 homologues contained a large heterochromatic region (LL), while in the remaining cells, one large and one small heterochromatic region (LS) was seen. As twin B and the mother exhibited an “SS” genotype, and only the father showed an “LS” genotype for this chromosome 9 polymorphism, the “LL” cell line was inferred to represent paternal uniparental disomy for chromosome 9. The presence of the LL cells was confirmed postnatally in chorionic villus cultures (stromal cells) from two separate placental biopsies, with the LL genotype observed in 20/20 and 25/30 cells, respectively.
DNA molecular testing of eight different loci spanning chromosome 9 was performed on cord blood and placental samples from case 1 and confirmed an excess of paternal alleles in the chorion and stroma, consistent with the presence of mosaicism for paternal uniparental isodisomy (UPiD) for chromosome 9 (table 1). However, the analysis of additional markers from other chromosomes also showed excess dosage of all paternal alleles, suggesting a paternal UPiD for all chromosomes, not only chromosome 9, in the LL lineage (table 1 and supplementary table 1). Thus the LL cells appeared to be derived exclusively from diploidisation of a single haploid paternal complement. The same haploid paternal complement of this androgenetic cell population also appeared to be present in the normal LS cells with biparental marker inheritance. The excess dosage of paternal alleles was not observed in DNA extracted from cord blood or two trophoblast biopsies in this case (fig 2).
Table 1 Microsatellite genotyping for case 1.
| Marker | Maternal blood | Twin 1B cord blood | Twin 1A cord blood, trophoblast | Twin 1A chorion, mesenchyme | Paternal blood |
|---|---|---|---|---|---|
| D2S237 | ab | nt | ad | ad/dd | cd |
| D6S260 | bd | cd | ad | ad/aa | ac |
| D7S691 | ac | nt | bc | bc/bb | bb |
| D9S286 | bd | ab | bc | bc/cc | ac |
| D9S257 | ab | bb | bc | bc/cc | bc |
| D9S162 | bb | bb | ab | ab/aa | ab |
| D9S304 | bc | bb | ac | ac/aa | ab |
| D9S165 | cc | ac | (b)c | (b)c | ab |
| D9S1788 | bb | bc | ab | ab/aa | ac |
| D9S152 | ac | ab | ab | ab/bb | bb |
| D9S176 | ac | bc | bc | bc/bb | bc |
| D12S87 | bc | ac | bb | bb | ab |
| D14S161 | ab | ab | ab | ab/bb | bb |
| AR | NT | NT | ab | ab/bb | NT |
(b)c indicates the maternal “c” allele is more intense than the “b” allele. This apparent excess is attributed to poor amplification of the paternal allele as the flanking markers (D9S304 and D9S1788) are at 0 cM on either side of D9S165 and do not show this pattern. nt, not tested.
The letters indicate the genotype inferred by allelic dosage. Results obtained from additional placental samples from twin 1A were identical to those obtained in cord. Two separate trophoblast and mesenchyme samples showed similar results (fig 2). NT, not tested.
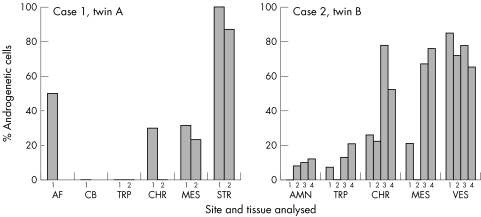
Figure 2 Distribution of androgenetic cells within each placenta. The estimated proportion of androgenetic cells is given for tissues isolated from two separate biopsy sites from case 1 and four biopsy sites from case 2. This proportion is estimated from the allele peak ratios for markers for D7S691, which was informative in both cases, had clearly separated bands and no evidence for allelic amplification bias. Only those samples with an estimate of <10% androgenetic cells (case 2: amnion 2, amnion 3, and trophoblast 1) did not show a consistent bias with all tested markers. The frequency of the “LL” genotype from cultured cells is used for the estimates in the amniotic fluid and stroma samples for case 1. AF, amniotic fluid; AMN, amnion; CB, cord blood; CHR, chorionic membrane; MES, chorionic mesenchyme; STR, stroma, derived from cultured chorionic villi; TRP, trophoblast; VES, enlarged chorionic vessel.
Amniocentesis results for case 2, twin A and twin B were normal (46,XY and 46, XX, respectively), as was CGH testing on a trophoblast sample taken from each placenta to rule out trisomy mosaicism. However, microsatellite loci tested from chromosomes 7, 9, 12, 16, and X all showed excess of paternal alleles in samples of chorion, an enlarged chorionic vessel, and chorionic villus mesenchyme from twin B (fig 1B, table 2, supplemental table 1). This was most apparent in the chorionic vessel samples, for which the maternal allele was barely detectable. Quantification of marker allelic ratios suggests that the androgenetic cells may have been present at very low levels within the trophoblast and amnion. Higher levels of the androgenetic cells were seen in other extra‐embryonic tissues (fig 2). FISH using a chromosome 4 probe was within the range expected for normal diploid tissue, thus excluding ploidy mosaicism as an explanation for the excess dosage of paternal alleles, and suggests the presence of an androgenetic diploid cell population.
Table 2 Microsatellite genotyping for case 2.
| Marker | Maternal blood | Twin 2A cord | Twin 2B cord, amnion, trophoblast | Twin 2B chorion, mesenchyme | Twin 2B chorionic vessel* |
|---|---|---|---|---|---|
| D7S691 | ab | ac | bc | bc/cc | cc |
| D7S976 | ab | aa | aa | aa | aa |
| D9S1788 | bc | bd | ac | ac/aa | aa |
| D9S257 | ab | ac | ac | ac/cc | cc |
| D12S87 | ab | bc | ac | ac/cc | cc |
| D16S409 | ac | cc | bc | bc/bb | bb |
| D16S515 | aa | aa | ab | ab/bb | bb |
| D16S3093 | ab | ac | ac | ac/cc | cc |
| AR | bc | nt | ab | ab/aa | nt |
| DXYS233 | ac | ad | bc | bc/bb | nt |
*The maternal allele was visible but very faint; nt, not tested.
The letters indicate the genotype as judged by allelic dosage. Additional placental samples from twin A yielded results that were identical to those obtained in cord from twin A; four sites from twin 2B placenta were sampled. The amnion, chorion, chorionic vessel, mesenchyme, and trophoblast from the four sampled sites all had similar findings, but with some variability in allelic ratios (fig 2).
Discussion
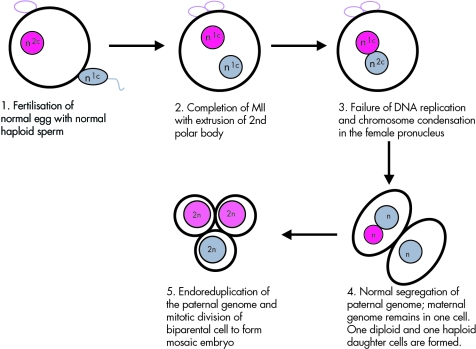
The mechanism of androgenetic/biparental mosaicism in the two cases presented here appears to have involved only a single egg and sperm nucleus, as is true for a similar case diagnosed as a CHM and a “parthenogenetic chimera” (gynogenetic/biparental mosaic, by our terminology) previously reported.14,15 Chimerism is excluded because the same haploid paternal complement contributed to both cell lineages. The simplest explanation would involve failure of replication of the maternal genome prior to the first cleavage, with normal replication and segregation of the paternal genome (fig 3). If the maternal chromosomes did not migrate to the metaphase plate due to delayed/failed breakdown of the pronuclear envelope, they would remain together in one daughter cell. Such a failed division would produce a diploid/haploid mosaic embryo, as is observed in 1–2% of human preimplantation embryos.16,17 This is analogous to the observed mechanism for formation of diploid/haploid embryos in mouse after delayed sperm incorporation into the oocyte.18 Endoreduplication of the haploid paternal‐only daughter cell could then occur to produce the diploid androgenetic lineage, while the female and male haploid complements would merge to form a daughter cell with normal biparental inheritance. Origin via fertilisation by a homozygous diploid sperm seems unlikely since there are no reports of triploidy arising in this manner.19
Figure 3 Proposed mechanism for the origin of androgenetic/biparental mosaicism. 2n represents a diploid chromosome complement; n represents a haploid chromosome complement; n1C represents an unreplicated haploid complement; and n2C represents a replicated haploid complement.
As both cases were found to involve the same mechanism, androgenetic/biparental mosaicism is likely a common cause of PMD. Furthermore, our model can explain the female preponderance of reported cases. If the androgenetic cell line has arisen by duplication of a haploid paternal genome, as in the two cases described here, then one would expect only females to be observed, since a 46,YY cell line would be non‐viable. The rare cases of PMD with a 46,XY genotype could arise by chimerism (fusion) of a normal embryo with a CHM derived from a diploid sperm or dispermy in early gestation. Interestingly, several reported cases of PMD had a coexisting twin CHM.9 The fact that both of the present cases were associated with normal dizygotic twins is probably coincidence, as most cases of PMD reported in the literature occur in singleton pregnancies and those that were associated with normal twins do not seem to differ in outcome.5,6,7,8,9 Furthermore, an additional case of androgenetic mosaicism was reported in an otherwise normal singleton pregnancy, although the placental findings were reported as that of a CHM and distinct from PMD.14 The phenotype of androgenetic mosaicism can presumably range from mild PMD, which may not even be diagnosed, to typical findings of a CHM, depending on the extent and distribution of the androgenetic lineage.
Interestingly, features of BWS are frequently reported in the fetus of pregnancies diagnosed with PMD.6,8,9 Case 1 presented here had liver cysts, the nature of which was not determined. BWS patients can develop hepatic tumours, and hepatic mesenchymal hamartoma leading to fetal death has been reported in conjunction with PMD in several instances.20,21 It is possible that loss of imprinting of the 11p15.5 genes alone could lead to PMD in some cases, particularly as a mouse model for BWS incorporating a null mutation for Cdkn1c/p57Kip2 and loss of Igf2 imprinting shows some placental abnormalities similar to those found in PMD, including a large placenta, disorganised labyrinthine layer (comparable to chorionic villi in humans), and fibrin cysts with accumulation of red blood cells.23 Alternatively, some cases of BWS may result from the presence of androgenetic cells in the fetal tissues and contribute to the 10–20% of BWS in which no abnormality is found.22
Comparison of these two human androgenetic mosaics to androgenetic mouse chimeras shows both similarities and differences. Mice chimeras artificially generated by aggregating normal and androgenetic blastomeres can lead to embryos that survive to term, but with differing phenotypes depending on the level of contribution and distribution of the uniparental cells.24,25,26 While competing with normal cells in chimeras, the androgenetic cells tend to contribute to the trophectoderm derived tissue of the placenta and to the yolk sac, but rarely to the chorionic mesoderm, amnion, or embryo proper.24,25 Chimeras in which androgenetic cells are found in the embryo proper rarely survive, and those that do have exhibited skeletal anomalies.26
As with the mouse chimeras, the androgenetic cells detected in the human cases described here were predominantly confined to the placenta. However, the androgenetic cells tended not to be present in the trophoblast and instead were confined predominantly to chorionic mesoderm, membrane, and vessels. In case 1A, androgenetic cells were observed in amniotic fluid and abnormal pathology was observed in the liver, suggesting that mosaicism was also present in the fetus. Similar to mouse androgenetic chimeras, there is a high intrauterine death rate of fetuses associated with PMD. However, skeletal anomalies were not identified in the present cases or in other births that have been associated with PMD. Any differences in comparison to the mouse chimeras may reflect differences in imprinting of genes in humans and mice. For example, the Igf2r, Mash2/Ascl2, Xist, and Esx1 genes are imprinted in mouse, but the corresponding human genes are either not imprinted or have a less clear imprinting status.27,28,29,30 There are also clear differences in expression patterns for some imprinted genes between mice and humans. For example, Peg3 is expressed in mouse spongiotrophoblast, secondary giant cells, and the labyrinth, whereas in humans PEG3 is only expressed in villus cytotrophoblast and no other placental cells.31
As we evaluated only term placentas, it is possible that androgenetic cells did contribute to the early, undifferentiated cytotrophoblast but are less able to differentiate and persist to term in this lineage. The hyper‐proliferation of the villus trophoblast observed in CHMs may be an indirect consequence of failure of these cells to differentiate down other pathways due to lack of key maternally expressed genes. In the androgenetic mosaics, biparental trophoblast cells should differentiate normally into invasive trophoblast or undergo fusion into syncytiotrophoblast. The presence of these normally differentiated trophoblast subtypes may provide appropriate feedback signals to the undifferentiated villus cytotrophoblast that prevent further uncontrolled growth. A term placenta consists of few undifferentiated cytotrophoblast cells and thus would be expected to consist predominantly of mature trophoblast with a biparental genome in this scenario.
The observation that androgenetic cells can arise spontaneously and persist in viable human pregnancies is intriguing and raises many questions. It remains to be determined if this is the mechanism for all cases of PMD or if the aetiology of PMD is heterogeneous. Androgenetic mosaicism could also lead to miscarriage in some pregnancies, as one would only expect cases with relatively low levels of androgenetic cells to survive into late gestation. It may also contribute to unexplained isolated intrauterine growth restriction (as in case 2), as the placental features may not always be obvious or diagnosed. In addition to the clinical implications, these pregnancies offer a rare opportunity to study imprinted gene expression and developmental potential of androgenetic cells existing in viable pregnancies. The ability to diagnose PMD in utero, based on the unusual findings of a cystic placenta with a normal viable fetus, will facilitate further characterisation of this developmental disorder linking a specific placental dysplasia with androgenetic mosaicism and the functions of imprinted genes in human development.
Acknowledgements
We would like to thank Brenda Lomax for help with CGH and FISH protocols.
Abbreviations
BWS - Beckwith‐Wiedemann syndrome
CGH - comparative genomic hybridisation
CHM - complete hydatidiform mole
FISH - fluorescence in situ hybridisation
PMD - placental mesenchymal dysplasia
PHM - partial hydatidiform mole
UPiD - uniparental isodisomy
Footnotes
Funding for this project was from the Canadian Institute for Health Research (grant MOP 15667 to WPR)
Competing interests: none declared
Consent was received for the publication of these details
References
- 1.Fisher R A, Hodges M D. Genomic imprinting in gestational trophoblastic disease‐‐a review. Placenta 200324(Suppl A)S111–S118. [DOI] [PubMed] [Google Scholar]
- 2.Barton S C, Surani M A, Norris M L. Role of paternal and maternal genomes in mouse development. Nature 1984311(5984)374–376. [DOI] [PubMed] [Google Scholar]
- 3.Surani M A, Barton S C, Norris M L. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 1984308(5959)548–550. [DOI] [PubMed] [Google Scholar]
- 4.McFadden D E, Pantzar J T. Placental pathology of triploidy. Hum Pathol 1996271018–1020. [DOI] [PubMed] [Google Scholar]
- 5.Gibson B R, Muir‐Padilla J, Champeaux A, Suarez E S. Mesenchymal dysplasia of the placenta. Placenta 200425(7)671–672. [DOI] [PubMed] [Google Scholar]
- 6.Jauniaux E, Nicolaides K H, Hustin J. Perinatal features associated with placental mesenchymal dysplasia. Placenta 199718(8)701–706. [DOI] [PubMed] [Google Scholar]
- 7.Matsui H, Iitsuka Y, Yamazawa K, Tanaka N, Mitsuhashi A, Seki K, Sekiya S. Placental mesenchymal dysplasia initially diagnosed as partial mole. Pathol Int 200353(11)810–813. [DOI] [PubMed] [Google Scholar]
- 8.Ohyama M, Kojyo T, Gotoda H, Sato T, Ijiri R, Tanaka Y. Mesenchymal dysplasia of the placenta. Pathol Int 200050(9)759–764. [DOI] [PubMed] [Google Scholar]
- 9.Paradinas F J, Sebire N J, Fisher R A, Rees H C, Foskett M, Seckl M J, Newlands E S. Pseudo‐partial moles: placental stem vessel hydrops and the association with Beckwith‐Wiedemann syndrome and complete moles. Histopathology 200139(5)447–454. [DOI] [PubMed] [Google Scholar]
- 10.Khong T Y. Placental vascular development and neonatal outcome. Semin Neonatol 20049(4)255–263. [DOI] [PubMed] [Google Scholar]
- 11.Lestou V S, Lomax B L, Barrett I J, Kalousek D K. Screening of human placentas for chromosomal mosaicism using comparative genomic hybridization. Teratology 199959(5)325–330. [DOI] [PubMed] [Google Scholar]
- 12.Lomax B K, Kalousek D, Kuchinka B, Barret I, Harrison K, Safavi H. The utilization of interphase cytogenetic analysis for the detection of mosaicism. Hum Genet 199493243–247. [DOI] [PubMed] [Google Scholar]
- 13.Beever C L.Skewed X chromosome inactivation in women experiencing recurrent pregnancy loss. Department of Medical Genetics, Vancouver: University of British Columbia, 2002
- 14.Makrydimas G, Sebire N J, Thornton S E, Zagorianakou N, Lolis D, Fisher R A. Complete hydatidiform mole and normal live birth: a novel case of confined placental mosaicism: case report. Hum Reprod 200217(9)2459–2463. [DOI] [PubMed] [Google Scholar]
- 15.Strain L, Warner J P, Johnston T, Bonthron D T. A human parthenogenetic chimaera. Nat Genet 199511164–169. [DOI] [PubMed] [Google Scholar]
- 16.Munne S, Weier H U, Grifo J, Cohen J. Chromosome mosaicism in human embryos. Biol Reprod 199451373–379. [DOI] [PubMed] [Google Scholar]
- 17.Harper J C, Coonen E, Handyside A H, Winston R M, Hopman A H, Delhanty J D. Mosaicism of autosomes and sex chromosomes in morphologically normal, monospermic preimplantation human embryos. Prenat Diagn 19951541–49. [DOI] [PubMed] [Google Scholar]
- 18.Maleszewski M, Borsuk E, Koziak K, Maluchnik D, Tarkowski A K. Delayed sperm incorporation into parthenogenetic mouse eggs: sperm nucleus transformation and development of resulting embryos. Mol Reprod Dev 199954(3)303–310. [DOI] [PubMed] [Google Scholar]
- 19.McFadden D E, Jiang R, Langlois S, Robinson W P. Dispermy‐‐origin of diandric triploidy: brief communication. Hum Reprod 200217(12)3037–3038. [DOI] [PubMed] [Google Scholar]
- 20.Kitano Y, Ruchelli E, Weiner S, Adzick N S. Hepatic mesenchymal hamartoma associated with mesenchymal stem villous hyperplasia of the placenta. Fetal Diagn Ther 200015(3)134–138. [DOI] [PubMed] [Google Scholar]
- 21.Laberge J M, Patenaude Y, Desilets V, Cartier L, Khalife S, Jutras L, Chen M F. Large hepatic mesenchymal hamartoma leading to mid‐trimester fetal demise. Fetal Diagn Ther 200520(2)141–145. [DOI] [PubMed] [Google Scholar]
- 22.Weksberg R, Smith A C, Squire J, Sadowski P. Beckwith‐Wiedemann syndrome demonstrates a role for epigenetic control of normal development. Hum Mol Genet 200312R61–R68. [DOI] [PubMed] [Google Scholar]
- 23.Caspary T, Cleary M A, Perlman E J, Zhang P, Elledge S J, Tilghman S M. Oppositely imprinted genes p57(Kip2) and igf2 interact in a mouse model for Beckwith‐Wiedemann syndrome. Genes Dev 199913(23)3115–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomson J A, Solter D. The developmental fate of androgenetic, parthenogenetic, and gynogenetic cells in chimeric gastrulating mouse embryos. Genes Dev 19882(10)1344–1351. [DOI] [PubMed] [Google Scholar]
- 25.Surani M A, Barton S C, Howlett S K, Norris M L. Influence of chromosomal determinants on development of androgenetic and parthenogenetic cells. Development 1988103(1)171–178. [PubMed] [Google Scholar]
- 26.Mann J R, Stewart C L. Development to term of mouse androgenetic aggregation chimeras. Development 1991113(4)1325–1333. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa O, McNoe L A, Eccles M R, Morison I M, Reeve A E. Human insulin‐like growth factor type I and type II receptors are not imprinted. Hum Mol Genet 19932(12)2163–2165. [DOI] [PubMed] [Google Scholar]
- 28.Westerman B A, Poutsma A, Looijenga L H, Wouters D, van Wijk I J, Oudejans C B. The Human Achaete Scute Homolog 2 gene contains two promotors, generating overlapping transcripts and encoding two proteins with different nuclear localization. Placenta 200122(6)511–518. [DOI] [PubMed] [Google Scholar]
- 29.Zeng S M, Yankowitz J. X‐inactivation patterns in human embryonic and extra‐embryonic tissues. Placenta 200324(2–3)270–275. [DOI] [PubMed] [Google Scholar]
- 30.Grati F R, Sirchia S M, Gentilin B, Rossella F, Ramoscelli L, Antonazzo P, Cavallari U, Bulfamante G, Cetin I, Simoni G, Miozzo M. Biparental expression of ESX1L gene in placentas from normal and intrauterine growth‐restricted pregnancies. Eur J Hum Genet 200412(4)272–278. [DOI] [PubMed] [Google Scholar]
- 31.Hiby S E, Lough M, Keverne E B, Surani M A, Loke Y W, King A. Paternal monoallelic expression of PEG3 in the human placenta. Hum Mol Genet 200110(10)1093–1100. [DOI] [PubMed] [Google Scholar]
- 32.Henderson K G, Shaw T E, Barnett I J, Telenius A H P, Wilson R D, Kalousek D K. Distribution of mosaicism in human placenta. Hum Genet 199697650–654. [DOI] [PubMed] [Google Scholar]